Vitamin D3 Ameliorates DNA Damage Caused by Developmental Exposure to Endocrine Disruptors in the Uterine Myometrial Stem Cells of Eker Rats
Abstract
1. Introduction
2. Materials and Methods
2.1. Uterine Fibroid Animal Model
2.2. Isolation of Eker Rat MMSCs, Cell Culture, and Treatment
2.3. Reagents and Antibodies
2.4. Immunohistochemistry (IHC) Analysis
2.5. Western Blot Analysis
2.6. Flow Cytometry Analysis
2.7. RNA Isolation and Quantitative Real-Time PCR
2.8. Cell Proliferation (MTT) Assay
2.9. Statistical Analyses
3. Results
3.1. Developmental DES Exposure Increased DNA Damage by Altering the DNA Repair Pathway in MMSCs
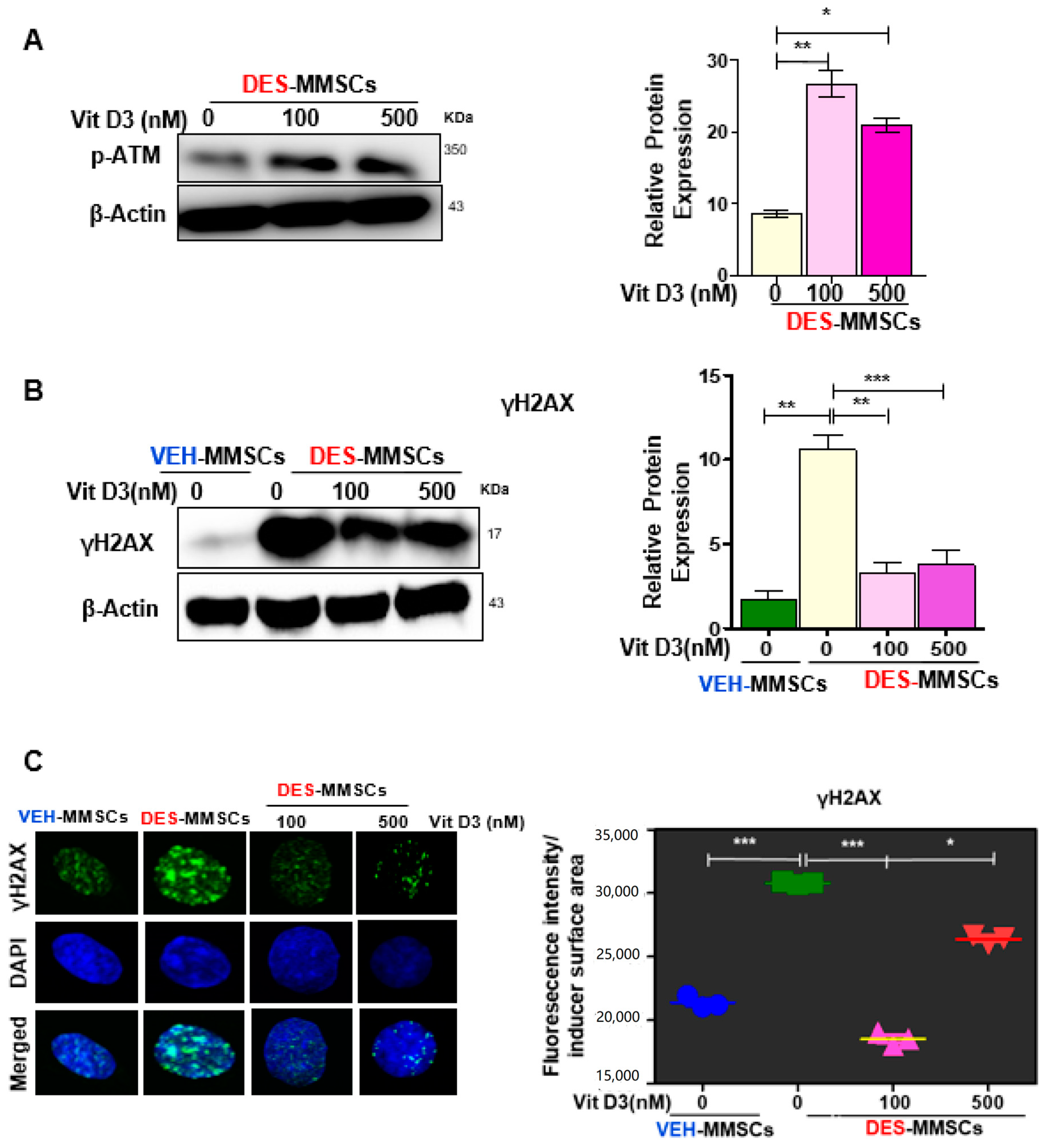
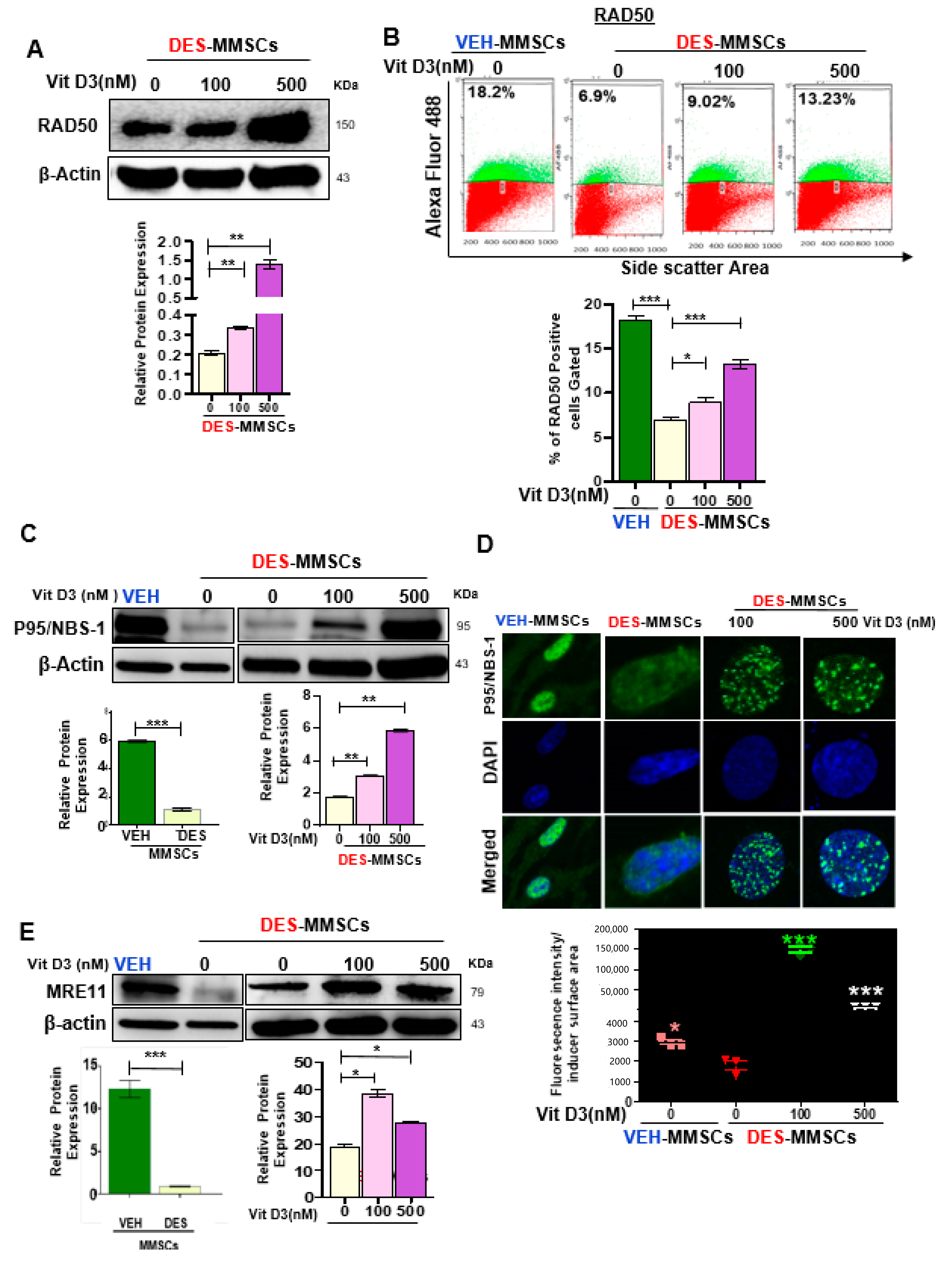
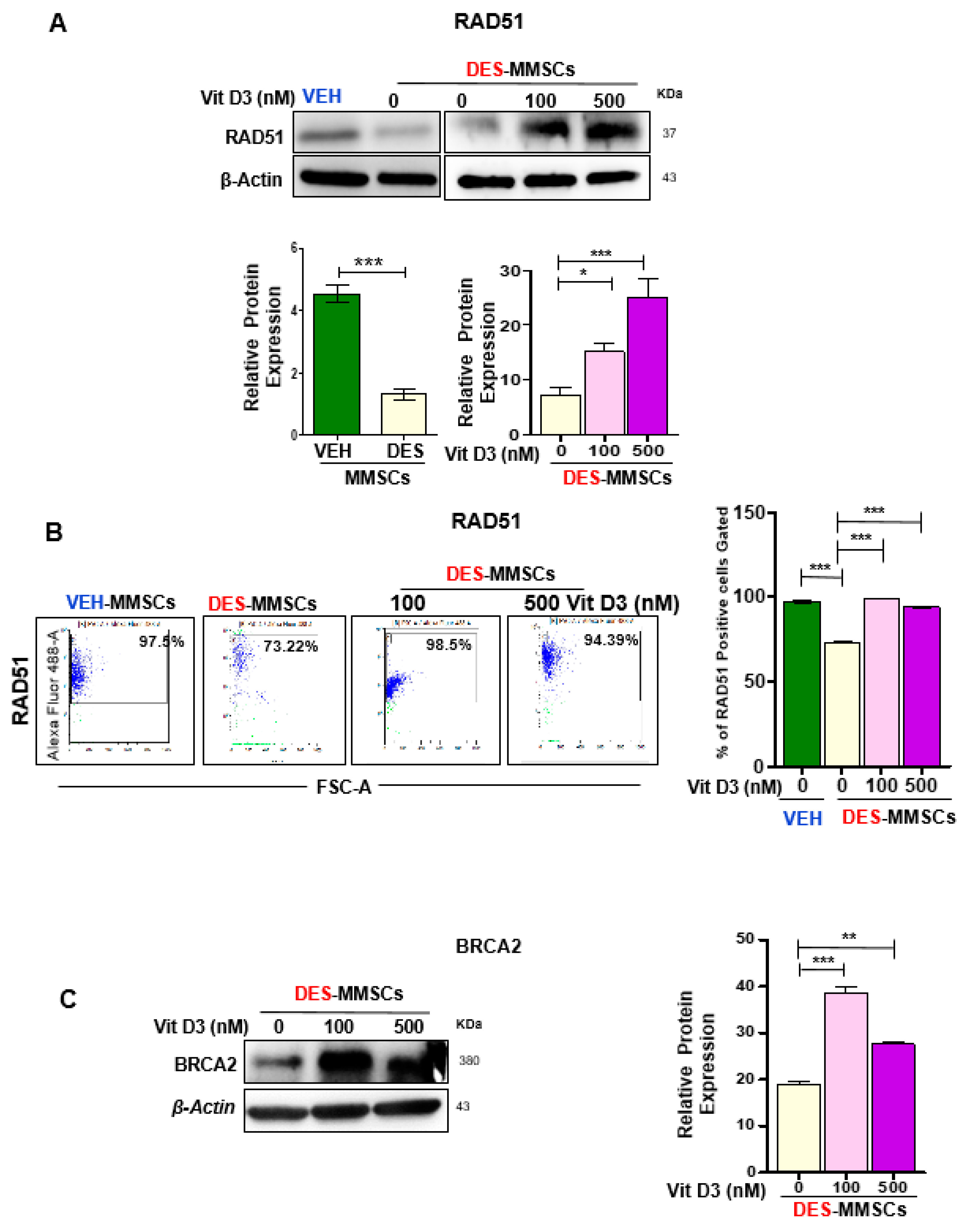
3.2. VitD3 Attenuated DES-Exposure-Induced DNA Damage in MMSCs and Upregulated the Expression of Protein Phosphokinases
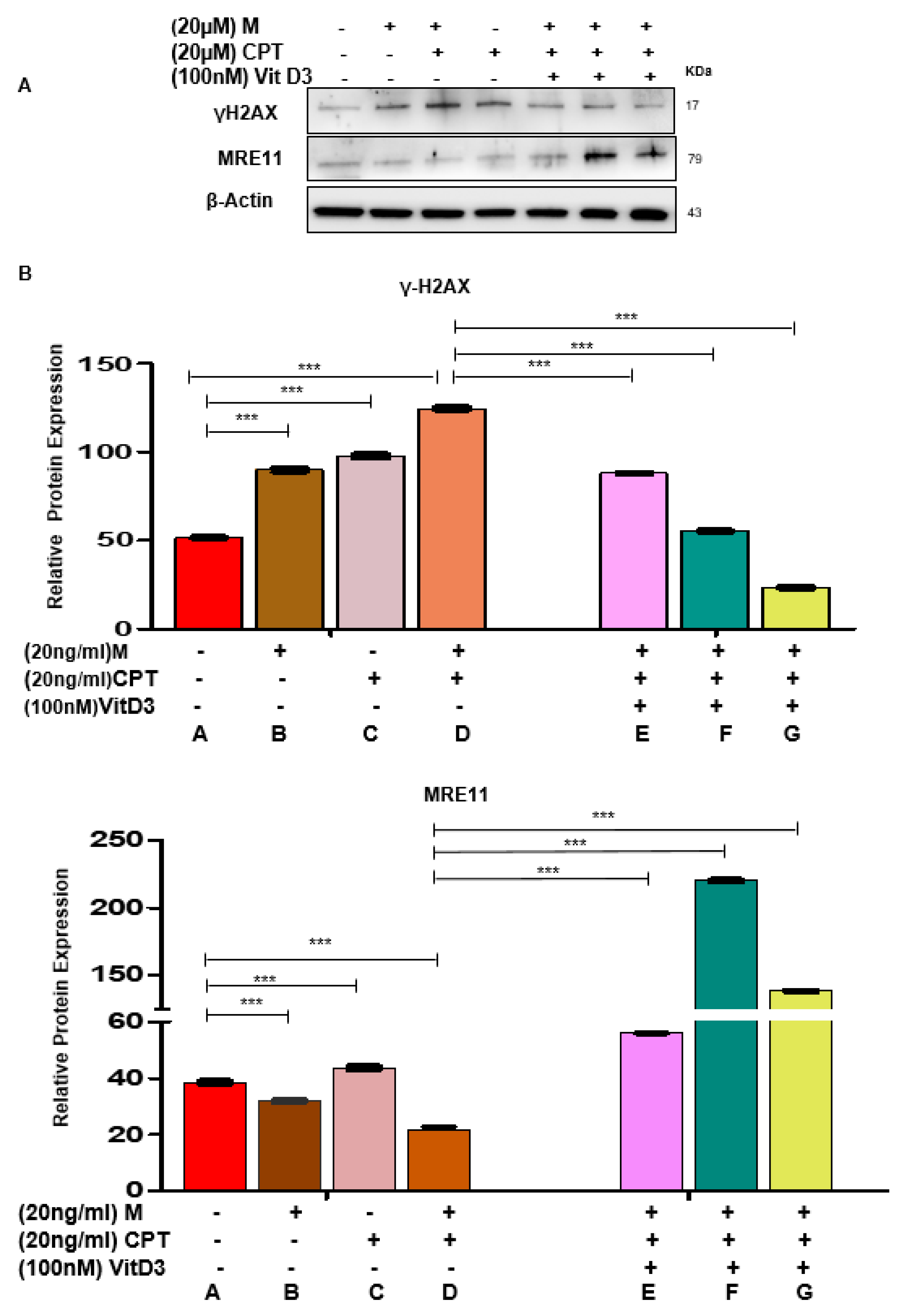
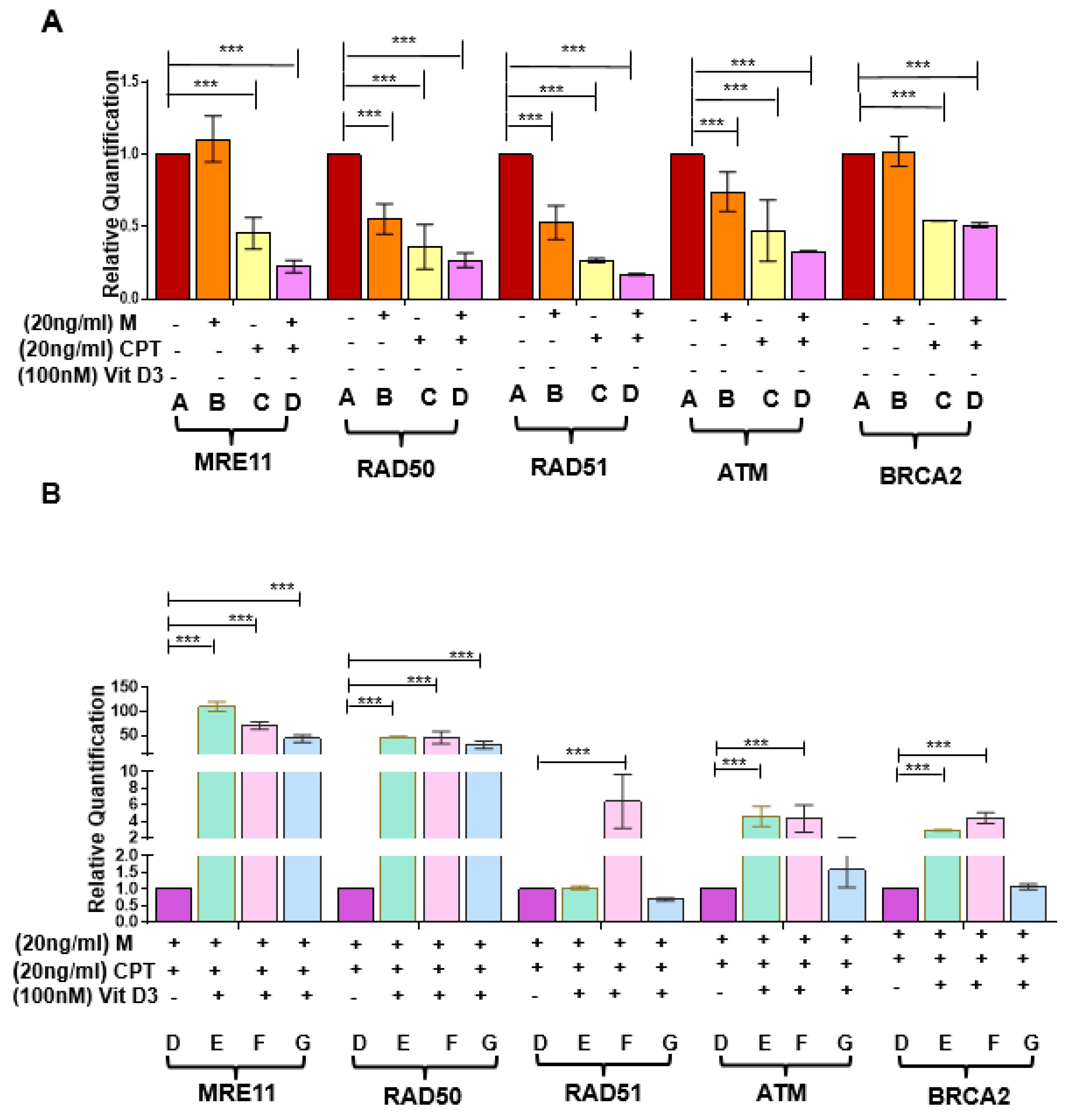
3.3. VitD3 Attenuated Mirin- and CPT-Induced DNA Damage in DES-MMSCs
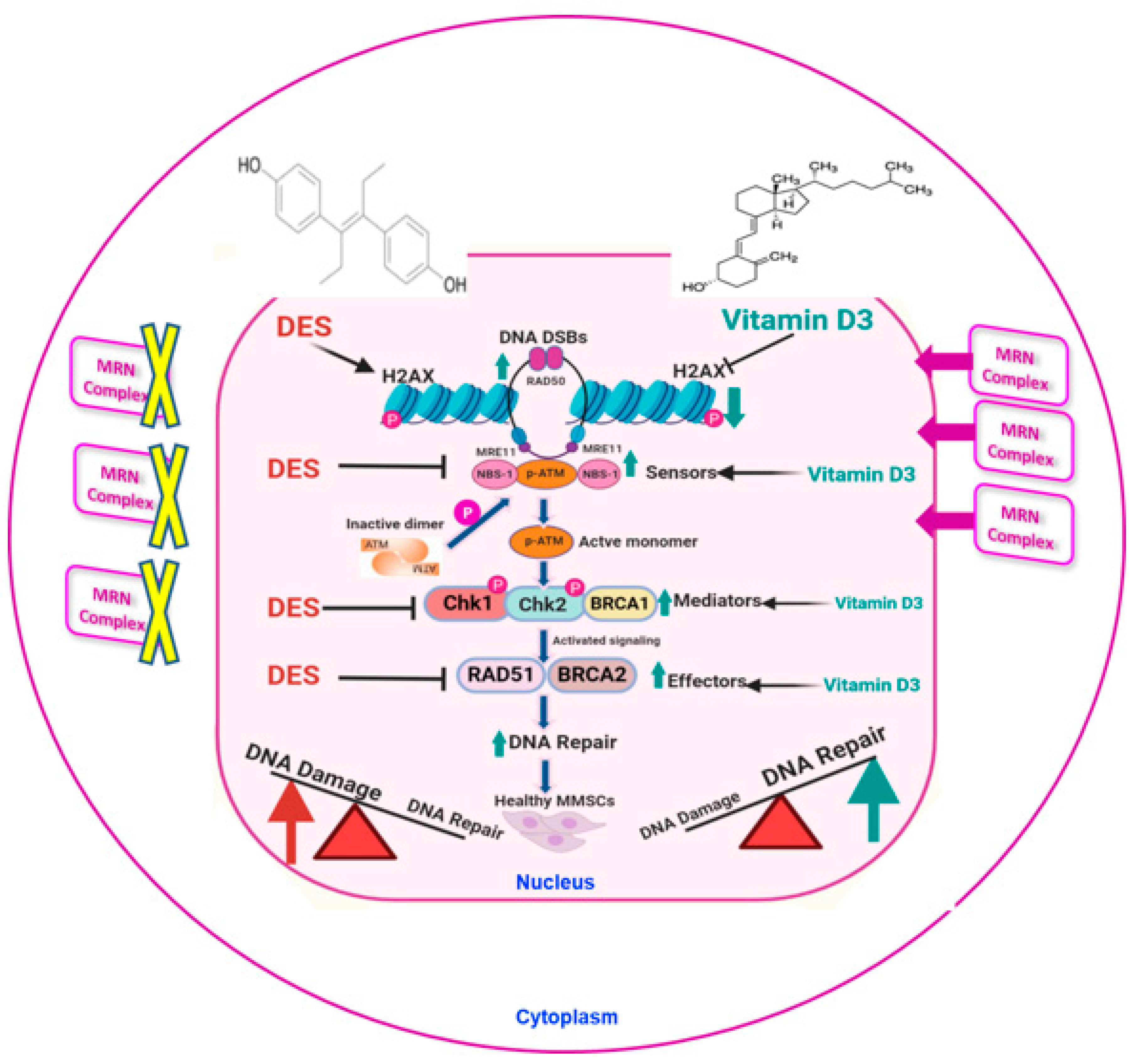
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bulun, S.E. Uterine fibroids. N. Engl. J. Med. 2013, 369, 1344–1355. [Google Scholar] [CrossRef] [PubMed]
- Stewart, E.A.; Laughlin-Tommaso, S.K.; Catherino, W.H.; Lalitkumar, S.; Gupta, D.; Vollenhoven, B. Uterine fibroids. Nat. Rev. Dis. Primers 2016, 2, 16043. [Google Scholar] [CrossRef]
- Elkafas, H.; El Andaloussi, A.; Al-Hendy, A. Emergence of uterine fibroid driver mutations in perturbed myometrial stem cells: The role of hypovitaminosis D. Fertil. Steril. 2017, 108, e390. [Google Scholar] [CrossRef][Green Version]
- Cardozo, E.R.; Clark, A.D.; Banks, N.K.; Henne, M.B.; Stegmann, B.J.; Segars, J.H. The estimated annual cost of uterine leiomyomata in the United States. Am. J. Obstet. Gynecol. 2012, 206, 211.e211–211.e219. [Google Scholar] [CrossRef] [PubMed]
- Fernung, L.P.; Yang, Q.; Sakamuro, D.; Kumari, A.; Mas, A.; Al-Hendy, A. Endocrine disruptor exposure during development increases incidence of uterine fibroids by altering DNA repair in myometrial stem cells. Biol. Reprod. 2018, 99, 735–748. [Google Scholar]
- Segars, J.H.; Parrott, E.C.; Nagel, J.D.; Guo, X.C.; Gao, X.; Birnbaum, L.S.; Pinn, V.W.; Dixon, D. Proceedings from the Third National Institutes of Health International Congress on Advances in Uterine Leiomyoma Research: Comprehensive review, conference summary and future recommendations. Hum. Reprod. Update 2014, 20, 309–333. [Google Scholar] [CrossRef]
- Yang, Q.; Diamond, M.P.; Al-Hendy, A. Early life adverse environmental exposures increase the risk of uterine fibroid development: Role of epigenetic regulation. Front. Pharmacol. 2016, 7, 40. [Google Scholar] [CrossRef]
