Valproic Acid Significantly Improves CRISPR/Cas9-Mediated Gene Editing
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of the RNP Complexes and Donor Vector
2.2. Cell Culture
2.3. Mouse Embryo Culture and Microinjection of One-Cell Stage Embryos
2.4. Surveyor Assay
2.5. Western Blot
2.6. Sanger Sequencing and PCR Analysis
2.7. Immunohistochemical Analysis
2.8. Bioinformatics Analysis
2.9. Statistical Analysis
3. Results
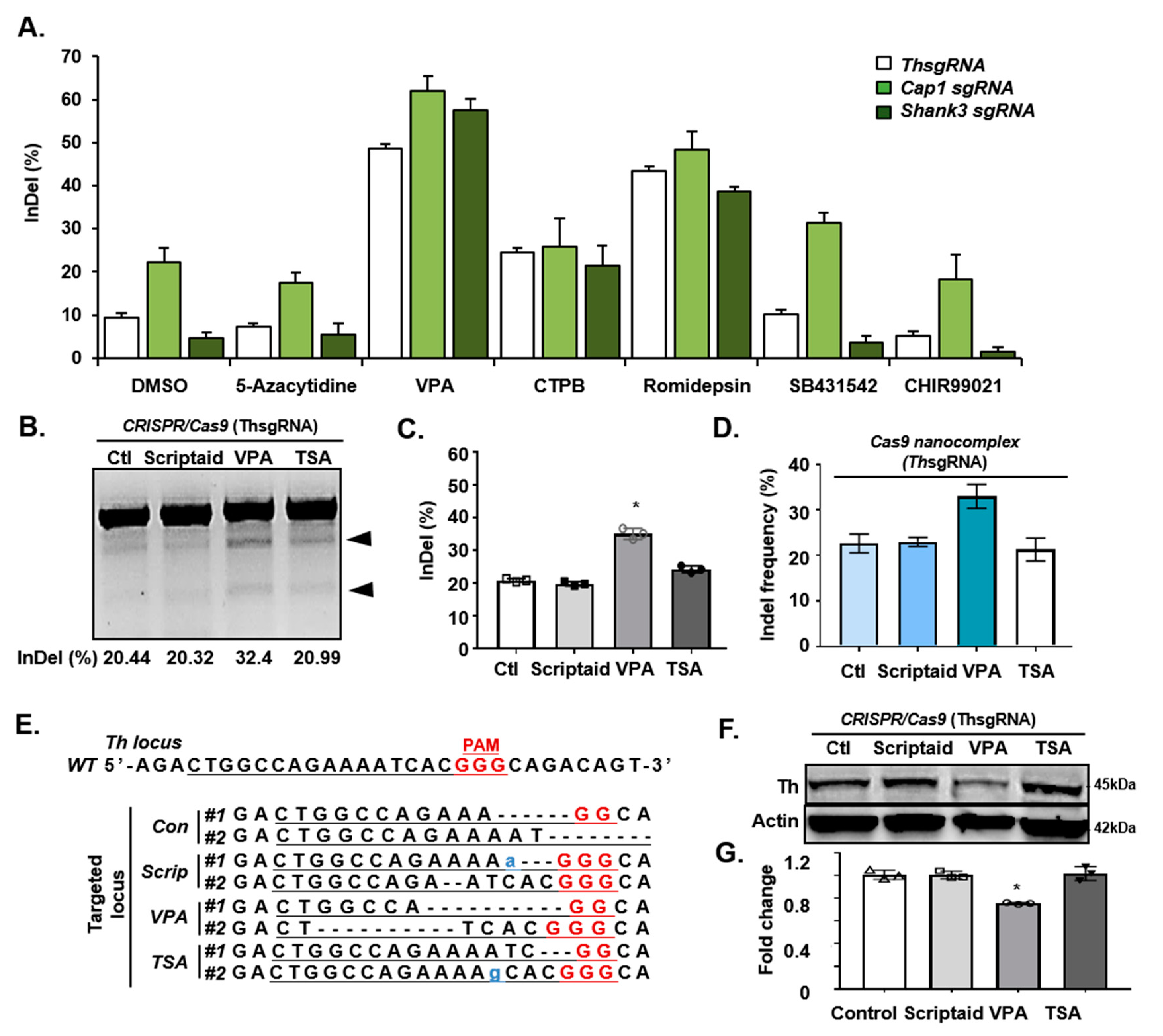
3.1. Effect of Valproic Acid (VPA) on CRISPR/Cas9-Mediated Gene Targeting In Vitro
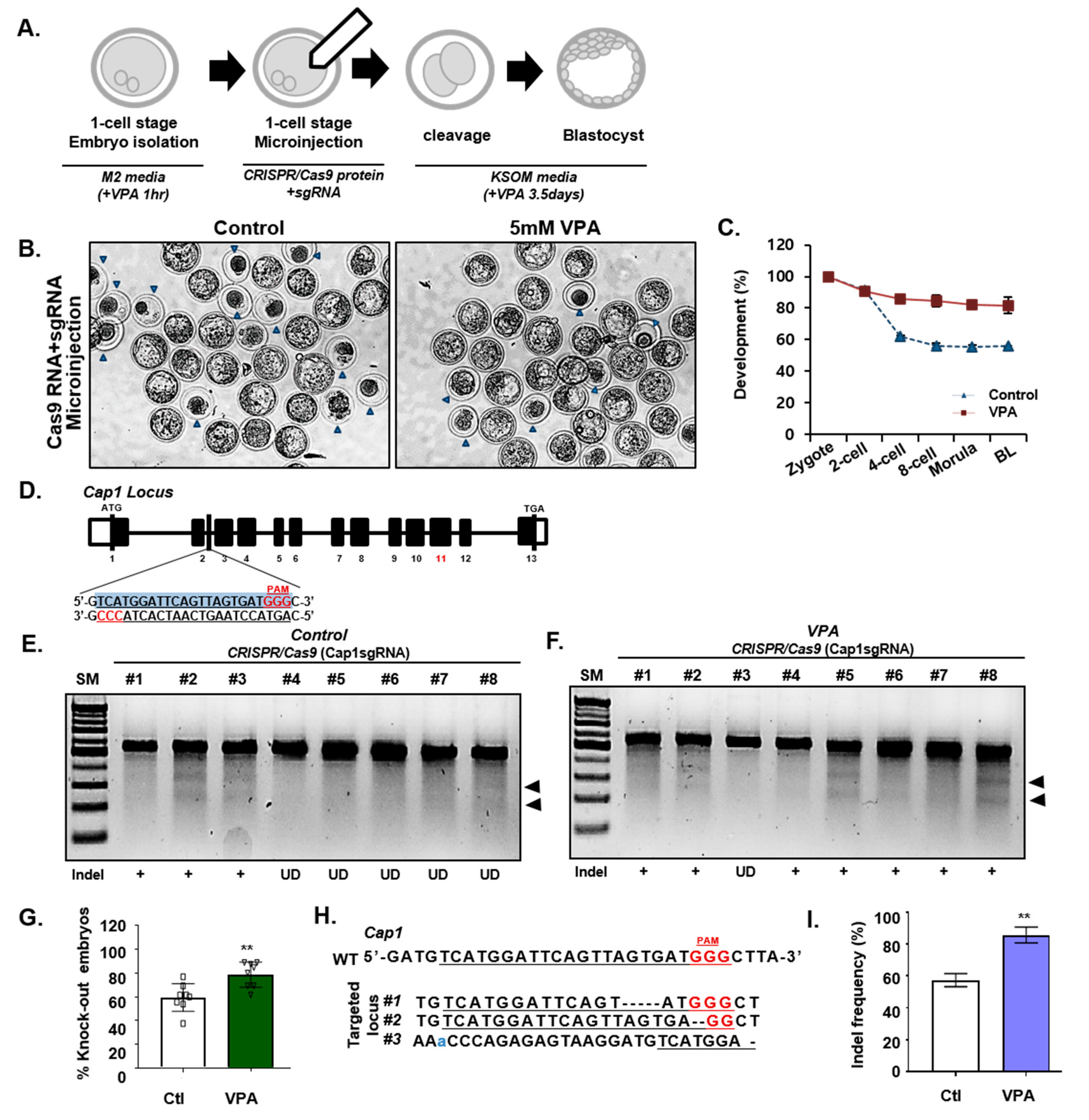
3.2. Effect of VPA on CRISPR/Cas9-Mediated NHEJ in Mouse Embryos
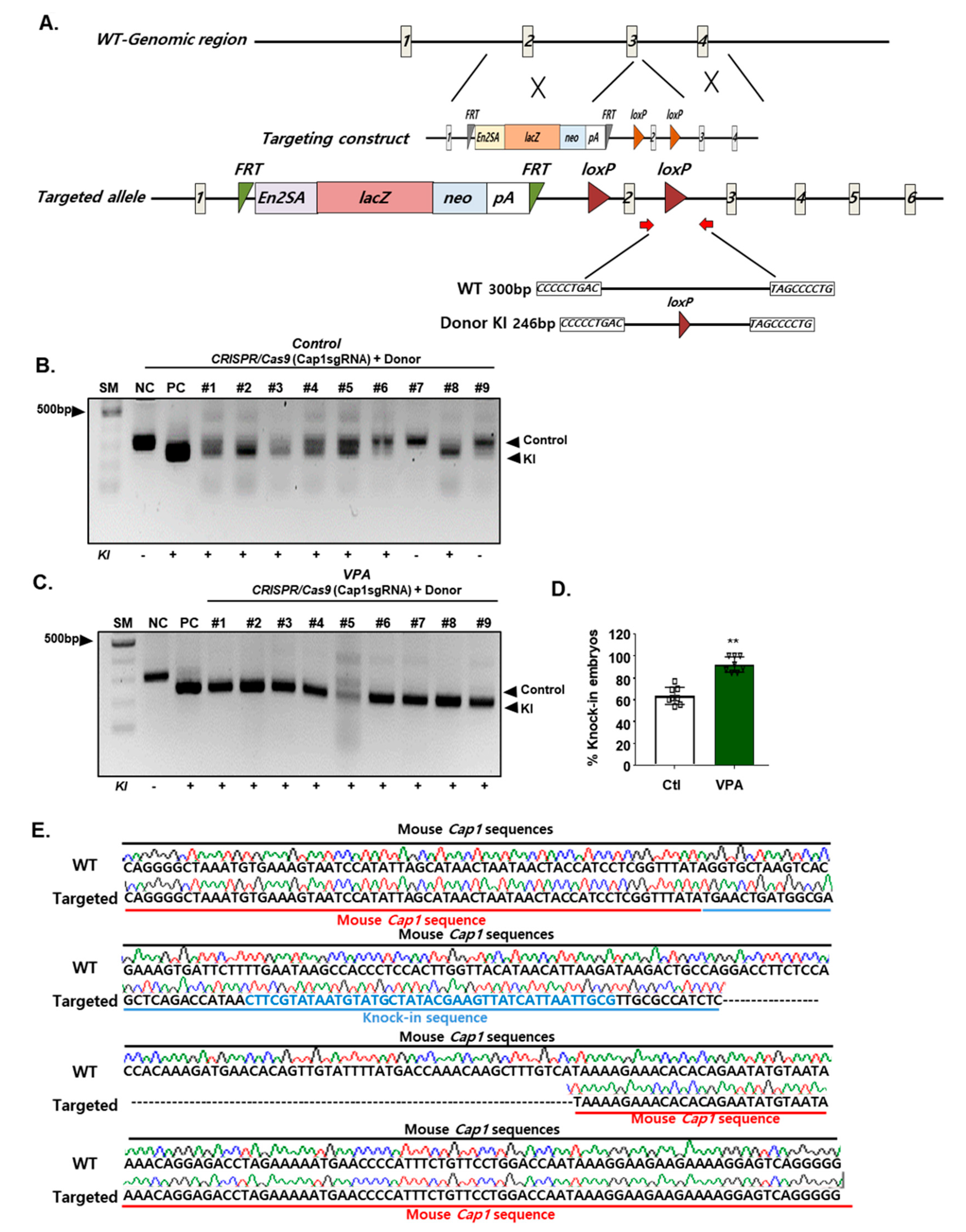
3.3. Effect of VPA on CRISPR/Cas9-Mediated HDR in Mouse Embryos
3.4. Mechanism of VPA in the CRISPR/Cas9-Based Efficient Gene Editing
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Gaj, T.; Gersbach, C.A.; Barbas, C.F. ZFN, TALEN and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Wiedenheft, B.; Sternberg, S.H.; Doudna, J.A. RNA-guided genetic silencing systems in bacteria and archaea. Nature 2012, 482, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Friedland, A.E.; Tzur, Y.B.; Esvelt, K.M.; Colaiacovo, M.P.; Church, G.M.; Calarco, J.A. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 2013, 10, 741–743. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Perez-Pinera, P.; Kocak, D.D.; Vockley, C.M.; Adler, A.F.; Kabadi, A.M.; Polstein, L.R.; Thakore, P.I.; Glass, K.A.; Ousterout, D.G.; Leong, K.W.; et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods 2013, 10, 973–976. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Scott, D.A.; Kriz, A.J.; Chiu, A.C.; Hsu, P.D.; Dadon, D.B.; Cheng, A.W.; Trevino, A.E.; Konermann, S.; Chen, S.; et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. 2014, 32, 670–676. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Potter, J.; Kumar, S.; Zou, Y.; Quintanilla, R.; Sridharan, M.; Carte, J.; Chen, W.; Roark, N.; Ranganathan, S.; et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol. 2015, 208, 44–53. [Google Scholar] [CrossRef]
