Closing the Gap: Membrane Contact Sites in the Regulation of Autophagy
Abstract
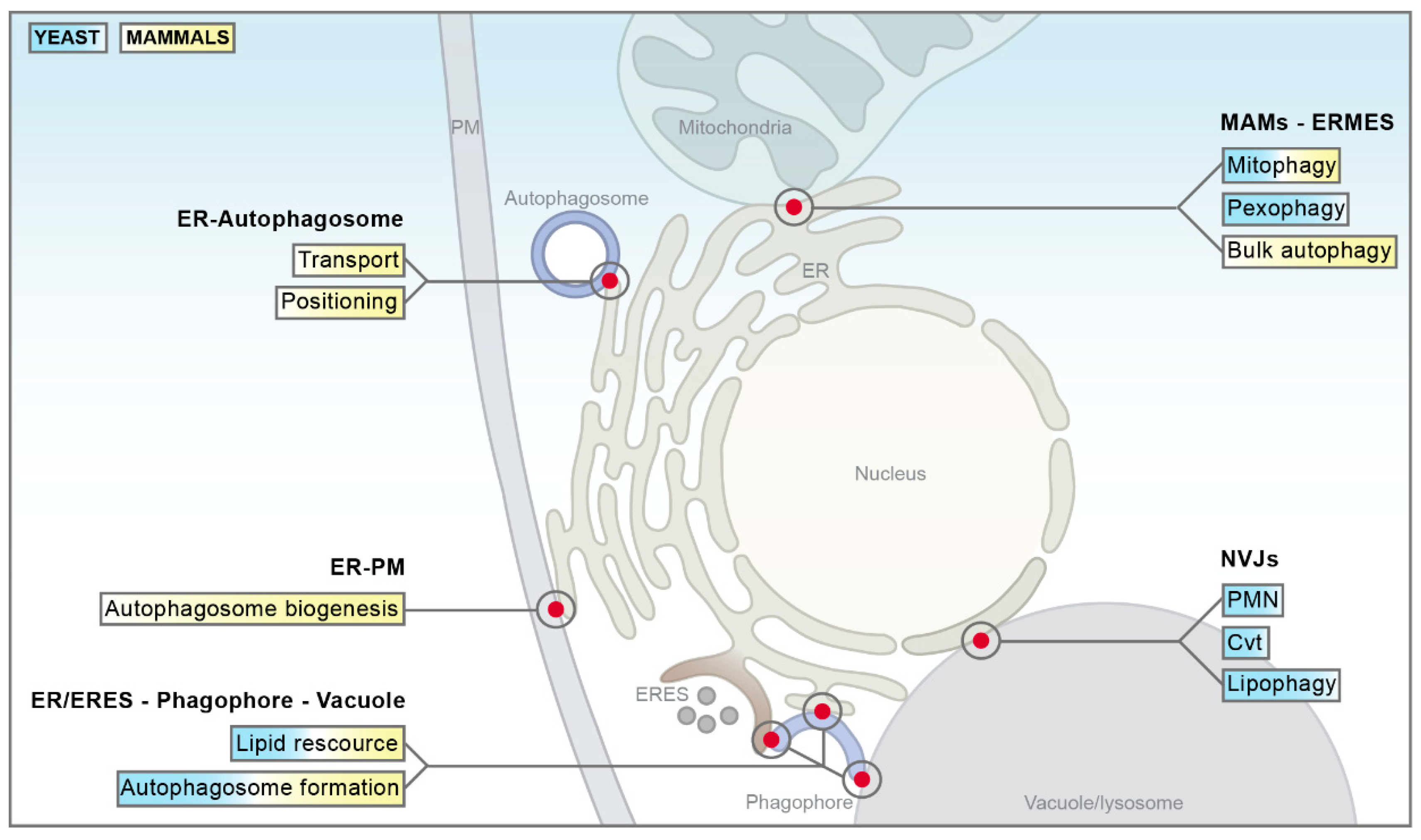
1. Introduction
2. Membrane Contact Sites at a Glimpse
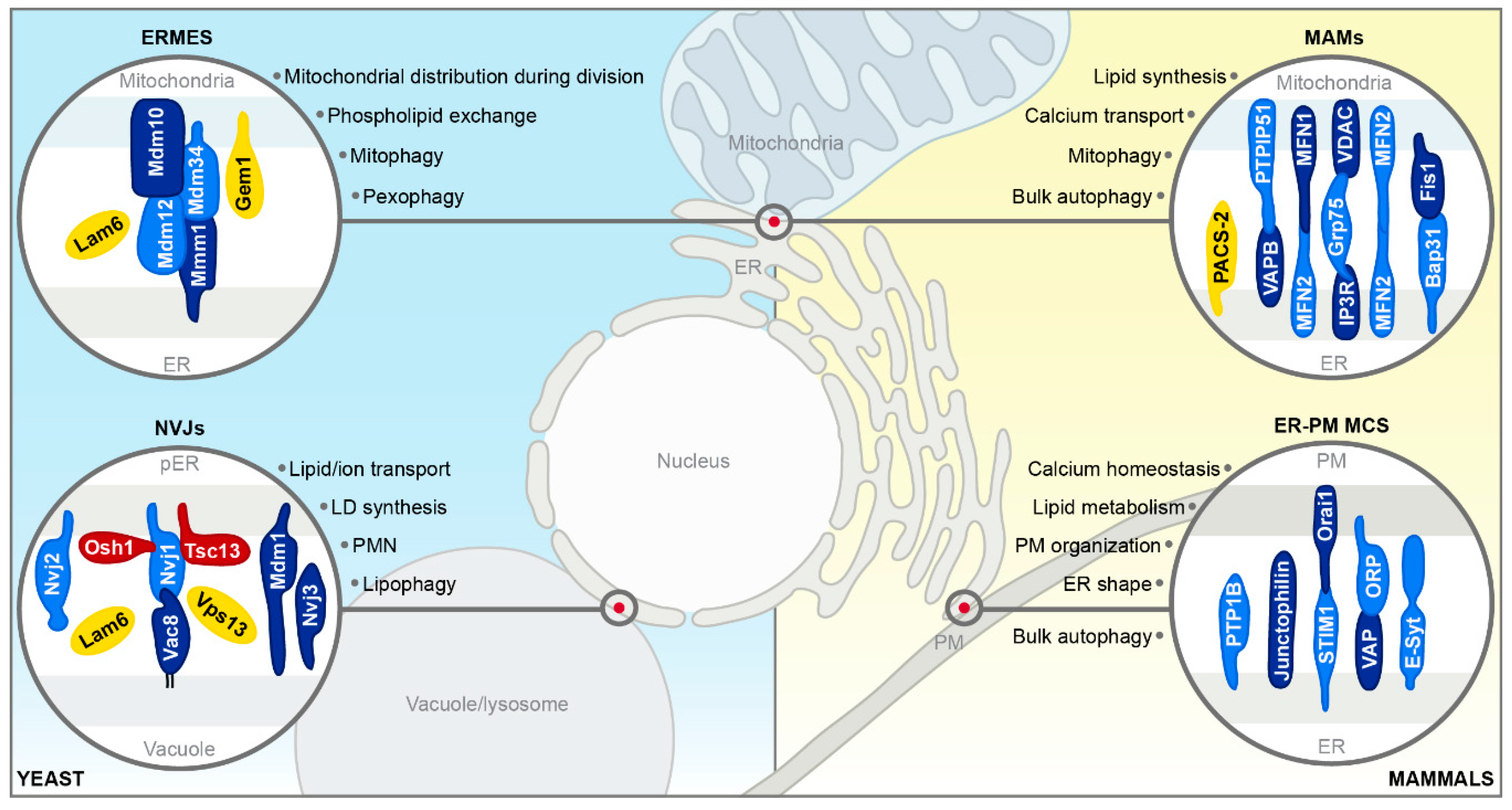
2.1. ER–Mitochondria Encounter Structure in Yeast
2.2. ER–Mitochondria Contact Sites in Mammals
2.3. ER–Plasma Membrane Contact Sites in Mammals
2.4. Nucleus–Vacuole Junctions in Yeast
3. The Nexus between Membrane Contact Sites and Autophagy
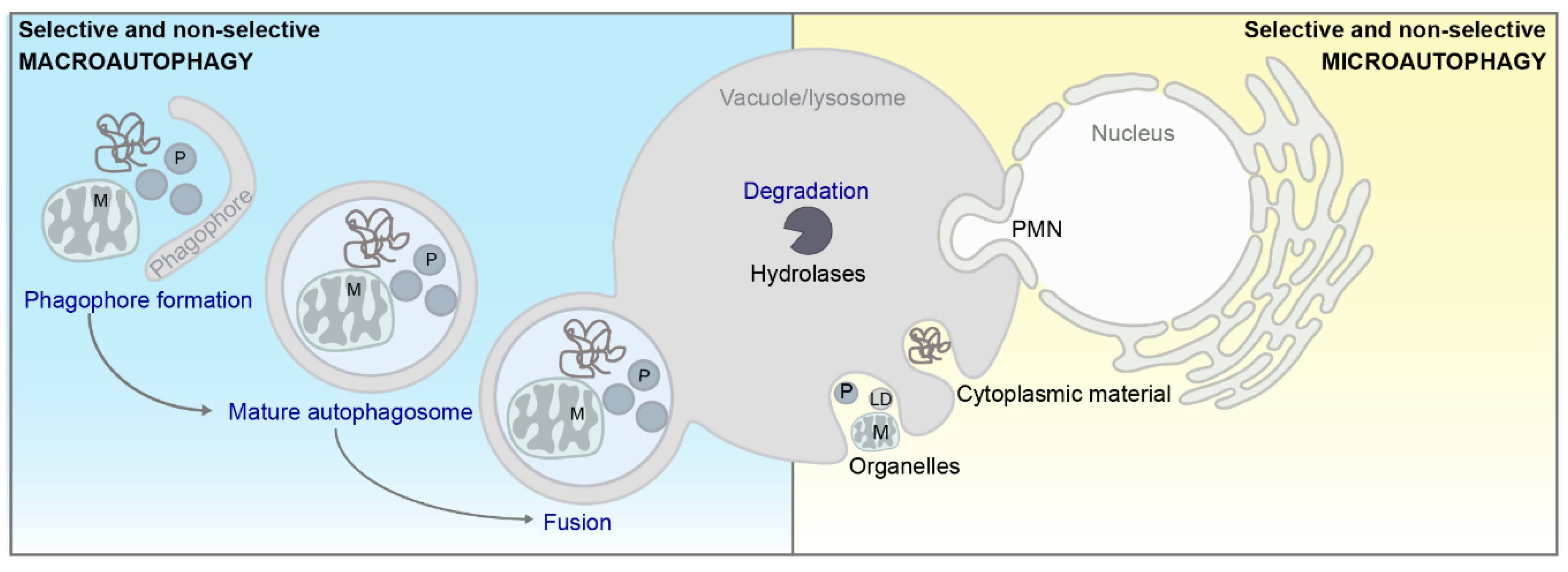
3.1. Autophagy en Passant
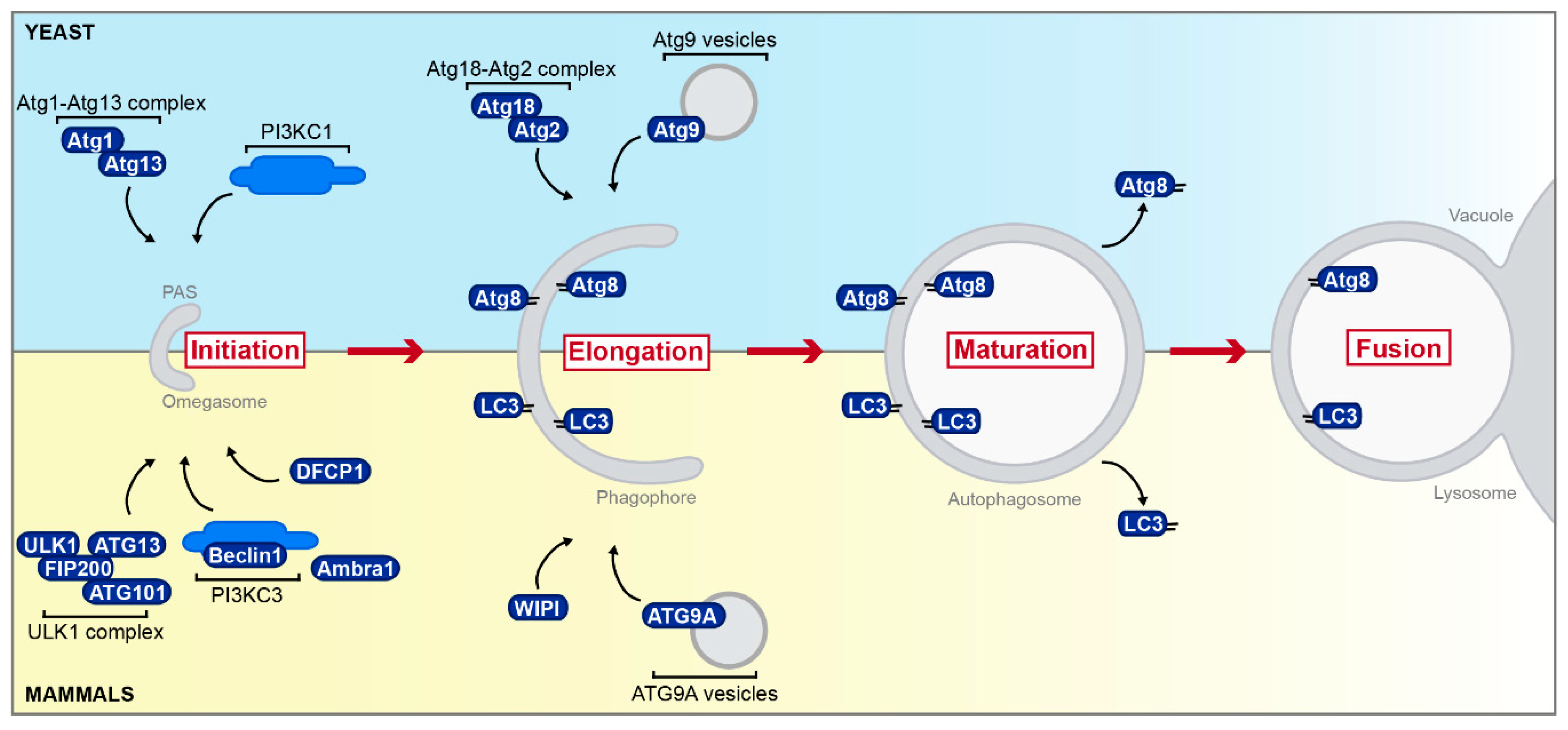
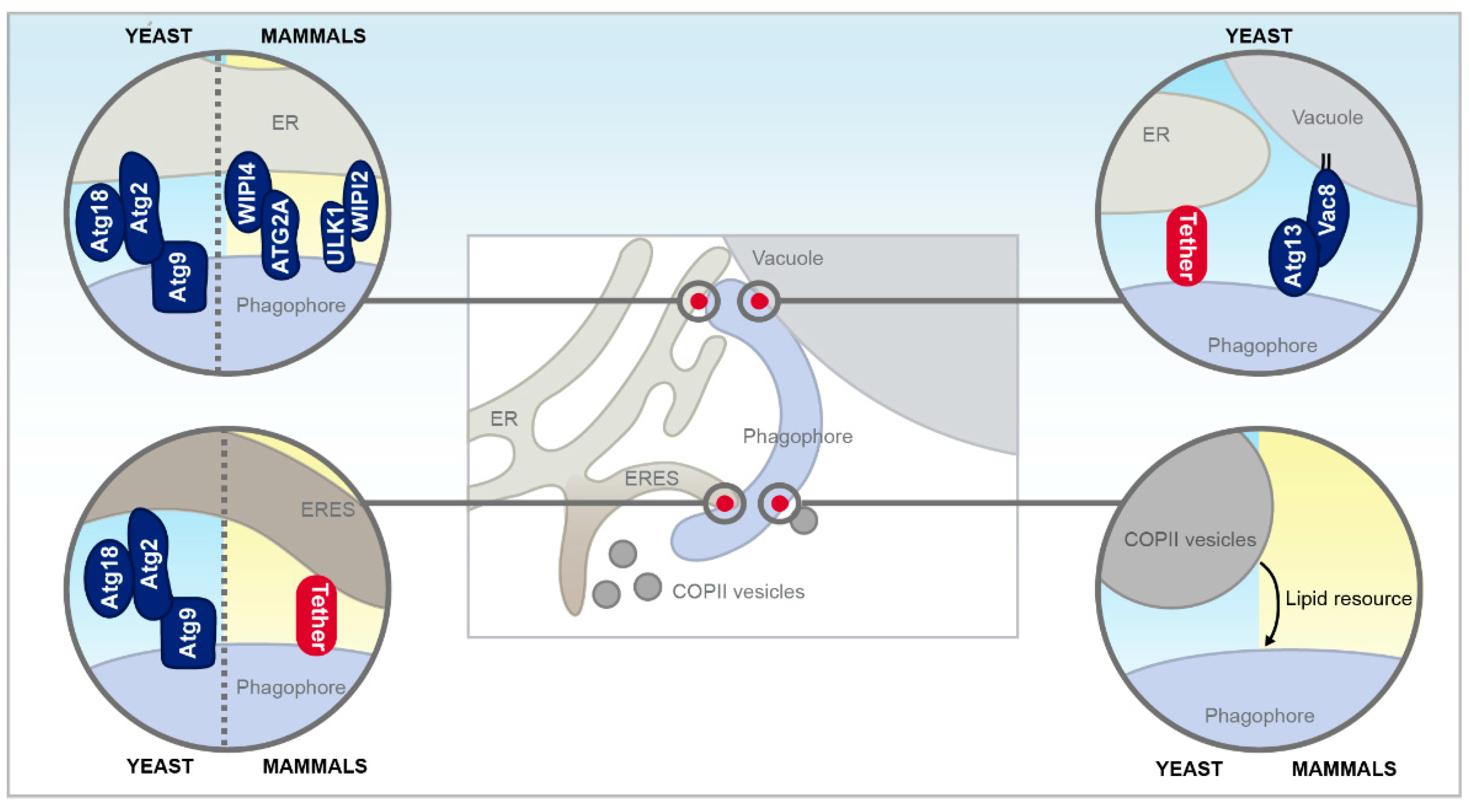
3.2. Membrane Contact Sites with the Growing Phagophore
3.3. ER–Mitochondria Contacts as Hubs for Autophagy
3.4. VMP1, the ‘Jack of All Trades’ in Mammalian Autophagy
3.5. Nucleus–Vacuole Junctions in Yeast Selective Autophagy
4. Conclusions and Outlook
Author Contributions
Funding
Conflicts of Interest
References
- Scorrano, L.; De Matteis, M.A.; Emr, S.; Giordano, F.; Hajnóczky, G.; Kornmann, B.; Lackner, L.L.; Levine, T.P.; Pellegrini, L.; Reinisch, K.; et al. Coming together to define membrane contact sites. Nat. Commun. 2019, 10, 1287. [Google Scholar] [CrossRef]
- Gottschling, D.E.; Nyström, T. The upsides and downsides of organelle interconnectivity. Cell 2017, 169, 24–34. [Google Scholar] [CrossRef]
- Cohen, S.; Valm, A.M.; Lippincott-Schwartz, J. Interacting organelles. Curr. Opin. Cell Biol. 2018, 53, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Gatta, A.T.; Levine, T.P. Piecing together the patchwork of contact sites. Trends Cell Biol. 2017, 27, 214–229. [Google Scholar] [CrossRef] [PubMed]
- Prinz, W.A. Bridging the gap: Membrane contact sites in signaling, metabolism, and organelle dynamics. J. Cell Biol. 2014, 205, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Phillips, M.J.; Voeltz, G.K. Structure and function of ER membrane contact sites with other organelles. Nat. Rev. Mol. Cell Biol. 2015, 17, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Carvalho, P.; Voeltz, G.K. Here, there, and everywhere: The importance of ER membrane contact sites. Science 2018, 361, eaan5835. [Google Scholar] [CrossRef]
- Medina, D.L.; Di Paola, S.; Peluso, I.; Armani, A.; De Stefani, D.; Venditti, R.; Montefusco, S.; Rosato, A.S.; Prezioso, C.; Forrester, A.; et al. Lysosomal calcium signaling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 2015, 17, 288–299. [Google Scholar] [CrossRef]
- Decuypere, J.-P.; Bultynck, G.; Parys, J.B. A dual role for Ca2+ in autophagy regulation. Cell Calcium 2011, 50, 242–250. [Google Scholar] [CrossRef]
- Dall’Armi, C.; Devereaux, K.A.; Di Paolo, G. The role of lipids in the control of autophagy. Curr. Biol. 2013, 23, R33–R45. [Google Scholar] [CrossRef]
- Li, D.; Song, J.-Z.; Li, H.; Shan, M.-H.; Liang, Y.; Zhu, J.; Xie, Z. Storage lipid synthesis is necessary for autophagy induced by nitrogen starvation. FEBS Lett. 2014, 589, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Reggiori, F.; Klionsky, D.J. Autophagic processes in yeast: Mechanism, machinery and regulation. Genetics 2013, 194, 341–361. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef] [PubMed]
- Uttenweiler, A.; Mayer, A. Microautophagy in the yeast saccharomyces cerevisiae. Methods Mol. Biol. 2008, 445, 245–259. [Google Scholar]
- Li, W.-W.; Li, J.; Bao, J. Microautophagy: Lesser-known self-eating. Cell. Mol. Life Sci. 2012, 69, 1125–1136. [Google Scholar] [CrossRef]
- Mijaljica, D.; Prescott, M.; Devenish, R.J. Microautophagy in mammalian cells: Revisiting a 40-year-old conundrum. Autophagy 2011, 7, 673–682. [Google Scholar] [CrossRef]
- Rubinsztein, D.C.; Mariño, G.; Kroemer, G. Autophagy and aging. Cell 2011, 146, 682–695. [Google Scholar] [CrossRef]
- Boya, P.; Reggiori, F.; Codogno, P. Emerging regulation and functions of autophagy. Nat. Cell Biol. 2013, 15, 713–720. [Google Scholar] [CrossRef]
- Bento, C.F.; Renna, M.; Ghislat, G.; Puri, C.; Ashkenazi, A.; Vicinanza, M.; Menzies, F.M.; Rubinsztein, D.C. Mammalian autophagy: How does it work? Annu. Rev. Biochem. 2016, 85, 685–713. [Google Scholar] [CrossRef]
- Hurley, J.H.; Young, L.N. Mechanisms of autophagy initiation. Annu. Rev. Biochem. 2017, 86, 225–244. [Google Scholar] [CrossRef]
- Reggiori, F.; Ungermann, C. Autophagosome maturation and fusion. J. Mol. Biol. 2017, 429, 486–496. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Klionsky, D.J. An overview of macroautophagy in yeast. J. Mol. Biol. 2016, 428, 1681–1699. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; He, D.; Yao, Z.; Klionsky, D.J. The machinery of macroautophagy. Cell Res. 2013, 24, 24–41. [Google Scholar] [CrossRef] [PubMed]
- Oku, M.; Sakai, Y. Three distinct types of microautophagy based on membrane dynamics and molecular machineries. BioEssays 2018, 40, 1800008. [Google Scholar] [CrossRef] [PubMed]
- Yorimitsu, T.; Klionsky, D.J. Autophagy: Molecular machinery for self-eating. Cell Death Differ. 2005, 12, 1542–1552. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Klionsky, D.J. Eaten alive: A history of macroautophagy. Nat. Cell Biol. 2010, 12, 814–822. [Google Scholar] [CrossRef]
- Gatica, D.; Lahiri, V.; Klionsky, D.J. Cargo recognition and degradation by selective autophagy. Nat. Cell Biol. 2018, 20, 233–242. [Google Scholar] [CrossRef]
- Kiššová, I.B.; Salin, B.; Schaeffer, J.; Bhatia, S.; Manon, S.; Camougrand, N. Selective and non-selective autophagic degradation of mitochondria in yeast. Autophagy 2007, 3, 329–336. [Google Scholar] [CrossRef]