- Mas, A.; Stone, L.; O’Connor, P.M.; Yang, Q.; Kleven, D.; Simon, C.; Walker, C.L.; Al-Hendy, A. Developmental Exposure to Endocrine Disruptors Expands Murine Myometrial Stem Cell Compartment as a Prerequisite to Leiomyoma Tumorigenesis. Stem Cells 2017, 35, 666–678. [Google Scholar] [CrossRef]
- Li, Y.; Hamilton, K.J.; Lai, A.Y.; Burns, K.A.; Li, L.; Wade, P.A.; Korach, K.S. Diethylstilbestrol (DES)-stimulated hormonal toxicity is mediated by ERalpha alteration of target gene methylation patterns and epigenetic modifiers (DNMT3A, MBD2, and HDAC2) in the mouse seminal vesicle. Environ. Health Perspect. 2014, 122, 262–268. [Google Scholar] [CrossRef]
- Prusinski, L.; Al-Hendy, A.; Yang, Q. Developmental exposure to endocrine disrupting chemicals alters the epigenome: Identification of reprogrammed targets. Gynecol. Obstet. Res. 2016, 3, 1–6. [Google Scholar] [CrossRef]
- Diamanti-Kandarakis, E.; Bourguignon, J.P.; Giudice, L.C.; Hauser, R.; Prins, G.S.; Soto, A.M.; Zoeller, R.T.; Gore, A.C. Endocrine-disrupting chemicals: An Endocrine Society scientific statement. Endocr. Rev. 2009, 30, 293–342. [Google Scholar] [CrossRef]
- McWilliams, M.M.; Chennathukuzhi, V.M. Recent advances in uterine fibroid etiology. Semin. Reprod. Med. 2017, 35, 181–189. [Google Scholar] [CrossRef] [PubMed]
- D’Aloisio, A.A.; Baird, D.D.; DeRoo, L.A.; Sandler, D.P. Association of intrauterine and early-life exposures with diagnosis of uterine leiomyomata by 35 years of age in the Sister Study. Environ. Health Perspect. 2010, 118, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Steiner, A.Z.; D’Aloisio, A.A.; DeRoo, L.A.; Sandler, D.P.; Baird, D.D. Association of intrauterine and early-life exposures with age at menopause in the Sister Study. Am. J. Epidemiol. 2010, 172, 140–148. [Google Scholar] [CrossRef] [PubMed]
- D’Aloisio, A.A.; Baird, D.D.; DeRoo, L.A.; Sandler, D.P. Early-life exposures and early-onset uterine leiomyomata in black women in the Sister Study. Environ. Health Perspect. 2012, 120, 406–412. [Google Scholar] [CrossRef]
- Reed, C.E.; Fenton, S.E. Exposure to diethylstilbestrol during sensitive life stages: A legacy of heritable health effects. Birth Defects Res. Part C Embryo Today Rev. 2013, 99, 134–146. [Google Scholar] [CrossRef] [PubMed]
- Watkins, O. Diethylstilbestrol in the prevention and treatment of complications of pregnancy. Am. J. Obstet. Gynecol. 1948, 56, 821–834. [Google Scholar] [CrossRef]
- Walker, C.L.; Hunter, D.; Everitt, J.I. Uterine leiomyoma in the Eker rat: A unique model for important diseases of women. Genes Chromosomes Cancer 2003, 38, 349–356. [Google Scholar] [CrossRef]