- Konermann, S.; Brigham, M.D.; Trevino, A.E.; Joung, J.; Abudayyeh, O.O.; Barcena, C.; Hsu, P.D.; Habib, N.; Gootenberg, J.S.; Nishimasu, H.; et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2015, 517, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishna, S.; Kwaku Dad, A.B.; Beloor, J.; Gopalappa, R.; Lee, S.K.; Kim, H. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014, 24, 1020–1027. [Google Scholar] [CrossRef] [PubMed]
- Thakore, P.I.; D’Ippolito, A.M.; Song, L.; Safi, A.; Shivakumar, N.K.; Kabadi, A.M.; Reddy, T.E.; Crawford, G.E.; Gersbach, C.A. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods 2015, 12, 1143–1149. [Google Scholar] [CrossRef] [PubMed]
- Platt, R.J.; Chen, S.; Zhou, Y.; Yim, M.J.; Swiech, L.; Kempton, H.R.; Dahlman, J.E.; Parnas, O.; Eisenhaure, T.M.; Jovanovic, M.; et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 2014, 159, 440–455. [Google Scholar] [CrossRef] [PubMed]
- Chang, N.; Sun, C.; Gao, L.; Zhu, D.; Xu, X.; Zhu, X.; Xiong, J.W.; Xi, J.J. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013, 23, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Shen, B.; Cui, Y.; Chen, Y.; Wang, J.; Wang, L.; Kang, Y.; Zhao, X.; Si, W.; Li, W.; et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 2014, 156, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.; Xu, Y.; Zhang, X.; Ding, C.; Huang, R.; Zhang, Z.; Lv, J.; Xie, X.; Chen, Y.; Li, Y.; et al. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell 2015, 6, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Xin, J.; Fan, N.; Zou, Q.; Huang, J.; Ouyang, Z.; Zhao, Y.; Zhao, B.; Liu, Z.; Lai, S.; et al. Generation of CRISPR/Cas9-mediated gene-targeted pigs via somatic cell nuclear transfer. Cell Mol. Life Sci 2015, 72, 1175–1184. [Google Scholar] [CrossRef] [PubMed]
- Long, C.; McAnally, J.R.; Shelton, J.M.; Mireault, A.A.; Bassel-Duby, R.; Olson, E.N. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science 2014, 345, 1184–1188. [Google Scholar] [CrossRef] [PubMed]