- Mukaiyama, H.; Oku, M.; Baba, M.; Samizo, T.; Hammond, A.T.; Glick, B.S.; Kato, N.; Sakai, Y. Paz2 and 13 other PAZ gene products regulate vacuolar engulfment of peroxisomes during micropexophagy. Genes Cells 2002, 7, 75–90. [Google Scholar] [CrossRef]
- Krick, R.; Mühe, Y.; Prick, T.; Bredschneider, M.; Bremer, S.; Wenzel, D.; Eskelinen, E.-L.; Thumm, M. Piecemeal microautophagy of the nucleus: Genetic and morphological traits. Autophagy 2009, 5, 270–272. [Google Scholar] [CrossRef]
- Roberts, P.; Moshitch-Moshkovitz, S.; Kvam, E.; O’Toole, E.; Winey, M.; Goldfarb, D.S. Piecemeal microautophagy of nucleus in saccharomyces cerevisiae. Mol. Biol. Cell 2003, 14, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Mijaljica, D.; Prescott, M.; Devenish, R.J. The intricacy of nuclear membrane dynamics during nucleophagy. Nucleus 2010, 1, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Mijaljica, D.; Devenish, R.J. Nucleophagy at a glance. J. Cell Sci. 2013, 126, 4325–4330. [Google Scholar] [CrossRef]
- Wang, C.-W.; Miao, Y.-H.; Chang, Y.-S. A sterol-enriched vacuolar microdomain mediates stationary phase lipophagy in budding yeast. J. Cell Biol. 2014, 206, 357–366. [Google Scholar] [CrossRef]
- Singh, R.; Cuervo, A.M. Lipophagy: Connecting autophagy and lipid metabolism. Int. J. Cell Biol. 2012, 2012, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, J.A.; Schessner, J.P.; Bircham, P.W.; Tsuji, T.; Funaya, C.; Pajonk, O.; Schaeff, K.; Ruffini, G.; Papagiannidis, D.; Knop, M.; et al. ESCRT machinery mediates selective microautophagy of endoplasmic reticulum in yeast. EMBO J. 2019, 39, e102586. [Google Scholar] [CrossRef] [PubMed]
- Müller, O.; Sattler, T.; Flötenmeyer, M.; Schwarz, H.; Plattner, H.; Mayer, A. Autophagic tubes: Vacuolar invaginations involved in lateral membrane sorting and inverse vesicle budding. J. Cell Biol. 2000, 151, 519–528. [Google Scholar] [CrossRef]
- Ping, H.A.; Kraft, L.M.; Chen, W.; Nilles, A.E.; Lackner, L.L. Num1 anchors mitochondria to the plasma membrane via two domains with different lipid binding specificities. J. Cell Biol. 2016, 213, 513–524. [Google Scholar] [CrossRef]
- Poston, C.N.; Duong, E.; Cao, Y.; Bazemore-Walker, C.R. Proteomic analysis of lipid raft-enriched membranes isolated from internal organelles. Biochem. Biophys. Res. Commun. 2011, 415, 355–360. [Google Scholar] [CrossRef]
- Garofalo, T.; Matarrese, P.; Manganelli, V.; Marconi, M.; Tinari, A.; Gambardella, L.; Faggioni, A.; Misasi, R.; Sorice, M.; Malorni, W. Evidence for the involvement of lipid rafts localized at the ER–mitochondria associated membranes in autophagosome formation. Autophagy 2016, 12, 917–935. [Google Scholar] [CrossRef]
- Szabadkai, G.; Bianchi, K.; Várnai, P.; De Stefani, D.; Wieckowski, M.R.; Cavagna, D.; Nagy, A.I.; Balla, T.; Rizzuto, R. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J. Cell Biol. 2006, 175, 901–911. [Google Scholar] [CrossRef]
- Giordano, F.; Saheki, Y.; Idevall-Hagren, O.; Colombo, S.F.; Pirruccello-Straub, M.; Milosevic, I.; Gracheva, E.O.; Bagriantsev, S.N.; Borgese, N.; De Camilli, P. PI(4,5)P(2)-dependent and Ca(2+)-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell 2013, 153, 1494–1509. [Google Scholar] [CrossRef]
- Park, C.Y.; Hoover, P.J.; Mullins, F.M.; Bachhawat, P.; Covington, E.D.; Raunser, S.; Walz, T.; Garcia, K.C.; Dolmetsch, R.; Lewis, R.S. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to orai1. Cell 2009, 136, 876–890. [Google Scholar] [CrossRef] [PubMed]
- Henne, W.M.; Zhu, L.; Balogi, Z.; Stefan, C.; Pleiss, J.A.; Emr, S.D. Mdm1/Snx13 is a novel ER–endolysosomal interorganelle tethering protein. J. Cell Biol. 2015, 210, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Roberts, P.; Chen, Y.; Kvam, E.; Shulga, N.; Huang, K.; Lemmon, S.; Goldfarb, D.S. Nucleus–vacuole junctions in saccharomyces cerevisiae are formed through the direct interaction of vac8p with nvj1p. Mol. Biol. Cell 2000, 11, 2445–2457. [Google Scholar] [CrossRef] [PubMed]
- Vance, J.E. Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 1990, 265, 7248–7256. [Google Scholar]
- Rizzuto, R.; Pinton, P.; Carrington, W.; Fay, F.S.; Fogarty, K.E.; Lifshitz, L.M.; Tuft, R.A.; Pozzan, T. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 1998, 280, 1763–1766. [Google Scholar] [CrossRef]
- Pichler, H.; Gaigg, B.; Hrastnik, C.; Achleitner, G.; Kohlwein, S.D.; Zellnig, G.; Perktold, A.; Daum, G. A subfraction of the yeast endoplasmic reticulum associates with the plasma membrane and has a high capacity to synthesize lipids. Eur. J. Biochem. 2001, 268, 2351–2361. [Google Scholar] [CrossRef]
- Wu, M.M.; Buchanan, J.; Luik, R.M.; Lewis, R.S. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J. Cell Biol. 2006, 174, 803–813. [Google Scholar] [CrossRef]
- Peretti, D.; Dahan, N.; Shimoni, E.; Hirschberg, K.; Lev, S. Coordinated lipid transfer between the endoplasmic reticulum and the golgi complex requires the VAP proteins and is essential for golgi-mediated transport. Mol. Biol. Cell 2008, 19, 3871–3884. [Google Scholar] [CrossRef]
- Shima, T.; Kirisako, H.; Nakatogawa, H. COPII vesicles contribute to autophagosomal membranes. J. Cell Biol. 2019, 218, 1503–1510. [Google Scholar] [CrossRef] [PubMed]
- Mari, M.; Tooze, S.A.; Reggiori, F. The puzzling origin of the autophagosomal membrane. Biol. Rep. 2011, 3, 25. [Google Scholar] [CrossRef] [PubMed]