- Cook, J.D.; Davis, B.J.; Cai, S.-L.; Barrett, J.C.; Conti, C.J.; Walker, C.L. Interaction between genetic susceptibility and early-life environmental exposure determines tumor-suppressor-gene penetrance. Proc. Natl. Acad. Sci. USA 2005, 102, 8644–8649. [Google Scholar] [CrossRef]
- Mas, A.; Cervelló, I.; Gil-Sanchis, C.; Faus, A.; Ferro, J.; Pellicer, A.; Simón, C. Identification and characterization of the human leiomyoma side population as putative tumor-initiating cells. Fertil. Steril. 2012, 98, 741–751.e746. [Google Scholar] [CrossRef]
- Rothkamm, K.; Lobrich, M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc. Natl. Acad. Sci. USA 2003, 100, 5057–5062. [Google Scholar] [CrossRef] [PubMed]
- Ono, M.; Qiang, W.; Serna, V.A.; Yin, P.; Navarro, A.; Monsivais, D.; Kakinuma, T.; Dyson, M.; Druschitz, S.; Unno, K. Role of stem cells in human uterine leiomyoma growth. PLoS ONE 2012, 7, e36935. [Google Scholar] [CrossRef] [PubMed]
- Elkafas, H.; Qiwei, Y.; Al-Hendy, A. Origin of Uterine Fibroids: Conversion of Myometrial Stem Cells to Tumor-Initiating Cells. Semin. Reprod. Med. 2017, 35, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Fernung, L.P.; Al-Hendy, A.; Yang, Q. A Preliminary Study: Human Fibroid Stro-1(+)/CD44(+) Stem Cells Isolated From Uterine Fibroids Demonstrate Decreased DNA Repair and Genomic Integrity Compared to Adjacent Myometrial Stro-1(+)/CD44(+) Cells. Reprod Sci. 2019, 26, 619–638. [Google Scholar] [CrossRef]
- Yang, Q.; Nair, S.; Laknaur, A.; Ismail, N.; Diamond, M.P.; Al-Hendy, A. The Polycomb Group Protein EZH2 Impairs DNA Damage Repair Gene Expression in Human Uterine Fibroids. Biol. Reprod 2016, 94, 69. [Google Scholar] [CrossRef]
- Bikle, D. Vitamin D: Production, Metabolism, and Mechanisms of Action. In Endotext; Feingold, K.R., Anawalt, B., Boyce, A., Chrousos, G., Dungan, K., Grossman, A., Hershman, J.M., Kaltsas, G., Koch, C., Kopp, P., Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar]
- Vanoirbeek, E.; Krishnan, A.; Eelen, G.; Verlinden, L.; Bouillon, R.; Feldman, D.; Verstuyf, A. The anti-cancer and anti-inflammatory actions of 1, 25 (OH) 2D3. Best Pract. Res. Clin. Endocrinol. Metab. 2011, 25, 593–604. [Google Scholar] [CrossRef]
- Wilmanski, T.; Buhman, K.; Donkin, S.S.; Burgess, J.R.; Teegarden, D. 1α, 25-dihydroxyvitamin D inhibits de novo fatty acid synthesis and lipid accumulation in metastatic breast cancer cells through down-regulation of pyruvate carboxylase. J. Nutr. Biochem. 2017, 40, 194–200. [Google Scholar] [CrossRef]
- Halder, S.K.; Sharan, C.; Al-Hendy, O.; Al-Hendy, A. Paricalcitol, a vitamin D receptor activator, inhibits tumor formation in a murine model of uterine fibroids. Reprod. Sci. 2014, 21, 1108–1119. [Google Scholar] [CrossRef]