- Schumann, K.; Lin, S.; Boyer, E.; Simeonov, D.R.; Subramaniam, M.; Gate, R.E.; Haliburton, G.E.; Ye, C.J.; Bluestone, J.A.; Doudna, J.A.; et al. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc. Natl. Acad. Sci. USA 2015, 112, 10437–10442. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Cao, J.; Xiong, M.; Petersen, A.J.; Dong, Y.; Tao, Y.; Huang, C.T.; Du, Z.; Zhang, S.C. Engineering Human Stem Cell Lines with Inducible Gene Knockout using CRISPR/Cas9. Cell Stem Cell 2015, 17, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Tsunekawa, Y.; Hernandez-Benitez, R.; Wu, J.; Zhu, J.; Kim, E.J.; Hatanaka, F.; Yamamoto, M.; Araoka, T.; Li, Z.; et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 2016, 540, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Liang, D.; Wang, Y.; Bai, M.; Tang, W.; Bao, S.; Yan, Z.; Li, D.; Li, J. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell 2013, 13, 659–662. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhu, X.; Hahm, H.S.; Wei, W.; Hao, E.; Hayek, A.; Ding, S. Revealing a core signaling regulatory mechanism for pluripotent stem cell survival and self-renewal by small molecules. Proc. Natl. Acad. Sci. USA 2010, 107, 8129–8134. [Google Scholar] [CrossRef] [PubMed]
- Borowiak, M.; Maehr, R.; Chen, S.; Chen, A.E.; Tang, W.; Fox, J.L.; Schreiber, S.L.; Melton, D.A. Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell 2009, 4, 348–358. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Staahl, B.T.; Alla, R.K.; Doudna, J.A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife 2014, 3, e04766. [Google Scholar] [CrossRef] [PubMed]
- Aksoy, Y.A.; Nguyen, D.T.; Chow, S.; Chung, R.S.; Guillemin, G.J.; Cole, N.J.; Hesselson, D. Chemical reprogramming enhances homology-directed genome editing in zebrafish embryos. Commun. Biol. 2019, 2, 198. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Scavuzzo, M.A.; Chmielowiec, J.; Sharp, R.; Bajic, A.; Borowiak, M. Enrichment of G2/M cell cycle phase in human pluripotent stem cells enhances HDR-mediated gene repair with customizable endonucleases. Sci. Rep. 2016, 6, 21264. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Yang, D.; Xu, J.; Zhu, T.; Chen, Y.E.; Zhang, J. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat. Commun. 2016, 7, 10548. [Google Scholar] [CrossRef] [PubMed]
