Exploring Extracellular Vesicles Biogenesis in Hypothalamic Cells through a Heavy Isotope Pulse/Trace Proteomic Approach
Abstract
1. Introduction
2. Materials and Methods
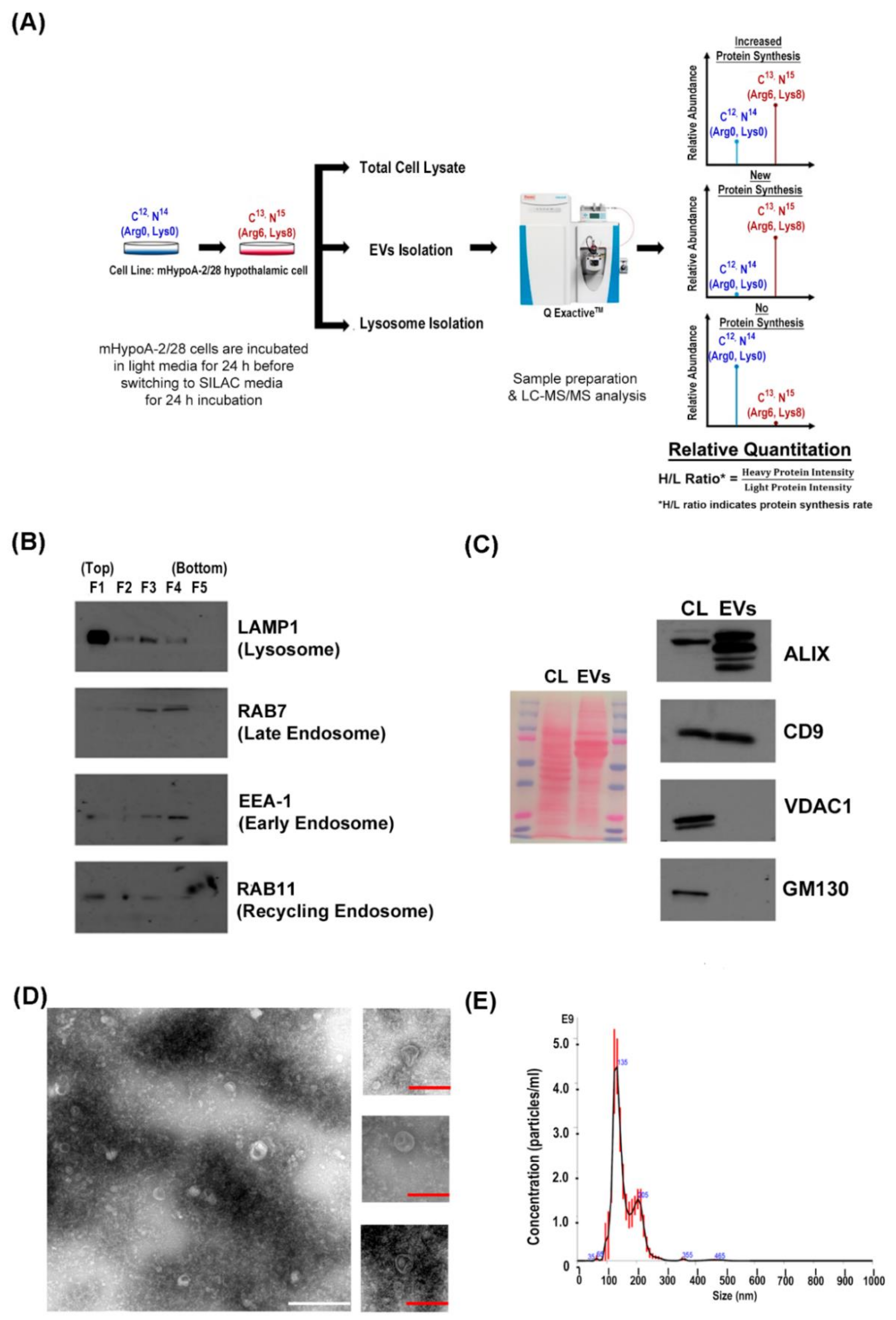
2.1. Cell Culture and pSILAC Treatment
2.2. EVs Isolation
2.3. Lysosome Enrichment
2.4. In-solution Digestion and HPLC Fractionation
2.5. In Gel Digestion
2.6. LC-MS/MS Analysis
2.7. Database Search
2.8. Western Blot Analysis
2.9. Nanoparticle Tracking Analysis
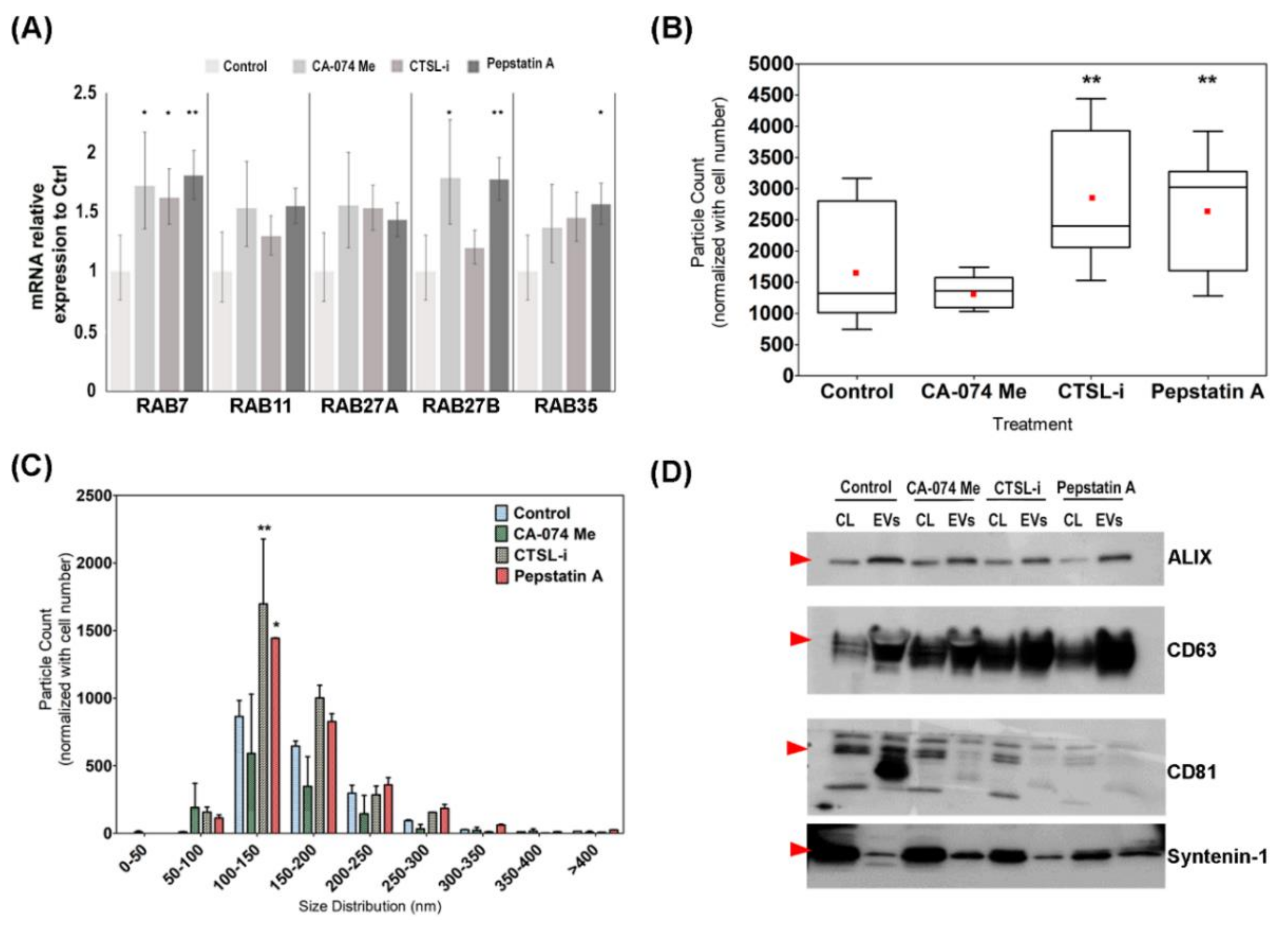
2.10. Cathepsin Inhibition Assay
2.11. Proteinase K Assay
2.12. RNA Isolation and RT PCR
2.13. Transmission Electron Microscopy
3. Results
3.1. Mass Spectrometric Identification and Quantification of Newly Synthesized Proteins in the Sub-proteome of mHypoA 2/28 Adult Hypothalamus Cell
3.2. Extracellular Vesicles Biogenesis is an Active Process that Required Newly Synthesized Protein
3.3. Hierarchical Clustering Analysis Revealed the Preferential Localization of Newly Synthesized Cathepsin Proteins into the EVs
3.4. Chemical Inhibition of Cathepsin D Promote EVs Secretion and Alter EVs Content
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Tan, H.S.; Datta, A.; Lai, R.C.; Zhang, H.; Meng, W.; Lim, S.K.; Sze, S.K. Hypoxic Tumor Cell Modulates Its Microenvironment to Enhance Angiogenic and Metastatic Potential by Secretion of Proteins and Exosomes. Mol. Cell. Proteomics 2010, 9, 1085–1099. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef] [PubMed]
- Eitan, E.; Suire, C.; Zhang, S.; Mattson, M.P. Impact of lysosome status on extracellular vesicle content and release. Ageing Res. Rev. 2016, 32, 65–74. [Google Scholar] [CrossRef]
- Miranda, A.M.; Lasiecka, Z.M.; Xu, Y.; Neufeld, J.; Shahriar, S.; Simoes, S.; Chan, R.B.; Oliveira, T.G.; Small, S.A.; Di Paolo, G. Neuronal lysosomal dysfunction releases exosomes harboring APP C-terminal fragments and unique lipid signatures. Nat. Commun. 2018, 9, 291. [Google Scholar] [CrossRef]
- Palmulli, R.; van Niel, G. To be or not to be... Secreted as exosomes, a balance finely tuned by the mechanisms of biogenesis. Essays Biochem. 2018, 62, 177–191. [Google Scholar] [CrossRef]
- Raiborg, C.; Stenmark, H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 2009, 458, 445–452. [Google Scholar] [CrossRef]
- Colombo, M.; Moita, C.; van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Thery, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013, 126, 5553–5565. [Google Scholar] [CrossRef]
- Buschow, S.I.; Nolte-’t Hoen, E.N.M.; Van Niel, G.; Pols, M.S.; Ten Broeke, T.; Lauwen, M.; Ossendorp, F.; Melief, C.J.M.; Raposo, G.; Wubbolts, R.; et al. MHC II in Dendritic Cells is Targeted to Lysosomes or T Cell-Induced Exosomes Via Distinct Multivesicular Body Pathways. Traffic 2009, 10, 1528–1542. [Google Scholar] [CrossRef]