- Sharan, C.; Halder, S.K.; Thota, C.; Jaleel, T.; Nair, S.; Al-Hendy, A. Vitamin D inhibits proliferation of human uterine leiomyoma cells via catechol-O-methyltransferase. Fertil. Steril. 2011, 95, 247–253. [Google Scholar] [CrossRef]
- Brakta, S.; Diamond, J.S.; Al-Hendy, A.; Diamond, M.P.; Halder, S.K. Role of vitamin D in uterine fibroid biology. Fertil. Steril. 2015, 104, 698–706. [Google Scholar] [CrossRef]
- Ali, M.; Shahin, S.M.; Sabri, N.A.; Al-Hendy, A.; Yang, Q. Hypovitaminosis D exacerbates the DNA damage load in human uterine fibroids, which is ameliorated by vitamin D3 treatment. Acta Pharmacol. Sin. 2019, 40, 957–970. [Google Scholar]
- Widlansky, M.E.; Jensen, D.M.; Wang, J.; Liu, Y.; Geurts, A.M.; Kriegel, A.J.; Liu, P.; Ying, R.; Zhang, G.; Casati, M.; et al. miR-29 contributes to normal endothelial function and can restore it in cardiometabolic disorders. EMBO Mol. Med. 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Al-Hendy, A.; Diamond, M.P.; Boyer, T.G.; Halder, S.K. Vitamin D3 Inhibits Wnt/beta-Catenin and mTOR Signaling Pathways in Human Uterine Fibroid Cells. J. Clin. Endocrinol. Metabol. 2016, 101, 1542–1551. [Google Scholar] [CrossRef] [PubMed]
- Halder, S.K.; Osteen, K.G.; Al-Hendy, A. 1, 25-dihydroxyvitamin d3 reduces extracellular matrix-associated protein expression in human uterine fibroid cells. Biol. Reprod. 2013, 89, 150–151. [Google Scholar] [CrossRef]
- Graziano, S.; Johnston, R.; Deng, O.; Zhang, J.; Gonzalo, S. Vitamin D/vitamin D receptor axis regulates DNA repair during oncogene-induced senescence. Oncogene 2016, 35, 5362–5376. [Google Scholar] [CrossRef]
- Da Silva Machado, C.; Venancio, V.P.; Aissa, A.F.; Hernandes, L.C.; de Mello, M.B.; Del Lama, J.E.C.; Marzocchi-Machado, C.M.; Bianchi, M.L.P.; Antunes, L.M.G. Vitamin D3 deficiency increases DNA damage and the oxidative burst of neutrophils in a hypertensive rat model. Mutat. Res./Genet. Toxicol. Environ. Mutagenesis 2016, 798, 19–26. [Google Scholar] [CrossRef]
- Elhusseini, H.; Elkafas, H.; Abdelaziz, M.; Halder, S.; Atabiekov, I.; Eziba, N.; Ismail, N.; El Andaloussi, A.; Al-Hendy, A. Diet-induced vitamin D deficiency triggers inflammation and DNA damage profile in murine myometrium. Int. J. Women’s Health 2018, 10, 503. [Google Scholar] [CrossRef]
- Ali, M.; Shahin, S.M.; Sabri, N.A.; Al-Hendy, A.; Yang, Q. 1, 25 Dihydroxyvitamin D3 Enhances the Antifibroid Effects of Ulipristal Acetate in Human Uterine Fibroids. Reprod. Sci. 2018, 26, 812–828. [Google Scholar] [CrossRef]
- Yoshida, A.; Ueda, T.; Wano, Y.; Nakamura, T. DNA damage and cell killing by camptothecin and its derivative in human leukemia HL-60 cells. Jpn. J. Cancer Res. 1993, 84, 566–573. [Google Scholar] [CrossRef]