- Andrejeva, G.; Gowan, S.; Lin, G.; Fong, A.-C.L.W.T.; Shamsaei, E.; Parkes, H.G.; Mui, J.; Raynaud, F.I.; Asad, Y.; Vizcay-Barrena, G.; et al. De novo phosphatidylcholine synthesis is required for autophagosome membrane formation and maintenance during autophagy. Autophagy 2019, 1–17. [Google Scholar] [CrossRef]
- Rubinsztein, D.C.; Shpilka, T.; Elazar, Z. Mechanisms of autophagosome biogenesis. Curr. Biol. 2012, 22, R29–R34. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg-Bord, M.; Shai, N.; Schuldiner, M.; Bohnert, M. A tether is a tether is a tether: Tethering at membrane contact sites. Dev. Cell 2016, 39, 395–409. [Google Scholar] [CrossRef] [PubMed]
- Kornmann, B.; Currie, E.; Collins, S.; Schuldiner, M.; Nunnari, J.; Weissman, J.S.; Walter, P. An ER–mitochondria tethering complex revealed by a synthetic biology screen. Science 2009, 325, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Dimmer, K.S.; Rapaport, D. Mitochondrial contact sites as platforms for phospholipid exchange. Biochim. Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2017, 1862, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Stroud, D.A.; Oeljeklaus, S.; Wiese, S.; Bohnert, M.; Lewandrowski, U.; Sickmann, A.; Guiard, B.; van der Laan, M.; Warscheid, B.; Wiedemann, N. Composition and topology of the endoplasmic reticulum–mitochondria encounter structure. J. Mol. Biol. 2011, 413, 743–750. [Google Scholar] [CrossRef]
- Murley, A.; Lackner, L.L.; Osman, C.; West, M.; Voeltz, G.K.; Walter, P.; Nunnari, J. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. eLife 2013, 2. [Google Scholar] [CrossRef]
- Böckler, S.; Westermann, B. Mitochondrial ER contacts are crucial for mitophagy in yeast. Dev. Cell 2014, 28, 450–458. [Google Scholar] [CrossRef]
- Kornmann, B.; Osman, C.; Walter, P. The conserved GTPase Gem1 regulates endoplasmic reticulum-mitochondria connections. Proc. Natl. Acad. Sci. USA 2011, 108, 14151–14156. [Google Scholar] [CrossRef] [PubMed]
- Elbaz-Alon, Y.; Eisenberg-Bord, M.; Shinder, V.; Stiller, S.B.; Shimoni, E.; Wiedemann, N.; Geiger, T.; Schuldiner, M. Lam6 regulates the extent of contacts between organelles. Cell Rep. 2015, 12, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Elbaz-Alon, Y.; Rosenfeld-Gur, E.; Shinder, V.; Futerman, A.H.; Geiger, T.; Schuldiner, M. A dynamic interface between vacuoles and mitochondria in yeast. Dev. Cell 2014, 30, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Hönscher, C.; Mari, M.; Auffarth, K.; Bohnert, M.; Griffith, J.; Geerts, W.J.C.; van der Laan, M.; Cabrera, M.; Reggiori, F.; Ungermann, C. Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev. Cell 2014, 30, 86–94. [Google Scholar] [CrossRef]
- Wideman, J.G. The ubiquitous and ancient ER membrane protein complex (EMC): Tether or not? F1000Research 2015, 4, 624. [Google Scholar] [CrossRef]
- Lahiri, S.; Chao, J.T.; Tavassoli, S.; Wong, A.K.O.; Choudhary, V.; Young, B.P.; Loewen, C.; Prinz, W.A. A conserved endoplasmic reticulum membrane protein complex (EMC) facilitates phospholipid transfer from the ER to mitochondria. PLoS Biol. 2014, 12, e1001969. [Google Scholar] [CrossRef]
- Herrera-Cruz, M.S.; Simmen, T. Of yeast, mice and men: MAMs come in two flavors. Biol. Direct 2017, 12, 3. [Google Scholar] [CrossRef]
- Giacomello, M.; Pellegrini, L. The coming of age of the mitochondria–ER contact: A matter of thickness. Cell Death Differ. 2016, 23, 1417–1427. [Google Scholar] [CrossRef]
- Wang, H.-J.; Guay, G.; Pogan, L.; Sauvé, R.; Nabi, I.R. Calcium regulates the association between mitochondria and a smooth subdomain of the endoplasmic reticulum. J. Cell Biol. 2000, 150, 1489–1498. [Google Scholar] [CrossRef]
- Vance, J.E. Newly made phosphatidylserine and phosphatidylethanolamine are preferentially translocated between rat liver mitochondria and endoplasmic reticulum. J. Biol. Chem. 1991, 266, 89–97. [Google Scholar]
- Csordás, G.; Várnai, P.; Golenár, T.; Roy, S.; Purkins, G.; Balla, T.; Hajnóczky, G. Imaging interorganelle contacts and local calcium dynamics at the ER–mitochondrial interface. Biophys. J. 2010, 98, 381a. [Google Scholar] [CrossRef]
- Hayashi, T.; Fujimoto, M. Detergent-resistant microdomains determine the localization of σ-1 receptors to the endoplasmic reticulum-mitochondria junction. Mol. Pharmacol. 2010, 77, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, M.; Hayashi, T.; Su, T.-P. The role of cholesterol in the association of endoplasmic reticulum membranes with mitochondria. Biochem. Biophys. Res. Commun. 2012, 417, 635–639. [Google Scholar] [CrossRef] [PubMed]
- Janikiewicz, J.; Szymański, J.; Malińska, D.; Patalas-Krawczyk, P.; Michalska, B.; Duszynski, J.; Giorgi, C.; Bonora, M.; Dobrzyń, A.; Wieckowski, M.R. Mitochondria-associated membranes in aging and senescence: Structure, function, and dynamics. Cell Death Dis. 2018, 9, 332. [Google Scholar] [CrossRef]
- Monteiro, J.P.; Oliveira, P.J.; Jurado, A.S. Mitochondrial membrane lipid remodeling in pathophysiology: A new target for diet and therapeutic interventions. Prog. Lipid Res. 2013, 52, 513–528. [Google Scholar] [CrossRef]
- De Vos, K.; Mórotz, G.M.; Stoica, R.; Tudor, E.L.; Lau, K.-F.; Ackerley, S.; Warley, A.; Shaw, P.J.; Miller, C.C.J. VAPB interacts with the mitochondrial protein PTPIP51 to regulate calcium homeostasis. Hum. Mol. Genet. 2012, 21, 1299–1311. [Google Scholar] [CrossRef]