- Baietti, M.F.; Zhang, Z.; Mortier, E.; Melchior, A.; Degeest, G.; Geeraerts, A.; Ivarsson, Y.; Depoortere, F.; Coomans, C.; Vermeiren, E.; et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell. Biol. 2012, 14, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Roucourt, B.; Meeussen, S.; Bao, J.; Zimmermann, P.; David, G. Heparanase activates the syndecan-syntenin-ALIX exosome pathway. Cell Res. 2015, 25, 412. [Google Scholar] [CrossRef] [PubMed]
- Stuffers, S.; Sem Wegner, C.; Stenmark, H.; Brech, A. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic 2009, 10, 925–937. [Google Scholar] [CrossRef] [PubMed]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brugger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef]
- Chairoungdua, A.; Smith, D.L.; Pochard, P.; Hull, M.; Caplan, M.J. Exosome release of beta-catenin: A novel mechanism that antagonizes Wnt signaling. J. Cell Biol. 2010, 190, 1079–1091. [Google Scholar] [CrossRef]
- Tricarico, C.; Clancy, J.; D’Souza-Schorey, C. Biology and biogenesis of shed microvesicles. Small GTPases 2016, 8, 220–232. [Google Scholar] [CrossRef]
- Muralidharan-Chari, V.; Clancy, J.; Plou, C.; Romao, M.; Chavrier, P.; Raposo, G.; D’Souza-Schorey, C. ARF6-regulated shedding of tumor cell-derived plasma membrane microvesicles. Curr. Biol. 2009, 19, 1875–1885. [Google Scholar] [CrossRef]
- Wang, T.; Gilkes, D.M.; Takano, N.; Xiang, L.; Luo, W.; Bishop, C.J.; Chaturvedi, P.; Green, J.J.; Semenza, G.L. Hypoxia-inducible factors and RAB22A mediate formation of microvesicles that stimulate breast cancer invasion and metastasis. Proc. Natl. Acad. Sci. USA 2014, 111, E3234–E3242. [Google Scholar] [CrossRef]
- Bianco, F.; Perrotta, C.; Novellino, L.; Francolini, M.; Riganti, L.; Menna, E.; Saglietti, L.; Schuchman, E.H.; Furlan, R.; Clementi, E.; et al. Acid sphingomyelinase activity triggers microparticle release from glial cells. EMBO J. 2009, 28, 1043–1054. [Google Scholar] [CrossRef]
- Nabhan, J.F.; Hu, R.; Oh, R.S.; Cohen, S.N.; Lu, Q. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc. Natl. Acad. Sci. USA 2012, 109, 4146–4151. [Google Scholar] [CrossRef]
- Li, M.; Ramratnam, B. Proteomic Characterization of Exosomes from HIV-1-Infected Cells. Methods Mol. Biol. 2016, 1354, 311–326. [Google Scholar] [CrossRef] [PubMed]
- Whitham, M.; Parker, B.L.; Friedrichsen, M.; Hingst, J.R.; Hjorth, M.; Hughes, W.E.; Egan, C.L.; Cron, L.; Watt, K.I.; Kuchel, R.P.; et al. Extracellular Vesicles Provide a Means for Tissue Crosstalk during Exercise. Cell. Metab. 2018, 27, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Palmisano, G.; Jensen, S.S.; Le Bihan, M.C.; Laine, J.; McGuire, J.N.; Pociot, F.; Larsen, M.R. Characterization of membrane-shed microvesicles from cytokine-stimulated beta-cells using proteomics strategies. Mol. Cell. Proteomics 2012, 11, 230–243. [Google Scholar] [CrossRef] [PubMed]