- Petroni, M.; Sardina, F.; Infante, P.; Bartolazzi, A.; Locatelli, E.; Fabretti, F.; Di Giulio, S.; Capalbo, C.; Cardinali, B.; Coppa, A.; et al. MRE11 inhibition highlights a replication stress-dependent vulnerability of MYCN-driven tumors. Cell Death Dis. 2018, 9, 895. [Google Scholar] [CrossRef]
- Cook, J.D.; Davis, B.J.; Goewey, J.A.; Berry, T.D.; Walker, C.L. Identification of a sensitive period for developmental programming that increases risk for uterine leiomyoma in Eker rats. Reprod. Sci. 2007, 14, 121–136. [Google Scholar] [CrossRef]
- Eid, J.I.; Eissa, S.M.; El-Ghor, A.A. Bisphenol A induces oxidative stress and DNA damage in hepatic tissue of female rat offspring. J. Basic Appl. Zool. 2015, 71, 10–19. [Google Scholar] [CrossRef]
- Bernstein, C.; Prasad, A.R.; Nfonsam, V.; Bernstein, H. DNA Damage, DNA Repair and Cancer. In New Research Directions in DNA Repair; Intech Open: Rijeka, Croatia, 2013; pp. 413–465. [Google Scholar]
- Cannan, W.J.; Pederson, D.S. Mechanisms and consequences of double-strand DNA break formation in chromatin. J. Cell. Physiol. 2016, 231, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Gonzalo, S. Novel roles of 1α, 25 (OH) 2D3 on DNA repair provide new strategies for breast cancer treatment. J. Steroid Biochem. Mol. Biol. 2014, 144, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Mas, A.; Prusinski, L.; Yang, Q.; Diaz-Gimeno, P.; Stone, L.; Diamond, M.P.; Simón, C.; Al-Hendy, A. Role of Stro1+/CD44+ stem cells in myometrial physiology and uterine remodeling during pregnancy. Biol. Reprod. 2016, 96, 70–80. [Google Scholar] [CrossRef]
- Chen, L.; Yang, R.; Qiao, W.; Yuan, X.; Wang, S.; Goltzman, D.; Miao, D. 1, 25-Dihydroxy vitamin D prevents tumorigenesis by inhibiting oxidative stress and inducing tumor cellular senescence in mice. Int. J. Cancer 2018, 143, 368–382. [Google Scholar] [CrossRef]
- Trémezaygues, L.; Seifert, M.; Tilgen, W.; Reichrath, J. 1, 25-Dihydroxyvitamin D3 protects human keratinocytes against UV-B-induced damage: In vitro analysis of cell viability/proliferation, DNA-damage and repair. Dermatoendocrinology 2009, 1, 239–245. [Google Scholar] [CrossRef]
- Machado, C.D.S.; Ferro Aissa, A.; Ribeiro, D.L.; Antunes, L.M.G. Vitamin D supplementation alters the expression of genes associated with hypertension and did not induce DNA damage in rats. J. Toxicol. Environ. Health Part A 2019, 82, 299–313. [Google Scholar] [CrossRef]
- Stracker, T.H.; Morales, M.; Couto, S.S.; Hussein, H.; Petrini, J.H. The carboxy terminus of NBS1 is required for induction of apoptosis by the MRE11 complex. Nature 2007, 447, 218. [Google Scholar] [CrossRef]