- Cerveny, L.; Svecova, L.; Anzenbacherova, E.; Vrzal, R.; Staud, F.; Dvorak, Z.; Ulrichova, J.; Anzenbacher, P.; Pavek, P. Valproic acid induces CYP3A4 and MDR1 gene expression by activation of constitutive androstane receptor and pregnane X receptor pathways. Drug Metab. Dispos. 2007, 35, 1032–1041. [Google Scholar] [CrossRef] [PubMed]
- Lagneaux, L.; Gillet, N.; Stamatopoulos, B.; Delforge, A.; Dejeneffe, M.; Massy, M.; Meuleman, N.; Kentos, A.; Martiat, P.; Willems, L.; et al. Valproic acid induces apoptosis in chronic lymphocytic leukemia cells through activation of the death receptor pathway and potentiates TRAIL response. Exp. Hematol. 2007, 35, 1527–1537. [Google Scholar] [CrossRef] [PubMed]
- Gottlicher, M.; Minucci, S.; Zhu, P.; Kramer, O.H.; Schimpf, A.; Giavara, S.; Sleeman, J.P.; Lo Coco, F.; Nervi, C.; Pelicci, P.G.; et al. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001, 20, 6969–6978. [Google Scholar] [CrossRef] [PubMed]
- Huangfu, D.; Maehr, R.; Guo, W.; Eijkelenboom, A.; Snitow, M.; Chen, A.E.; Melton, D.A. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 2008, 26, 795–797. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.; Nakashima, K.; Kuwabara, T.; Mejia, E.; Gage, F.H. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc. Natl. Acad. Sci. USA 2004, 101, 16659–16664. [Google Scholar] [CrossRef] [PubMed]
- Shan, Z.; Feng-Nian, R.; Jie, G.; Ting, Z. Effects of valproic acid on proliferation, apoptosis, angiogenesis and metastasis of ovarian cancer in vitro and in vivo. Asian Pac. J. Cancer Prev. 2012, 13, 3977–3982. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ran, F.A.; Hsu, P.D.; Lin, C.Y.; Gootenberg, J.S.; Konermann, S.; Trevino, A.E.; Scott, D.A.; Inoue, A.; Matoba, S.; Zhang, Y.; et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013, 154, 1380–1389. [Google Scholar] [CrossRef] [PubMed]