- Schwanhausser, B.; Gossen, M.; Dittmar, G.; Selbach, M. Global analysis of cellular protein translation by pulsed SILAC. Proteomics 2009, 9, 205–209. [Google Scholar] [CrossRef]
- Young, L.; Sung, J.; Stacey, G.; Masters, J.R. Detection of Mycoplasma in cell cultures. Nat. Protocol. 2010, 5, 929–934. [Google Scholar] [CrossRef]
- Park, J.E.; Dutta, B.; Tse, S.W.; Gupta, N.; Tan, C.F.; Low, J.K.; Yeoh, K.W.; Kon, O.L.; Tam, J.P.; Sze, S.K. Hypoxia-induced tumor exosomes promote M2-like macrophage polarization of infiltrating myeloid cells and microRNA-mediated metabolic shift. Oncogene 2019, 38, 5158–5173. [Google Scholar] [CrossRef]
- Kall, L.; Canterbury, J.D.; Weston, J.; Noble, W.S.; MacCoss, M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 2007, 4, 923–925. [Google Scholar] [CrossRef]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis, 2 ed.; Springer: Cham, Switzerland, 2016. [Google Scholar]
- Willms, E.; Johansson, H.J.; Mäger, I.; Lee, Y.; Blomberg, K.E.M.; Sadik, M.; Alaarg, A.; Smith, C.I.E.; Lehtiö, J.; El Andaloussi, S.; et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci. Rep. 2016, 6, 22519. [Google Scholar] [CrossRef]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef]
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Biol. 2016, 428, 688–692. [Google Scholar] [CrossRef] [PubMed]
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.; Bond, V.C.; Borras, F.E.; Breakefield, X.; Budnik, V.; et al. Vesiclepedia: A compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012, 10, e1001450. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, D.; Isaacs, A.M. The role of ESCRT proteins in fusion events involving lysosomes, endosomes and autophagosomes. Biochem. Soc. Trans. 2010, 38, 1469–1473. [Google Scholar] [CrossRef] [PubMed]
- Jae, N.; McEwan, D.G.; Manavski, Y.; Boon, R.A.; Dimmeler, S. Rab7a and Rab27b control secretion of endothelial microRNA through extracellular vesicles. FEBS Lett. 2015, 589, 3182–3188. [Google Scholar] [CrossRef]
- Katoh, K.; Shibata, H.; Suzuki, H.; Nara, A.; Ishidoh, K.; Kominami, E.; Yoshimori, T.; Maki, M. The ALG-2-interacting protein Alix associates with CHMP4b, a human homologue of yeast Snf7 that is involved in multivesicular body sorting. J. Biol. Chem. 2003, 278, 39104–39113. [Google Scholar] [CrossRef]
- Martin-Serrano, J.; Yarovoy, A.; Perez-Caballero, D.; Bieniasz, P.D. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. PNAS USA 2003, 100, 12414–12419. [Google Scholar] [CrossRef]
- Christ, L.; Raiborg, C.; Wenzel, E.M.; Campsteijn, C.; Stenmark, H. Cellular Functions and Molecular Mechanisms of the ESCRT Membrane-Scission Machinery. Trends Biochem. Sci. 2017, 42, 42–56. [Google Scholar] [CrossRef]
- van Niel, G.; Charrin, S.; Simoes, S.; Romao, M.; Rochin, L.; Saftig, P.; Marks, M.S.; Rubinstein, E.; Raposo, G. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Develop. Cell 2011, 21, 708–721. [Google Scholar] [CrossRef]