- Stracker, T.H.; Theunissen, J.-W.F.; Morales, M.; Petrini, J.H. The Mre11 complex and the metabolism of chromosome breaks: The importance of communicating and holding things together. DNA Repair 2004, 3, 845–854. [Google Scholar] [CrossRef]
- Situ, Y.; Chung, L.; Lee, C.S.; Ho, V. MRN (MRE11-RAD50-NBS1) complex in human cancer and prognostic implications in colorectal cancer. Int. J. Mol. Sci. 2019, 20, 816. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.T.; Siddique, A.; Shahid, N.; Khokher, S.; Fatima, W. Breast cancer risk associated with genes encoding DNA repair MRN complex: A study from Punjab, Pakistan. Breast Cancer 2018, 25, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Olson, E.; Nievera, C.J.; Liu, E.; Lee, A.Y.-L.; Chen, L.; Wu, X. The Mre11 complex mediates the S-phase checkpoint through an interaction with replication protein A. Mol. Cell. Biol. 2007, 27, 6053–6067. [Google Scholar] [CrossRef]
- Ting, H.-J.; Yasmin-Karim, S.; Yan, S.-J.; Hsu, J.-W.; Lin, T.-H.; Zeng, W.; Messing, J.; Sheu, T.-J.; Bao, B.-Y.; Li, W.X. A positive feedback signaling loop between ATM and the vitamin D receptor is critical for cancer chemoprevention by vitamin D. Cancer Res. 2012, 72, 958–968. [Google Scholar] [CrossRef] [PubMed]
- Maréchal, A.; Zou, L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 2013, 5, a012716. [Google Scholar] [CrossRef] [PubMed]
- Martino, C.; Pandya, D.; Lee, R.; Levy, G.; Lo, T.; Lobo, S.; Frank, R.C. ATM-Mutated Pancreatic Cancer: Clinical and Molecular Response to Gemcitabine/Nab-Paclitaxel After Genome-Based Therapy Resistance. Pancreas 2020, 49, 143–147. [Google Scholar] [CrossRef]
- Lavin, M.F.; Kozlov, S.; Gatei, M.; Kijas, A.W. ATM-Dependent Phosphorylation of All Three Members of the MRN Complex: From Sensor to Adaptor. Biomolecules 2015, 5, 2877–2902. [Google Scholar] [CrossRef]
- Kijas, A.W.; Lim, Y.C.; Bolderson, E.; Cerosaletti, K.; Gatei, M.; Jakob, B.; Tobias, F.; Taucher-Scholz, G.; Gueven, N.; Oakley, G. ATM-dependent phosphorylation of MRE11 controls extent of resection during homology directed repair by signalling through Exonuclease 1. Nucleic Acids Res. 2015, 43, 8352–8367. [Google Scholar] [CrossRef]
- Halicka, H.D.; Zhao, H.; Li, J.; Traganos, F.; Studzinski, G.P.; Darzynkiewicz, Z. Attenuation of constitutive DNA damage signaling by 1, 25-dihydroxyvitamin D3. Aging (Albany N. Y.) 2012, 4, 270. [Google Scholar]
- Sørensen, C.S.; Hansen, L.T.; Dziegielewski, J.; Syljuåsen, R.G.; Lundin, C.; Bartek, J.; Helleday, T. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat. Cell Biol. 2005, 7, 195–201. [Google Scholar] [CrossRef]
- Yarden, R.I.; Metsuyanim, S.; Pickholtz, I.; Shabbeer, S.; Tellio, H.; Papa, M.Z. BRCA1-dependent Chk1 phosphorylation triggers partial chromatin disassociation of phosphorylated Chk1 and facilitates S-phase cell cycle arrest. Int. J. Biochem. Cell Biol. 2012, 44, 1761–1769. [Google Scholar] [CrossRef] [PubMed]