- Iwasawa, R.; Mahul-Mellier, A.-L.; Datler, C.; Pazarentzos, E.; Grimm, S. Fis1 and Bap31 bridge the mitochondria-ER interface to establish a platform for apoptosis induction. EMBO J. 2010, 30, 556–568. [Google Scholar] [CrossRef]
- Simmen, T.; Aslan, J.E.; Blagoveshchenskaya, A.D.; Thomas, L.; Wan, L.; Xiang, Y.; Feliciangeli, S.; Hung, C.-H.; Crump, C.; Thomas, G. PACS-2 controls endoplasmic reticulum–mitochondria communication and Bid-mediated apoptosis. EMBO J. 2005, 24, 717–729. [Google Scholar] [CrossRef]
- De Brito, O.M.; Scorrano, L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 2008, 456, 605–610. [Google Scholar] [CrossRef]
- Vance, J.E. MAM (mitochondria-associated membranes) in mammalian cells: Lipids and beyond. Biochim. Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2014, 1841, 595–609. [Google Scholar] [CrossRef]
- Aufschnaiter, A.; Kohler, V.; Diessl, J.; Peselj, C.; Carmona-Gutierrez, D.; Keller, W.; Büttner, S. Mitochondrial lipids in neurodegeneration. Cell Tissue Res. 2017, 367, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Williamson, C.D.; Wong, D.S.; Bullough, M.D.; Brown, K.; Hathout, Y.; Colberg-Poley, A.M. Quantitative proteomic analyses of human cytomegalovirus-induced restructuring of endoplasmic reticulum-mitochondrial contacts at late times of infection. Mol. Cell. Proteom. 2011, 10, M111 009936. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Rizzuto, R.; Hajnoczky, G.; Su, T.-P. MAM: More than just a housekeeper. Trends Cell Biol. 2009, 19, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Lynes, E.M.; Bui, M.; Yap, M.C.; Benson, M.D.; Schneider, B.; Ellgaard, L.; Berthiaume, L.; Simmen, T. Palmitoylated TMX and calnexin target to the mitochondria-associated membrane. EMBO J. 2011, 31, 457–470. [Google Scholar] [CrossRef] [PubMed]
- Area-Gomez, E.; Schon, E.A. Mitochondria-associated ER membranes and Alzheimer disease. Curr. Opin. Genet. Dev. 2016, 38, 90–96. [Google Scholar] [CrossRef]
- Arribas, M.R.; Yakhine-Diop, S.M.S.; Pedro, J.M.B.-S.; Gomez-Suaga, P.; Gómez-Sánchez, R.; Chacón, G.M.; Fuentes, J.M.; Polo, R.-A.G.; Niso-Santano, M. Mitochondria-Associated Membranes (MAMs): Overview and its role in parkinson’s disease. Mol. Neurobiol. 2017, 54, 6287–6303. [Google Scholar] [CrossRef]
- Arruda, A.P.; Pers, B.M.; Parlakgul, G.; Guney, E.; Inouye, K.; Hotamisligil, G.S. Chronic enrichment of hepatic endoplasmic reticulum–mitochondria contact leads to mitochondrial dysfunction in obesity. Nat. Med. 2014, 20, 1427–1435. [Google Scholar] [CrossRef]
- West, M.; Zurek, N.; Hoenger, A.; Voeltz, G.K. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J. Cell Biol. 2011, 193, 333–346. [Google Scholar] [CrossRef]
- Fernández-Busnadiego, R.; Saheki, Y.; De Camilli, P. Three-dimensional architecture of extended synaptotagmin-mediated endoplasmic reticulum–plasma membrane contact sites. Proc. Natl. Acad. Sci. USA 2015, 112, E2004–E2013. [Google Scholar] [CrossRef]
- Stefan, C.J.; Manford, A.; Emr, S.D. ER-PM connections: Sites of information transfer and inter-organelle communication. Curr. Opin. Cell Biol. 2013, 25, 434–442. [Google Scholar] [CrossRef]
- Burgoyne, T.; Patel, S.; Eden, E.R.; Burgoyne, T. Calcium signaling at ER membrane contact sites. Biochim. Biophys. Acta (BBA)—Mol. Cell Res. 2015, 1853, 2012–2017. [Google Scholar] [CrossRef] [PubMed]
- Saheki, Y.; Bian, X.; Schauder, C.M.; Sawaki, Y.; Surma, M.A.; Klose, C.; Pincet, F.; Reinisch, K.M.; De Camilli, P. Control of plasma membrane lipid homeostasis by the extended synaptotagmins. Nat. Cell Biol. 2016, 18, 504–515. [Google Scholar] [CrossRef] [PubMed]
- Murphy, S.E.; Levine, T.P. VAP, a versatile access point for the endoplasmic reticulum: Review and analysis of FFAT-like motifs in the VAPome. Biochim. Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2016, 1861, 952–961. [Google Scholar] [CrossRef] [PubMed]
- Venditti, R.; Rega, L.R.; Masone, M.C.; Santoro, M.; Polishchuk, E.; Sarnataro, D.; Paladino, S.; D’Auria, S.; Varriale, A.; Olkkonen, V.M.; et al. Molecular determinants of ER–Golgi contacts identified through a new FRET–FLIM system. J. Cell Biol. 2019, 218, 1055–1065. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.; Torta, F.; Masai, K.; Lucast, L.; Czapla, H.; Tanner, L.B.; Narayanaswamy, P.; Wenk, M.R.; Nakatsu, F.; De Camilli, P. PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER-plasma membrane contacts. Science 2015, 349, 428–432. [Google Scholar] [CrossRef]
- Anderie, I.; Schulz, I.; Schmid, A. Direct interaction between ER membrane-bound PTP1B and its plasma membrane-anchored targets. Cell. Signal. 2007, 19, 582–592. [Google Scholar] [CrossRef]
- Takeshima, H.; Hoshijima, M.; Song, L.-S. Ca²⁺ microdomains organized by junctophilins. Cell Calcium 2015, 58, 349–356. [Google Scholar] [CrossRef]
- Manford, A.; Stefan, C.J.; Yuan, H.L.; MacGurn, J.A.; Emr, S.D. ER-to-plasma membrane tethering proteins regulate cell signaling and er morphology. Dev. Cell 2012, 23, 1129–1140. [Google Scholar] [CrossRef]
- Schrader, M.; Godinho, L.F.; Costello, J.; Islinger, M. The different facets of organelle interplay—An overview of organelle interactions. Front. Cell Dev. Biol. 2015, 3, 9113. [Google Scholar] [CrossRef] [PubMed]