- Andreu, Z.; Yáñez-Mó, M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014, 5, 442. [Google Scholar] [CrossRef]
- Perez-Hernandez, D.; Gutierrez-Vazquez, C.; Jorge, I.; Lopez-Martin, S.; Ursa, A.; Sanchez-Madrid, F.; Vazquez, J.; Yanez-Mo, M. The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J. Biol. Chem. 2013, 288, 11649–11661. [Google Scholar] [CrossRef]
- Gould, S.; Fordjour, F.K.; Daaboul, G. A shared pathway of exosome biogenesis operates at plasma and endosome membranes. bioRxiv 2019. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Brix, K. Lysosomal Proteases. In Lysosomes; Saftig, P., Ed.; Springer US: Boston, MA, USA, 2005; pp. 50–59. [Google Scholar] [CrossRef]
- Sanman, L.E.; Bogyo, M. Activity-based profiling of proteases. Annu. Rev. Biochem. 2014, 83, 249–273. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Huo, C.; Qiao, Z.; Shang, Z.; Uzzaman, A.; Liu, S.; Jiang, X.; Fan, L.Y.; Ji, L.; Guan, X.; et al. Comparative Proteomic Analysis of Exosomes and Microvesicles in Human Saliva for Lung Cancer. J. Proteome. Res. 2018, 17, 1101–1107. [Google Scholar] [CrossRef]
- Downs, C.A.; Dang, V.D.; Johnson, N.M.; Denslow, N.D.; Alli, A.A. Hydrogen Peroxide Stimulates Exosomal Cathepsin B Regulation of the Receptor for Advanced Glycation End-Products (RAGE). J. Cell Biochem. 2018, 119, 599–606. [Google Scholar] [CrossRef]
- Urbanelli, L.; Magini, A.; Buratta, S.; Brozzi, A.; Sagini, K.; Polchi, A.; Tancini, B.; Emiliani, C. Signaling pathways in exosomes biogenesis, secretion and fate. Genes 2013, 4, 152–170. [Google Scholar] [CrossRef]
- Araki, N.; Yokota, S.; Takashima, Y.; Ogawa, K. The distribution of cathepsin D in two types of lysosomal or endosomal profiles of rat hepatocytes as revealed by combined immunocytochemistry and acid phosphatase enzyme cytochemistry. Exp. Cell Res. 1995, 217, 469–476. [Google Scholar] [CrossRef]
- Pillay, C.S.; Elliott, E.; Dennison, C. Endolysosomal proteolysis and its regulation. Biochem. J. 2002, 363, 417–429. [Google Scholar] [CrossRef]
- Boonen, M.; Staudt, C.; Gilis, F.; Oorschot, V.; Klumperman, J.; Jadot, M. Cathepsin D and its newly identified transport receptor SEZ6L2 can modulate neurite outgrowth. J. Cell Sci. 2016, 129, 557–568. [Google Scholar] [CrossRef]
- Markmann, S.; Thelen, M.; Cornils, K.; Schweizer, M.; Brocke-Ahmadinejad, N.; Willnow, T.; Heeren, J.; Gieselmann, V.; Braulke, T.; Kollmann, K. Lrp1/LDL Receptor Play Critical Roles in Mannose 6-Phosphate-Independent Lysosomal Enzyme Targeting. Traffic 2015, 16, 743–759. [Google Scholar] [CrossRef]
- Ahn, K.; Yeyeodu, S.; Collette, J.; Madden, V.; Arthur, J.; Li, L.; Erickson, A.H. An alternate targeting pathway for procathepsin L in mouse fibroblasts. Traffic 2002, 3, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Yeyeodu, S.; Ahn, K.; Madden, V.; Chapman, R.; Song, L.; Erickson, A.H. Procathepsin L self-association as a mechanism for selective secretion. Traffic 2000, 1, 724–737. [Google Scholar] [CrossRef] [PubMed]