- Hezroni, H.; Tzchori, I.; Davidi, A.; Mattout, A.; Biran, A.; Nissim-Rafinia, M.; Westphal, H.; Meshorer, E. H3K9 histone acetylation predicts pluripotency and reprogramming capacity of ES cells. Nucleus 2011, 2, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Hezroni, H.; Sailaja, B.S.; Meshorer, E. Pluripotency-related, valproic acid (VPA)-induced genome-wide histone H3 lysine 9 (H3K9) acetylation patterns in embryonic stem cells. J. Biol. Chem. 2011, 286, 35977–35988. [Google Scholar] [CrossRef] [PubMed]
- Kondo, T. Epigenetic alchemy for cell fate conversion. Curr. Opin. Genet. Dev. 2006, 16, 502–507. [Google Scholar] [CrossRef] [PubMed]
- Dashwood, R.H.; Ho, E. Dietary histone deacetylase inhibitors: From cells to mice to man. Semin. Cancer Biol. 2007, 17, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.P.; Blau, H.M. 5-Azacytidine permits gene activation in a previously noninducible cell type. Cell 1985, 40, 417–424. [Google Scholar] [CrossRef]
- Marchion, D.C.; Bicaku, E.; Daud, A.I.; Sullivan, D.M.; Munster, P.N. Valproic acid alters chromatin structure by regulation of chromatin modulation proteins. Cancer Res. 2005, 65, 3815–3822. [Google Scholar] [CrossRef] [PubMed]
- Mantelingu, K.; Reddy, B.A.; Swaminathan, V.; Kishore, A.H.; Siddappa, N.B.; Kumar, G.V.; Nagashankar, G.; Natesh, N.; Roy, S.; Sadhale, P.P.; et al. Specific inhibition of p300-HAT alters global gene expression and represses HIV replication. Chem. Biol. 2007, 14, 645–657. [Google Scholar] [CrossRef] [PubMed]
- Karthik, S.; Sankar, R.; Varunkumar, K.; Ravikumar, V. Romidepsin induces cell cycle arrest, apoptosis, histone hyperacetylation and reduces matrix metalloproteinases 2 and 9 expression in bortezomib sensitized non-small cell lung cancer cells. Biomed. Pharmacother. 2014, 68, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Inman, G.J.; Nicolas, F.J.; Callahan, J.F.; Harling, J.D.; Gaster, L.M.; Reith, A.D.; Laping, N.J.; Hill, C.S. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharm. 2002, 62, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Tan, L.; Yang, R.; Fang, B.; Qu, S.; Schulze, E.N.; Song, H.; Ying, Q.; Li, P. Pleiotropy of glycogen synthase kinase-3 inhibition by CHIR99021 promotes self-renewal of embryonic stem cells from refractory mouse strains. PLoS ONE 2012, 7, e35892. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.D.; Li, S.; Lu, Y.; Wang, W.; Liang, S.; Liu, X.; Jin, J.X.; Hong, Y.; Yan, C.G.; Yin, X.J. Valproic acid improved in vitro development of pig cloning embryos but did not improve survival of cloned pigs to adulthood. Theriogenology 2013, 79, 306–311.e1. [Google Scholar] [CrossRef] [PubMed]
- Song, B.S.; Yoon, S.B.; Sim, B.W.; Kim, Y.H.; Cha, J.J.; Choi, S.A.; Jeong, K.J.; Kim, J.S.; Huh, J.W.; Lee, S.R.; et al. Valproic acid enhances early development of bovine somatic cell nuclear transfer embryos by alleviating endoplasmic reticulum stress. Reprod. Fertil. Dev. 2014, 26, 432–440. [Google Scholar] [CrossRef]
- Xu, W.; Wang, Y.; Li, Y.; Wang, L.; Xiong, X.; Su, J.; Zhang, Y. Valproic acid improves the in vitro development competence of bovine somatic cell nuclear transfer embryos. Cell. Reprogram. 2012, 14, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Gurvich, N.; Tsygankova, O.M.; Meinkoth, J.L.; Klein, P.S. Histone deacetylase is a target of valproic acid-mediated cellular differentiation. Cancer Res. 2004, 64, 1079–1086. [Google Scholar] [CrossRef] [PubMed]
- Phiel, C.J.; Zhang, F.; Huang, E.Y.; Guenther, M.G.; Lazar, M.A.; Klein, P.S. Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J. Biol. Chem. 2001, 276, 36734–36741. [Google Scholar] [CrossRef] [PubMed]
- DeVane, C.L. Pharmacokinetics, drug interactions, and tolerability of valproate. Psychopharmacol. Bull. 2003, 37, 25–42. [Google Scholar] [PubMed]
- Valcourt, U.; Kowanetz, M.; Niimi, H.; Heldin, C.H.; Moustakas, A. TGF-beta and the Smad signaling pathway support transcriptomic reprogramming during epithelial-mesenchymal cell transition. Mol. Biol. Cell 2005, 16, 1987–2002. [Google Scholar] [CrossRef] [PubMed]