- Tavassoli, S.; Chao, J.T.; Young, B.P.; Cox, R.C.; Prinz, W.A.; De Kroon, A.I.P.M.; Loewen, C.J.R. Plasma membrane—Endoplasmic reticulum contact sites regulate phosphatidylcholine synthesis. EMBO Rep. 2013, 14, 434–440. [Google Scholar] [CrossRef]
- Wolf, W.; Kilic, A.; Schrul, B.; Lorenz, H.; Schwappach, B.; Seedorf, M. Yeast Ist2 recruits the endoplasmic reticulum to the plasma membrane and creates a ribosome-free membrane microcompartment. PLoS ONE 2012, 7, 39703. [Google Scholar] [CrossRef] [PubMed]
- Henne, W.M.; Hariri, H. Endoplasmic reticulum-vacuole contact sites “Bloom” with stress-induced lipid droplets. Contact 2018, 1, 251525641875611. [Google Scholar] [CrossRef] [PubMed]
- Krick, R.; Muehe, Y.; Prick, T.; Bremer, S.; Schlotterhose, P.; Eskelinen, E.-L.; Millen, J.; Goldfarb, D.S.; Thumm, M. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol. Biol. Cell 2008, 19, 4492–4505. [Google Scholar] [CrossRef] [PubMed]
- Dawaliby, R.; Mayer, A. Microautophagy of the nucleus coincides with a vacuolar diffusion barrier at nuclear–vacuolar junctions. Mol. Biol. Cell 2010, 21, 4173–4183. [Google Scholar] [CrossRef]
- Hariri, H.; Rogers, S.; Ugrankar, R.; Liu, Y.L.; Feathers, J.R.; Henne, W.M. Lipid droplet biogenesis is spatially coordinated at ER–vacuole contacts under nutritional stress. EMBO Rep. 2018, 19, 57–72. [Google Scholar] [CrossRef]
- Toulmay, A.; Prinz, W.A. A conserved membrane-binding domain targets proteins to organelle contact sites. J. Cell Sci. 2012, 125, 49–58. [Google Scholar] [CrossRef]
- Kohlwein, S.D.; Eder, S.; Oh, C.-S.; Martin, C.E.; Gable, K.; Bacikova, D.; Dunn, T.M. Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear–vacuolar interface insaccharomyces cerevisiae. Mol. Cell. Biol. 2001, 21, 109–125. [Google Scholar] [CrossRef]
- Levine, T.P.; Munro, S. Dual targeting of Osh1p, a yeast homologue of oxysterol-binding protein, to both the golgi and the nucleus–vacuole junction. Mol. Biol. Cell 2001, 12, 1633–1644. [Google Scholar] [CrossRef]
- Kvam, E.; Goldfarb, D.S. Nvj1p is the outer-nuclear-membrane receptor for oxysterol-binding protein homolog Osh1p in Saccharomyces cerevisiae. J. Cell Sci. 2004, 117, 4959–4968. [Google Scholar] [CrossRef]
- Kvam, E.; Gable, K.; Dunn, T.M.; Goldfarb, D.S. Targeting of Tsc13p to nucleus–vacuole junctions: A role for very-long-chain fatty acids in the biogenesis of microautophagic vesicles. Mol. Biol. Cell 2005, 16, 3987–3998. [Google Scholar] [CrossRef]
- Murley, A.; Sarsam, R.D.; Toulmay, A.; Yamada, J.; Prinz, W.A.; Nunnari, J. Ltc1 is an ER-localized sterol transporter and a component of ER–mitochondria and ER–vacuole contacts. J. Cell Biol. 2015, 209, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Lang, A.B.; Peter, A.T.J.; Walter, P.; Kornmann, B. ER–mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. J. Cell Biol. 2015, 210, 883–890. [Google Scholar] [CrossRef] [PubMed]
- Raiborg, C.; Wenzel, E.M.; Stenmark, H. ER–endosome contact sites: Molecular compositions and functions. EMBO J. 2015, 34, 1848–1858. [Google Scholar] [CrossRef] [PubMed]
- Eden, E.R. The formation and function of ER-endosome membrane contact sites. Biochim. Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2016, 1861, 874–879. [Google Scholar] [CrossRef]
- Suzuki, K.; Kirisako, T.; Kamada, Y.; Mizushima, N.; Noda, T.; Ohsumi, Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001, 20, 5971–5981. [Google Scholar] [CrossRef]
- Axe, E.L.; Walker, S.; Manifava, M.; Chandra, P.; Roderick, H.L.; Habermann, A.; Griffiths, G.; Ktistakis, N.T. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008, 182, 685–701. [Google Scholar] [CrossRef]
- Chen, Y.; Klionsky, D.J. The regulation of autophagy—Unanswered questions. J. Cell Sci. 2010, 124, 161–170. [Google Scholar] [CrossRef]
- Itakura, E.; Mizushima, N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 2010, 6, 764–776. [Google Scholar] [CrossRef]
- Nascimbeni, A.C.; Codogno, P.; Morel, E. Phosphatidylinositol-3-phosphate in the regulation of autophagy membrane dynamics. FEBS J. 2017, 284, 1267–1278. [Google Scholar] [CrossRef] [PubMed]
- Noda, T.; Kim, J.; Huang, W.-P.; Baba, M.; Tokunaga, C.; Ohsumi, Y.; Klionsky, D.J. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the cvt and autophagy pathways. J. Cell Biol. 2000, 148, 465–480. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Kakuta, S.; Watanabe, T.M.; Kitamura, A.; Sekito, T.; Kondo-Kakuta, C.; Ichikawa, R.; Kinjo, M.; Ohsumi, Y. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J. Cell Biol. 2012, 198, 219–233. [Google Scholar] [CrossRef]
- Orsi, A.; Razi, M.; Dooley, H.C.; Robinson, D.; Weston, A.; Collinson, L.M.; Tooze, S.A. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol. Biol. Cell 2012, 23, 1860–1873. [Google Scholar] [CrossRef]
- Proikas-Cezanne, T.; Takacs, Z.; Kohlbacher, O.; Dönnes, P. WIPI proteins: Essential PtdIns3P effectors at the nascent autophagosome. J. Cell Sci. 2015, 128, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Hou, Y.; Wang, J.; Chen, X.; Shao, Z.-M.; Yin, X.-M. Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 substrates. J. Biol. Chem. 2011, 286, 7327–7338. [Google Scholar] [CrossRef] [PubMed]
- Weidberg, H.; Shvets, E.; Shpilka, T.; Shimron, F.; Shinder, V.; Elazar, Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010, 29, 1792–1802. [Google Scholar] [CrossRef] [PubMed]