- Katunuma, N. Structure-based development of specific inhibitors for individual cathepsins and their medical applications. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2011, 87, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Blanc, L.; Vidal, M. New insights into the function of Rab GTPases in the context of exosomal secretion. Small GTPases 2018, 9, 95–106. [Google Scholar] [CrossRef]
- Vitelli, R.; Santillo, M.; Lattero, D.; Chiariello, M.; Bifulco, M.; Bruni, C.B.; Bucci, C. Role of the small GTPase Rab7 in the late endocytic pathway. J. Biol. Chem. 1997, 272, 4391–4397. [Google Scholar] [CrossRef]
- Greening, D.W.; Xu, R.; Gopal, S.K.; Rai, A.; Simpson, R.J. Proteomic insights into extracellular vesicle biology—defining exosomes and shed microvesicles. Expert Rev. Proteomics 2017, 14, 69–95. [Google Scholar] [CrossRef]
- Takahashi, S.; Kubo, K.; Waguri, S.; Yabashi, A.; Shin, H.W.; Katoh, Y.; Nakayama, K. Rab11 regulates exocytosis of recycling vesicles at the plasma membrane. J. Cell Sci. 2012, 125, 4049–4057. [Google Scholar] [CrossRef]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010, 12, 19–30. [Google Scholar] [CrossRef]
- Sato, M.; Sato, K.; Liou, W.; Pant, S.; Harada, A.; Grant, B.D. Regulation of endocytic recycling by C. elegans Rab35 and its regulator RME-4, a coated-pit protein. EMBO J. 2008, 27, 1183–1196. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Thery, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Gallart-Palau, X.; Serra, A.; Hase, Y.; Tan, C.F.; Chen, C.P.; Kalaria, R.N.; Sze, S.K. Brain-derived and circulating vesicle profiles indicate neurovascular unit dysfunction in early Alzheimer’s disease. Brain Pathol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Cheow, E.S.; Cheng, W.C.; Lee, C.N.; de Kleijn, D.; Sorokin, V.; Sze, S.K. Plasma-derived Extracellular Vesicles Contain Predictive Biomarkers and Potential Therapeutic Targets for Myocardial Ischemic (MI) Injury. Mol. Cell Proteom. 2016, 15, 2628–2640. [Google Scholar] [CrossRef]
- Ishidoh, K.; Kominami, E. Gene regulation and extracellular functions of procathepsin L. Biol. Chem. 1998, 379, 131–135. [Google Scholar] [PubMed]
- Alvarez-Erviti, L.; Seow, Y.; Schapira, A.H.; Gardiner, C.; Sargent, I.L.; Wood, M.J.; Cooper, J.M. Lysosomal dysfunction increases exosome-mediated alpha-synuclein release and transmission. Neurobiol. Dis. 2011, 42, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Kirschke, H.; Cathepsin, L. Handbook of Proteolytic Enzymes, 3rd ed.; Rawlings, N.D., Salvesen, G., Eds.; Academic Press: Cambridge, MA, USA, 2013; Volume 2, pp. 1808–1817. [Google Scholar]
- Man, S.M.; Kanneganti, T.-D. Regulation of lysosomal dynamics and autophagy by CTSB/cathepsin B. Autophagy 2016, 12, 2504–2505. [Google Scholar] [CrossRef] [PubMed]