- Buscemi, G.; Perego, P.; Carenini, N.; Nakanishi, M.; Chessa, L.; Chen, J.; Khanna, K.; Delia, D. Activation of ATM and Chk2 kinases in relation to the amount of DNA strand breaks. Oncogene 2004, 23, 7691. [Google Scholar] [CrossRef] [PubMed]
- Caldon, C.E. Estrogen signaling and the DNA damage response in hormone dependent breast cancers. Front Oncol. 2014, 4, 106. [Google Scholar] [CrossRef] [PubMed]
- Godin, S.K.; Sullivan, M.R.; Bernstein, K.A. Novel insights into RAD51 activity and regulation during homologous recombination and DNA replication. Biochem Cell Biol. 2016, 94, 407–418. [Google Scholar] [CrossRef]
- Mason, J.M.; Chan, Y.L.; Weichselbaum, R.W.; Bishop, D.K. Non-enzymatic roles of human RAD51 at stalled replication forks. Nat. Commun. 2019, 10, 4410. [Google Scholar] [CrossRef]
- D’Alessandro, G.; Whelan, D.R.; Howard, S.M.; Vitelli, V.; Renaudin, X.; Adamowicz, M.; Iannelli, F.; Jones-Weinert, C.W.; Lee, M.; Matti, V.; et al. BRCA2 controls DNA:RNA hybrid level at DSBs by mediating RNase H2 recruitment. Nat. Commun. 2018, 9, 5376. [Google Scholar] [CrossRef]
- Wasiewicz, T.; Piotrowska, A.; Wierzbicka, J.; Slominski, A.T.; Zmijewski, M.A. Antiproliferative activity of non-calcemic vitamin D analogs on human melanoma lines in relation to VDR and PDIA3 receptors. Int. J. Mol. Sci. 2018, 19, 2583. [Google Scholar] [CrossRef]
- Lin, Z.; Li, W. The Roles of Vitamin D and Its Analogs in Inflammatory Diseases. Curr. Top. Med. Chem. 2016, 16, 1242–1261. [Google Scholar] [CrossRef]
- Hengel, S.R.; Spies, M.A.; Spies, M. Small-molecule inhibitors targeting DNA repair and DNA repair deficiency in research and cancer therapy. Cell Chem. Biol. 2017, 24, 1101–1119. [Google Scholar] [CrossRef]
- Jayasooriya, R.G.P.T.; Dilshara, M.G.; Molagoda, I.M.N.; Park, C.; Park, S.R.; Lee, S.; Choi, Y.H.; Kim, G.-Y. Camptothecin induces G2/M phase arrest through the ATM-Chk2-Cdc25C axis as a result of autophagy-induced cytoprotection: Implications of reactive oxygen species. Oncotarget 2018, 9, 21744. [Google Scholar] [CrossRef] [PubMed]

| Antibody | Manufacturer | Catalog Number | Species, Monoclonal or Polyclonal | Application and Dilution |
|---|---|---|---|---|
| γH2AX | Cell Signaling Technology | 9718 | Rabbit, monoclonal | WB 1:1000 IF and F 1:200 |
| RAD51 | Abcam | AB133534 | Rabbit, monoclonal | WB 1:5000 F 1:1000 |
| RAD50 | Abcam | AB89 | Mouse, monoclonal | WB 1:5000 |
| NBS1 | Cell Signaling Technology | 14959 | Rabbit, monoclonal | WB 1:1000 F 1:400 |
| ATM | Abcam | Ab78 | Mouse, monoclonal | WB 1:1000 |