- Martens, S. No ATG8s, no problem? How LC3/GAB ARAP proteins contribute to autophagy. J. Cell Biol. 2016, 215, 761–763. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kirisako, T.; Ichimura, Y.; Okada, H.; Kabeya, Y.; Mizushima, N.; Yoshimori, T.; Ohsumi, M.; Takao, T.; Noda, T.; Ohsumi, Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 2000, 151, 263–275. [Google Scholar] [CrossRef]
- Ichimura, Y.; Kirisako, T.; Takao, T.; Satomi, Y.; Shimonishi, Y.; Ishihara, N.; Mizushima, N.; Tanida, I.; Kominami, E.; Ohsumi, M.; et al. A ubiquitin-like system mediates protein lipidation. Nature 2000, 408, 488–492. [Google Scholar] [CrossRef]
- Kabeya, Y.; Mizushima, N.; Yamamoto, A.; Oshitani-Okamoto, S.; Ohsumi, Y.; Yoshimori, T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 2004, 117, 2805–2812. [Google Scholar] [CrossRef]
- Puri, C.; Renna, M.; Bento, C.F.; Moreau, K.; Rubinsztein, D.C. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell 2013, 154, 1285–1299. [Google Scholar] [CrossRef]
- Fujita, N.; Itoh, T.; Omori, H.; Fukuda, M.; Noda, T.; Yoshimori, T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell 2008, 19, 2092–2100. [Google Scholar] [CrossRef]
- Satoo, K.; Noda, N.N.; Kumeta, H.; Fujioka, Y.; Mizushima, N.; Ohsumi, Y.; Inagaki, F. The structure of Atg4B–LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 2009, 28, 1341–1350. [Google Scholar] [CrossRef] [PubMed]
- Cebollero, E.; van der Vaart, A.; Zhao, M.; Rieter, E.; Klionsky, D.J.; Helms, J.B.; Reggiori, F. Phosphatidylinositol-3-phosphate clearance plays a key role in autophagosome completion. Curr. Biol. 2012, 22, 1545–1553. [Google Scholar] [CrossRef] [PubMed]
- Noda, T.; Fujita, N.; Yoshimori, T. The late stages of autophagy: How does the end begin? Cell Death Differ. 2009, 16, 984–990. [Google Scholar] [CrossRef] [PubMed]
- Epple, U.D.; Suriapranata, I.; Eskelinen, E.-L.; Thumm, M. Aut5/Cvt17p, a putative lipase essential for disintegration of autophagic bodies inside the vacuole. J. Bacteriol. 2001, 183, 5942–5955. [Google Scholar] [CrossRef] [PubMed]
- Razi, M.; Chan, E.Y.; Tooze, S.A. Early endosomes and endosomal coatomer are required for autophagy. J. Cell Biol. 2009, 185, 305–321. [Google Scholar] [CrossRef] [PubMed]
- Aufschnaiter, A.; Büttner, S. The vacuolar shapes of ageing: From function to morphology. Biochim. Biophys. Acta (BBA)—Mol. Cell Res. 2019, 1866, 957–970. [Google Scholar] [CrossRef]
- Jahreiss, L.; Menzies, F.M.; Rubinsztein, D.C. The itinerary of autophagosomes: From peripheral formation to kiss-and-run fusion with lysosomes. Traffic 2008, 9, 574–587. [Google Scholar] [CrossRef]
- Jeong, Y.; Simoneschi, D.; Keegan, S.; Melville, D.; Adler, N.S.; Saraf, A.; Florens, L.; Washburn, M.P.; Cavasotto, C.N.; Fenyö, D.; et al. The ULK1-FBXW5-SEC23B nexus controls autophagy. eLife 2018, 7, 7. [Google Scholar] [CrossRef]
- De Tito, S.; Hervás, J.H.; van Vliet, A.R.; Tooze, S.A. The golgi as an assembly line to the autophagosome. Trends Biochem. Sci. 2020. [Google Scholar] [CrossRef]
- Ge, L.; Melville, D.; Zhang, M.; Schekman, R. The ER–Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. eLife 2013, 2, 2. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.; Zhang, M.; Kenny, S.J.; Liu, D.; Maeda, M.; Saito, K.; Mathur, A.; Xu, K.; Schekman, R. Remodeling of ER-exit sites initiates a membrane supply pathway for autophagosome biogenesis. EMBO Rep. 2017, 18, 1586–1603. [Google Scholar] [CrossRef] [PubMed]
- Ravikumar, B.; Moreau, K.; Jahreiss, L.; Puri, C.; Rubinsztein, D.C. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 2010, 12, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Sánchez, R.; Rose, J.; Guimarães, R.; Mari, M.; Papinski, D.; Rieter, E.; Geerts, W.J.C.; Hardenberg, R.; Kraft, C.; Ungermann, C.; et al. Atg9 establishes Atg2-dependent contact sites between the endoplasmic reticulum and phagophores. J. Cell Biol. 2018, 217, 2743–2763. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.-X.; Li, Y.; Ding, Y.-H.; Liu, J.-J.; Zhang, M.-J.; Dong, M.-Q.; Wang, H.-W.; Yu, L. Architecture of the ATG2B-WDR45 complex and an aromatic Y/HF motif crucial for complex formation. Autophagy 2017, 13, 1870–1883. [Google Scholar] [CrossRef] [PubMed]
- Graef, M. Membrane tethering by the autophagy ATG2A-WIPI4 complex. Proc. Natl. Acad. Sci. USA 2018, 115, 10540–10541. [Google Scholar] [CrossRef]
- Chowdhury, S.; Otomo, C.; Leitner, A.; Ohashi, K.; Aebersold, R.; Lander, G.C.; Otomo, T. Insights into autophagosome biogenesis from structural and biochemical analyses of the ATG2A-WIPI4 complex. Proc. Natl. Acad. Sci. USA 2018, 115, E9792–E9801. [Google Scholar] [CrossRef]
- Zhao, Y.G.; Chen, Y.; Miao, G.; Zhao, H.; Qu, W.; Li, D.; Wang, Z.; Liu, N.; Li, L.; Chen, S.; et al. The ER-localized transmembrane protein EPG-3/VMP1 regulates SERCA activity to control ER-isolation membrane contacts for autophagosome formation. Mol. Cell 2017, 67, 974–989. [Google Scholar] [CrossRef]
- Suzuki, K.; Akioka, M.; Kondo-Kakuta, C.; Yamamoto, H.; Ohsumi, Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J. Cell Sci. 2013, 126, 2534–2544. [Google Scholar] [CrossRef] [PubMed]
- Graef, M.; Friedman, J.; Graham, C.; Babu, M.; Nunnari, J. ER exit sites are physical and functional core autophagosome biogenesis components. Mol. Biol. Cell 2013, 24, 2918–2931. [Google Scholar] [CrossRef]