- Lamore, S.D.; Wondrak, G.T. Autophagic-lysosomal dysregulation downstream of cathepsin B inactivation in human skin fibroblasts exposed to UVA. Photochem. Photobiol. Sci. 2012, 11, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, S.A.; Pérez-González, R.; Sharma, A.; Huang, F.-K.; Alldred, M.J.; Pawlik, M.; Kaur, G.; Ginsberg, S.D.; Neubert, T.A.; Levy, E. Enhanced exosome secretion in Down syndrome brain—A protective mechanism to alleviate neuronal endosomal abnormalities. Acta. Neuropathol. Commun. 2017, 5, 65. [Google Scholar] [CrossRef]
- Laurent-Matha, V.; Derocq, D.; Prebois, C.; Katunuma, N.; Liaudet-Coopman, E. Processing of human cathepsin D is independent of its catalytic function and auto-activation: Involvement of cathepsins L and B. J. Biochem. 2006, 139, 363–371. [Google Scholar] [CrossRef]
- Kishimoto, Y.; Hiraiwa, M.; O’Brien, J.S. Saposins: Structure, function, distribution, and molecular genetics. J. Lipid Res. 1992, 33, 1255–1267. [Google Scholar]
- Rijnboutt, S.; Aerts, H.M.; Geuze, H.J.; Tager, J.M.; Strous, G.J. Mannose 6-phosphate-independent membrane association of cathepsin D, glucocerebrosidase, and sphingolipid-activating protein in HepG2 cells. J. Biol. Chem. 1991, 266, 4862–4868. [Google Scholar]
- Gopalakrishnan, M.M.; Grosch, H.-W.; Locatelli-Hoops, S.; Werth, N.; Smolenová, E.; Nettersheim, M.; Sandhoff, K.; Hasilik, A. Purified recombinant human prosaposin forms oligomers that bind procathepsin D and affect its autoactivation. Biochem. J. 2004, 383, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Azuma, N.; O’Brien, J.S.; Moser, H.W.; Kishimoto, Y. Stimulation of acid ceramidase activity by saposin D. Arch. Biochem. Biophys. 1994, 311, 354–357. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, S.; Martin, B.M.; Kishimoto, Y.; O’Brien, J.S. Saposin D: A sphingomyelinase activator. Biochem. Biophys. Res. Commun. 1988, 156, 403–410. [Google Scholar] [CrossRef]
- Heinrich, M.; Wickel, M.; Winoto-Morbach, S.; Schneider-Brachert, W.; Weber, T.; Brunner, J.; Saftig, P.; Peters, C.; Kronke, M.; Schutze, S. Ceramide as an activator lipid of cathepsin D. Adv. Exp. Med. Biol. 2000, 477, 305–315. [Google Scholar] [CrossRef]


| (A) | |||
| Cell Lysate | |||
| Gene Name | Protein Name | H/L Ratio | Unique Peptide |
| CHMP2A | Charged multivesicular body protein 2A | 5.67 | 5 |
| HGS | Hepatocyte growth factor-regulated tyrosine kinase substrate | 1.84 | 12 |
| PDCD6IP | Alix | 1.61 | 48 |
| RAB7A | Ras-related protein RAB-7a | 1.55 | 19 |
| SDC1 | Syndecan 1 | 100 | 3 |
| SDCBP | Syntenin-1 | 24.62 | 5 |
| TSG101 | Tumor susceptibility gene 101 | 1.51 | 10 |
| VPS4A | Vacuolar protein sorting-associated protein 4A | 3.2 | 5 |
| VPS4B | Vacuolar protein sorting-associated protein 4B | 2.13 | 7 |
| (B) | |||
| EVs | |||