| p-ATM | Abcam | AB36810 | Mouse, monoclonal | WB 1:1000 F 1µg for 106 cells |
| BRCA2 | Bioss Antibodies | Bs-1210R | Rabbit, polyclonal | WB 1:100–1000 |
| Anti-β actin | Sigma Biochemicals, | A5316 | Mouse, monoclonal | WB 1:5000 |
| Alexa Fluor 488 Conjugate | Cell Signaling Technology | 4408 | Anti-Mouse IgG (H + L), F(ab’)2 Fragment | IF and F 1:500–1:2000 |
| Alexa Fluor 488 Conjugate | Cell Signaling Technology | 4412 | Anti-Rabbit IgG (H + L), F(ab’)2 Fragment | IF and F 1:500–1:2000 |
| Gene Symbol | Gene Description | Forward Primer Sequence | Reverse Primer Sequence |
|---|---|---|---|
| Chk1 | Chk1 checkpoint homolog | GGATGAGAAGATACTGGTTGACTT | TCGCTGAGCTTCCCTTTAATC |
| Chk2 | Chk2 checkpoint homolog | GAATTGTTTGACCGTGTGGTG | GTCCCGATGTATGATCCCATTT |
| ATM | Ataxia Telangiectasia mutated | GCAGTCATCATGCAGACCTATC | GTATCAGAGAACCGCGCTAATG |
| MRE11 | MRE11meiotic recombination 11 | TTCTTAAGGAGCGCCACATC | CCTCTCGAACTTCATCGTCTTC |
| RAD50 | RAD50 | GCTCGGATATGCTCTTGATG | GAACTGGAGAAGCTGAGTAAAG |
| NBS1 | Nijmegen breakage syndrome protein 1 | CAGAAGAGGAAGTGTTGGAAGAG | CATCCATCCTGGGCTTCTTT |
| RAD51 | RAD51 | TGCCCTTTACAGAACAGACTAC | ACCACTGCTACACCAAACTC |
| BRCA1 | Breast cancer 1, early onset | GAATCCTGTAAGCCAGAATCCA | CCTTCTCATTCCTGACTCCTTATC |
| BRCA2 | Breast cancer 2, early onset | GCAGATGGTGGATGGCTAAT | TTAGAGACCCAGACGCTAGAA |
| 18S | 18S Ribosomal RNA | CACGGACAGGATTGACAGATT | GAGTCTCGTTCGTTATCGGAATTA |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elkafas, H.; Ali, M.; Elmorsy, E.; Kamel, R.; Thompson, W.E.; Badary, O.; Al-Hendy, A.; Yang, Q. Vitamin D3 Ameliorates DNA Damage Caused by Developmental Exposure to Endocrine Disruptors in the Uterine Myometrial Stem Cells of Eker Rats. Cells 2020, 9, 1459. https://doi.org/10.3390/cells9061459
Elkafas H, Ali M, Elmorsy E, Kamel R, Thompson WE, Badary O, Al-Hendy A, Yang Q. Vitamin D3 Ameliorates DNA Damage Caused by Developmental Exposure to Endocrine Disruptors in the Uterine Myometrial Stem Cells of Eker Rats. Cells. 2020; 9(6):1459. https://doi.org/10.3390/cells9061459
Chicago/Turabian StyleElkafas, Hoda, Mohamed Ali, Engy Elmorsy, Rehab Kamel, Winston E. Thompson, Osama Badary, Ayman Al-Hendy, and Qiwei Yang. 2020. "Vitamin D3 Ameliorates DNA Damage Caused by Developmental Exposure to Endocrine Disruptors in the Uterine Myometrial Stem Cells of Eker Rats" Cells 9, no. 6: 1459. https://doi.org/10.3390/cells9061459
APA StyleElkafas, H., Ali, M., Elmorsy, E., Kamel, R., Thompson, W. E., Badary, O., Al-Hendy, A., & Yang, Q. (2020). Vitamin D3 Ameliorates DNA Damage Caused by Developmental Exposure to Endocrine Disruptors in the Uterine Myometrial Stem Cells of Eker Rats. Cells, 9(6), 1459. https://doi.org/10.3390/cells9061459