- Fujioka, Y.; Alam, J.M.; Noshiro, D.; Mouri, K.; Ando, T.; Okada, Y.; May, A.I.; Knorr, R.L.; Suzuki, K.; Ohsumi, Y.; et al. Phase separation organizes the site of autophagosome formation. Nature 2020, 578, 301–305. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, Y.; Hu, J.; Xiao, J.; Qu, L.; Wang, Z.; Ma, D.; Chen, Y. A novel ER-localized transmembrane protein, EMC6, interacts with RAB5A and regulates cell autophagy. Autophagy 2013, 9, 150–163. [Google Scholar] [CrossRef]
- Nascimbeni, A.C.; Giordano, F.; Dupont, N.; Grasso, D.; Vaccaro, M.; Codogno, P.; Morel, E. ER–plasma membrane contact sites contribute to autophagosome biogenesis by regulation of local PI 3P synthesis. EMBO J. 2017, 36, 2018–2033. [Google Scholar] [CrossRef] [PubMed]
- Tábara, L.-C.; Escalante, R. VMP1 establishes ER-microdomains that regulate membrane contact sites and autophagy. PLoS ONE 2016, 11, e0166499. [Google Scholar] [CrossRef] [PubMed]
- Hamasaki, M.; Furuta, N.; Matsuda, A.; Nezu, A.; Yamamoto, A.; Fujita, N.; Oomori, H.; Noda, T.; Haraguchi, T.; Hiraoka, Y.; et al. Autophagosomes form at ER–mitochondria contact sites. Nature 2013, 495, 389–393. [Google Scholar] [CrossRef] [PubMed]
- Wijdeven, R.H.; Janssen, H.; Nahidiazar, L.; Janssen, L.; Jalink, K.; Berlin, I.; Neefjes, J. Cholesterol and ORP1L-mediated ER contact sites control autophagosome transport and fusion with the endocytic pathway. Nat. Commun. 2016, 7, 11808. [Google Scholar] [CrossRef]
- Böckler, S.; Westermann, B. ER–mitochondria contacts as sites of mitophagosome formation. Autophagy 2014, 10, 1346–1347. [Google Scholar] [CrossRef]
- Belgareh-Touzé, N.; Cavellini, L.; Cohen, M.M. Ubiquitination of ERMES components by the E3 ligase Rsp5 is involved in mitophagy. Autophagy 2017, 13, 114–132. [Google Scholar] [CrossRef]
- Kanki, T.; Furukawa, K.; Yamashita, S.-I. Mitophagy in yeast: Molecular mechanisms and physiological role. Biochim. Biophys. Acta (BBA)—Mol. Cell Res. 2015, 1853, 2756–2765. [Google Scholar] [CrossRef]
- Kanki, T.; Klionsky, D.J. Mitophagy in yeast occurs through a selective mechanism. J. Biol. Chem. 2008, 283, 32386–32393. [Google Scholar] [CrossRef]
- Hamacher-Brady, A.; Brady, N.R. Mitophagy programs: Mechanisms and physiological implications of mitochondrial targeting by autophagy. Cell. Mol. Life Sci. 2016, 73, 775–795. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wen, X.; Klionsky, D.J. Endoplasmic reticulum–mitochondria contacts are required for pexophagy in saccharomyces cerevisiae. Contact 2019, 2, 251525641882158. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.; Liu, X.; Feng, Y.; Klionsky, D.J. The progression of peroxisomal degradation through autophagy requires peroxisomal division. Autophagy 2014, 10, 652–661. [Google Scholar] [CrossRef] [PubMed]
- Motley, A.M.; Nuttall, J.M.; Hettema, E.H. Pex3-anchored Atg36 tags peroxisomes for degradation inSaccharomyces cerevisiae. EMBO J. 2012, 31, 2852–2868. [Google Scholar] [CrossRef] [PubMed]
- Kondapalli, C.; Kazlauskaite, A.; Zhang, N.; Woodroof, H.I.; Campbell, D.G.; Gourlay, R.; Burchell, L.; Walden, H.; Macartney, T.J.; Deák, M.; et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012, 2, 120080. [Google Scholar] [CrossRef] [PubMed]
- Aerts, L.; Craessaerts, K.; De Strooper, B.; Morais, V.A. PINK1 kinase catalytic activity is regulated by phosphorylation on serines 228 and 402. J. Biol. Chem. 2015, 290, 2798–2811. [Google Scholar] [CrossRef]
- Trempe, J.-F.; Sauvé, V.; Grenier, K.; Seirafi, M.; Tang, M.Y.; Ménade, M.; Al-Abdul-Wahid, S.; Krett, J.; Wong, K.; Kozlov, G.; et al. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science 2013, 340, 1451–1455. [Google Scholar] [CrossRef]
- Geisler, S.; Holmström, K.M.; Skujat, D.; Fiesel, F.C.; Rothfuss, O.C.; Kahle, P.J.; Springer, W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010, 12, 119–131. [Google Scholar] [CrossRef]
- Gelmetti, V.; De Rosa, P.; Torosantucci, L.; Marini, E.S.; Romagnoli, A.; Di Rienzo, M.; Arena, G.; Vignone, M.; Fimia, G.M.; Valente, E.M. PINK1 and BECN1 relocalize at mitochondria-associated membranes during mitophagy and promote ER–mitochondria tethering and autophagosome formation. Autophagy 2017, 13, 654–669. [Google Scholar] [CrossRef]
- Moulis, M.; Grousset, E.; Faccini, J.; Richetin, K.; Thomas, G.; Vindis, C. The multifunctional sorting protein PACS-2 controls mitophagosome formation in human vascular smooth muscle cells through mitochondria-ER contact sites. Cells 2019, 8, 638. [Google Scholar] [CrossRef]
- Tang, Z.; Takahashi, Y.; He, H.; Hattori, T.; Chen, C.; Liang, X.; Chen, H.; Young, M.M.; Wang, H.-G. TOM40 targets Atg2 to mitochondria-associated er membranes for phagophore expansion. Cell Rep. 2019, 28, 1744–1757. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.N.; Tang, B.L. Location and membrane sources for autophagosome formation—From ER–mitochondria contact sites to Golgi-endosome-derived carriers. Mol. Membr. Biol. 2013, 30, 394–402. [Google Scholar] [CrossRef] [PubMed]
- Hailey, D.W.; Rambold, A.S.; Satpute-Krishnan, P.; Mitra, K.; Sougrat, R.; Kim, P.K.; Lippincott-Schwartz, J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 2010, 141, 656–667. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Yang, P.; Huang, X.; Hu, W.; Guo, B.; Wu, F.; Lin, L.; Kovács, A.L.; Yu, L.; Zhang, H. The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev. Cell 2011, 21, 343–357. [Google Scholar] [CrossRef]
- Tian, Y.; Li, Z.; Hu, W.; Ren, H.; Tian, E.; Zhao, Y.; Lu, Q.; Huang, X.; Yang, P.; Li, X.; et al. elegans screen identifies autophagy genes specific to multicellular organisms. Cell 2010, 141, 1042–1055. [Google Scholar] [CrossRef]