| Gene Name | Protein Name | H/L Ratio | Unique Peptide |
| PDCD6IP | ALIX | 1.64 | 21 |
| RAB7A | Ras-related protein RAB-7a | 5.64 | 5 |
| SDCBP | Syntenin-1 | 2.73 | 7 |
| (A) | |||||||
| ESCRT-0 | |||||||
| EVs | Lysosome | Cell Lysate | |||||
| Gene Name | Protein Name | H/L Ratio | Unique Peptide | H/L Ratio | Unique Peptide | H/L Ratio | Unique Peptide |
| HGS | Hepatocyte growth factor-regulated tyrosine kinase substrate | - | - | 100 | 2 | 1.81 | 12 |
| STAM1 | Signal transducing adapter molecule 1 | - | - | - | - | 1.45 | 13 |
| STAM2 | Signal transducing adapter molecule 2 | - | - | - | - | 2.67 | 12 |
| (B) | |||||||
| ESCRT-I | |||||||
| EVs | Lysosome | Cell Lysate | |||||
| Gene Name | Protein Name | H/L Ratio | Unique Peptide | H/L Ratio | Unique Peptide | H/L Ratio | Unique Peptide |
| MVB12A | Multivesicular body subunit 12A | 0.01 | 1 | - | - | 2.93 | 5 |
| MVB12B | Multivesicular body subunit 12B | - | - | - | - | 7.76 | 3 |
| TSG101 * | Tumor susceptibility gene 101 | 3.61 | 1 | 2.47 | 5 | 1.51 | 10 |
| UBAP1 | Ubiquitin-associated protein 1 | - | - | - | - | 1.60 | 11 |
| VPS28 | Vacuolar protein sorting-associated protein 28 | 2.61 | 3 | - | - | 1.25 | 11 |
| VPS37A | Vacuolar protein sorting-associated protein 37A | - | - | - | - | 2.03 | 7 |
| VPS37B | Vacuolar protein sorting-associated protein 37B | - | - | - | - | 3.40 | 5 |
| VPS37C | Vacuolar protein sorting-associated protein 37C | - | - | - | - | 2.19 | 6 |
| (C) | |||||||
| ESCRT-II | |||||||
| EVs | Lysosome | Cell Lysate | |||||
| Gene Name | Protein Name | H/L Ratio | Unique Peptide | H/L Ratio | Unique Peptide | H/L Ratio | Unique Peptide |
| VPS25 | Vacuolar protein sorting-associated protein 25 | - | - | - | - | 0.70 | 9 |
| VPS36 | Vacuolar protein sorting-associated protein 36 | - | - | - | - | 2.84 | 12 |
| (D) | |||||||
| ESCRT-III | |||||||
| EVs | Lysosome | Cell Lysate | |||||
| Gene Name | Protein Name | H/L Ratio | Unique Peptide | H/L Ratio | Unique Peptide | H/L Ratio | Unique Peptide |
| CHMP1A | Charged multivesicular body protein 1A | - | - | - | - | 0.01 | 2 |
| CHMP1B | Charged multivesicular body protein 1B | - | - | - | - | 2.11 | 1 |
| CHMP2A | Charged multivesicular body protein 2A | - | - | - | - | 5.67 | 5 |
| CHMP2B | Charged multivesicular body protein 2B | - | - | - | - | 1.18 | 3 |
| CHMP3 | Charged multivesicular body protein 3 | - | - | - | - | 2 | 4 |
| CHMP4B | Charged multivesicular body protein 4B | 2.15 | 1 | 100 | 1 | 1.89 | 5 |
| CHMP4C | Charged multivesicular body protein 4C | - | - | - | - | 21.31 | 4 |
| CHMP5 | Charged multivesicular body protein 5 | - | - | - | - | 2.14 | 7 |
| CHMP6 | Charged multivesicular body protein 6 | - | - | - | - | 1.98 | 3 |
| CHMP7 | Charged multivesicular body protein 7 | - | - | - | - | 16.08 | 2 |
| IST1 | IST1 homolog | - | - | 0.48 | 2 | 1.59 | 8 |
| (E) | |||||||
| ESCRT-Accessory Proteins | |||||||