- Huangfu, D.; Osafune, K.; Maehr, R.; Guo, W.; Eijkelenboom, A.; Chen, S.; Muhlestein, W.; Melton, D.A. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat. Biotechnol. 2008, 26, 1269–1275. [Google Scholar] [CrossRef] [PubMed]

| Treatment | Targeting System | Blastocysts /Injected Zygotes | Targeted Blastocysts/Total | InDel Mutation Efficiency (%) |
|---|---|---|---|---|
| Control | Cas9 protein + Cap1 sgRNA | 34/60 | 21/34 | 61.8 |
| VPA | Cas9 protein + Cap1 sgRNA | 43/50 | 32/43 | 74.4 |
| VPA | Cas9 protein + Cap1 sgRNA | 45/50 | 36/45 | 80 |
| Control | Cas9 protein + Lphn2 sgRNA | 35/60 | 20/35 | 57.1 |
| VPA | Cas9 protein + Lphn2 sgRNA | 53/60 | 49/53 | 92.5 |
| VPA | Cas9 protein + Lphn2 sgRNA | 55/60 | 48/55 | 87.3 |
| Treatment | Targeting System | Blastocysts /Injected Zygotes | Targeted Blastocysts/Total | Knock-In Efficiency (%) |
|---|---|---|---|---|
| Control | Cas9 protein + Cap1 sgRNA + Cap1 Donor | 29/55 | 18/29 | 62.1 |
| Control | Cas9 protein + Cap1 sgRNA + Cap1 Donor | 25/50 | 25/39 | 64.1 |
| VPA | Cas9 protein + Cap1 sgRNA + Cap1 Donor | 32/40 | 32/34 | 94.1 |
| VPA | Cas9 protein + Cap1 sgRNA + Cap1 Donor | 45/55 | 45/50 | 90 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, H.; Shin, J.; Choi, H.; Cho, B.; Kim, J. Valproic Acid Significantly Improves CRISPR/Cas9-Mediated Gene Editing. Cells 2020, 9, 1447. https://doi.org/10.3390/cells9061447
Park H, Shin J, Choi H, Cho B, Kim J. Valproic Acid Significantly Improves CRISPR/Cas9-Mediated Gene Editing. Cells. 2020; 9(6):1447. https://doi.org/10.3390/cells9061447
Chicago/Turabian StylePark, Hanseul, Jaein Shin, Hwan Choi, Byounggook Cho, and Jongpil Kim. 2020. "Valproic Acid Significantly Improves CRISPR/Cas9-Mediated Gene Editing" Cells 9, no. 6: 1447. https://doi.org/10.3390/cells9061447
APA StylePark, H., Shin, J., Choi, H., Cho, B., & Kim, J. (2020). Valproic Acid Significantly Improves CRISPR/Cas9-Mediated Gene Editing. Cells, 9(6), 1447. https://doi.org/10.3390/cells9061447