- Bissa, B.; Deretic, V. Autophagosome formation: Cutting the gordian knot at the ER. Curr. Biol. 2018, 28, R347–R349. [Google Scholar] [CrossRef]
- Vaccaro, M.; Ropolo, A.; Grasso, D.; Iovanna, J.L. A novel mammalian trans-membrane protein reveals an alternative initiation pathway for autophagy. Autophagy 2008, 4, 388–390. [Google Scholar] [CrossRef]
- Molejon, M.I.; Ropolo, A.; Vaccaro, M. VMP1 is a new player in the regulation of the autophagy-specific phosphatidylinositol 3-kinase complex activation. Autophagy 2013, 9, 933–935. [Google Scholar] [CrossRef]
- Zhao, Y.G.; Liu, N.; Miao, G.; Chen, Y.; Zhao, H.; Zhang, H. The ER contact proteins VAPA/B interact with multiple autophagy proteins to modulate autophagosome biogenesis. Curr. Biol. 2018, 28, 1234–1245. [Google Scholar] [CrossRef]
- Kvam, E.; Goldfarb, D. Nucleus–vacuole junctions in yeast: Anatomy of a membrane contact site. Biochem. Soc. Trans. 2006, 34, 340–342. [Google Scholar] [CrossRef]
- Jeong, H.; Park, J.; Kim, H.-I.; Lee, M.; Ko, Y.-J.; Lee, S.; Jun, Y.; Lee, C. Mechanistic insight into the nucleus–vacuole junction based on the Vac8p–Nvj1p crystal structure. Proc. Natl. Acad. Sci. USA 2017, 114, E4539–E4548. [Google Scholar] [CrossRef]
- Wang, Y.-X.; Catlett, N.L.; Weisman, L.S. Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole. J. Cell Biol. 1998, 140, 1063–1074. [Google Scholar] [CrossRef] [PubMed]
- Scott, S.V.; Nice, D.C.; Nau, J.J.; Weisman, L.S.; Kamada, Y.; Keizer-Gunnink, I.; Funakoshi, T.; Veenhuis, M.; Ohsumi, Y.; Klionsky, D.J. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J. Biol. Chem. 2000, 275, 25840–25849. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Kim, H.-I.; Jeong, H.; Lee, M.; Jang, S.H.; Yoon, S.Y.; Kim, H.; Park, Z.-Y.; Jun, Y.; Lee, C. Quaternary structures of Vac8 differentially regulate the Cvt and PMN pathways. Autophagy 2019, 1–16. [Google Scholar] [CrossRef]
- Ward, C.; Martinez-Lopez, N.; Otten, E.; Carroll, B.; Maetzel, R.; Singh, R.; Sarkar, S.; Korolchuk, V.I. Autophagy, lipophagy and lysosomal lipid storage disorders. Biochim. Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2016, 1861, 269–284. [Google Scholar] [CrossRef]
- Kounakis, K.; Chaniotakis, M.; Markaki, M.; Tavernarakis, N. Emerging roles of lipophagy in health and disease. Front. Cell Dev. Biol. 2019, 7, 185. [Google Scholar] [CrossRef]
- Van Zutphen, T.; Todde, V.; de Boer, R.; Kreim, M.; Hofbauer, H.F.; Wolinski, H.; Veenhuis, M.; van der Klei, I.J.; Kohlwein, S.D. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 2014, 25, 290–301. [Google Scholar] [CrossRef]
- Li, Z.; Schulze, R.J.; Weller, S.G.; Krueger, E.W.; Schott, M.B.; Zhang, X.; Casey, C.A.; Liu, J.; Stöckli, J.; James, D.E.; et al. A novel Rab10-EHBP1-EHD2 complex essential for the autophagic engulfment of lipid droplets. Sci. Adv. 2016, 2, e1601470. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Zhao, X.; Song, Y.; Cheng, H.; Zhou, R. Nuclear autophagy: An evolutionarily conserved mechanism of nuclear degradation in the cytoplasm. Autophagy 2016, 12, 1973–1983. [Google Scholar] [CrossRef] [PubMed]
- Rocha, N.; Kuijl, C.; van der Kant, R.; Janssen, L.; Houben, D.; Janssen, H.; Zwart, W.; Neefjes, J. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7–RILP–p150Glued and late endosome positioning. J. Cell Biol. 2009, 185, 1209–1225. [Google Scholar] [CrossRef] [PubMed]
- Bento, C.F.; Ashkenazi, A.; Jimenez-Sanchez, M.; Rubinsztein, D.C. The Parkinson’s disease-associated genes ATP13A2 and SYT11 regulate autophagy via a common pathway. Nat. Commun. 2016, 7, 11803. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.C.; Kim, S.; Peng, W.; Krainc, D. Regulation and function of mitochondria-lysosome membrane contact sites in cellular homeostasis. Trends Cell Biol. 2019, 29, 500–513. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.C.; Ysselstein, D.; Krainc, D. Mitochondria–lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature 2018, 554, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Nair, U.; Klionsky, D.J. Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell 2008, 19, 3290–3298. [Google Scholar] [CrossRef] [PubMed]
- Schütter, M.; Giavalisco, P.; Brodesser, S.; Graef, M. Local fatty acid channeling into phospholipid synthesis drives phagophore expansion during autophagy. Cell 2020, 180, 135–149. [Google Scholar] [CrossRef]
- Hansen, M.; Rubinsztein, D.C.; Walker, D. Autophagy as a promoter of longevity: Insights from model organisms. Nat. Rev. Mol. Cell Biol. 2018, 19, 579–593. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kohler, V.; Aufschnaiter, A.; Büttner, S. Closing the Gap: Membrane Contact Sites in the Regulation of Autophagy. Cells 2020, 9, 1184. https://doi.org/10.3390/cells9051184
Kohler V, Aufschnaiter A, Büttner S. Closing the Gap: Membrane Contact Sites in the Regulation of Autophagy. Cells. 2020; 9(5):1184. https://doi.org/10.3390/cells9051184
Chicago/Turabian StyleKohler, Verena, Andreas Aufschnaiter, and Sabrina Büttner. 2020. "Closing the Gap: Membrane Contact Sites in the Regulation of Autophagy" Cells 9, no. 5: 1184. https://doi.org/10.3390/cells9051184
APA StyleKohler, V., Aufschnaiter, A., & Büttner, S. (2020). Closing the Gap: Membrane Contact Sites in the Regulation of Autophagy. Cells, 9(5), 1184. https://doi.org/10.3390/cells9051184