| EVs | Lysosome | Cell Lysate | |||||
| Gene Name | Protein Name | H/L Ratio | Unique Peptide | H/L Ratio | Unique Peptide | H/L Ratio | Unique Peptide |
| PDCD6IP | ALIX | 5.64 | 21 | 1.93 | 23 | 1.61 | 48 |
| VPS4A | Vacuolar protein sorting-associated protein 4A | - | - | - | - | 3.2 | 5 |
| VPS4B | Vacuolar protein sorting-associated protein 4B | - | - | 9.86 | 2 | 2.13 | 7 |
| VTA1 | Vacuolar protein sorting-associated protein VTA1 homolog | - | - | 8.00 | 4 | 1.18 | 7 |
| (A) | |||||||
| ALIX-Syntenin-Syndecan Axis | |||||||
| EVs | Lysosome | Cell Lysate | |||||
| Gene Name | Protein Name | H/L Ratio | Unique Peptide | H/L Ratio | Unique Peptide | H/L Ratio | Unique Peptide |
| PDCD6IP | ALIX | 5.64 | 21 | 1.93 | 23 | 1.61 | 48 |
| SSDCBP | Syntenin-1 | 2.73 | 7 | 10.8 | 7 | 24.62 | 5 |
| SDC1 | Syndecan 1 | - | - | - | - | 100 | 3 |
| (B) | |||||||
| Tetraspanin-enriched Domain | |||||||
| EVs | Lysosome | Cell Lysate | |||||
| Gene Name | Protein Name | H/L Ratio | Unique Peptide | H/L Ratio | Unique Peptide | H/L Ratio | Unique Peptide |
| CD9 | CD9 | 2.40 | 2 | 3.41 | 3 | 2.13 | 4 |
| CD63 | CD63 | 0.19 | 1 | 0.73 | 5 | 1.53 | 4 |
| CD81 | CD81 | 10.34 | 3 | 30.61 | 2 | 7.43 | 3 |
| (C) | |||||||
| Ceramide-based EVs Biogenesis | |||||||
| EVs | Lysosome | Cell Lysate | |||||
| Gene Name | Protein Name | H/L Ratio | Unique Peptide | H/L Ratio | Unique Peptide | H/L Ratio | Unique Peptide |
| SMPD1 | Sphingomyelin phosphodiesterase | 0.01 | 1 | - | - | 16.24 | 1 |
| SMPD2 | Sphingomyelin phosphodiesterase 2 | - | - | - | - | 1.56 | 7 |
| SMPD3 | Sphingomyelin phosphodiesterase 3 | - | - | - | - | 0.88 | 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, C.F.; Teo, H.S.; Park, J.E.; Dutta, B.; Tse, S.W.; Leow, M.K.-S.; Wahli, W.; Sze, S.K. Exploring Extracellular Vesicles Biogenesis in Hypothalamic Cells through a Heavy Isotope Pulse/Trace Proteomic Approach. Cells 2020, 9, 1320. https://doi.org/10.3390/cells9051320
Tan CF, Teo HS, Park JE, Dutta B, Tse SW, Leow MK-S, Wahli W, Sze SK. Exploring Extracellular Vesicles Biogenesis in Hypothalamic Cells through a Heavy Isotope Pulse/Trace Proteomic Approach. Cells. 2020; 9(5):1320. https://doi.org/10.3390/cells9051320
Chicago/Turabian StyleTan, Chee Fan, Hui San Teo, Jung Eun Park, Bamaprasad Dutta, Shun Wilford Tse, Melvin Khee-Shing Leow, Walter Wahli, and Siu Kwan Sze. 2020. "Exploring Extracellular Vesicles Biogenesis in Hypothalamic Cells through a Heavy Isotope Pulse/Trace Proteomic Approach" Cells 9, no. 5: 1320. https://doi.org/10.3390/cells9051320
APA StyleTan, C. F., Teo, H. S., Park, J. E., Dutta, B., Tse, S. W., Leow, M. K.-S., Wahli, W., & Sze, S. K. (2020). Exploring Extracellular Vesicles Biogenesis in Hypothalamic Cells through a Heavy Isotope Pulse/Trace Proteomic Approach. Cells, 9(5), 1320. https://doi.org/10.3390/cells9051320








