Calcium-Free and Cytochalasin B Treatment Inhibits Blastomere Fusion in 2-Cell Stage Embryos for the Generation of Floxed Mice via Sequential Electroporation
Abstract
1. Introduction
2. Materials and Methods
2.1. Mouse
2.2. Preparation of Embryos
2.3. Preparation of Cas9, crRNA, tracrRNA and ssODN
2.4. Electroporation
2.5. Inhibition of Blastomere Fusion
2.6. Assay for KI and Floxed Allele
2.7. Statistical Analysis
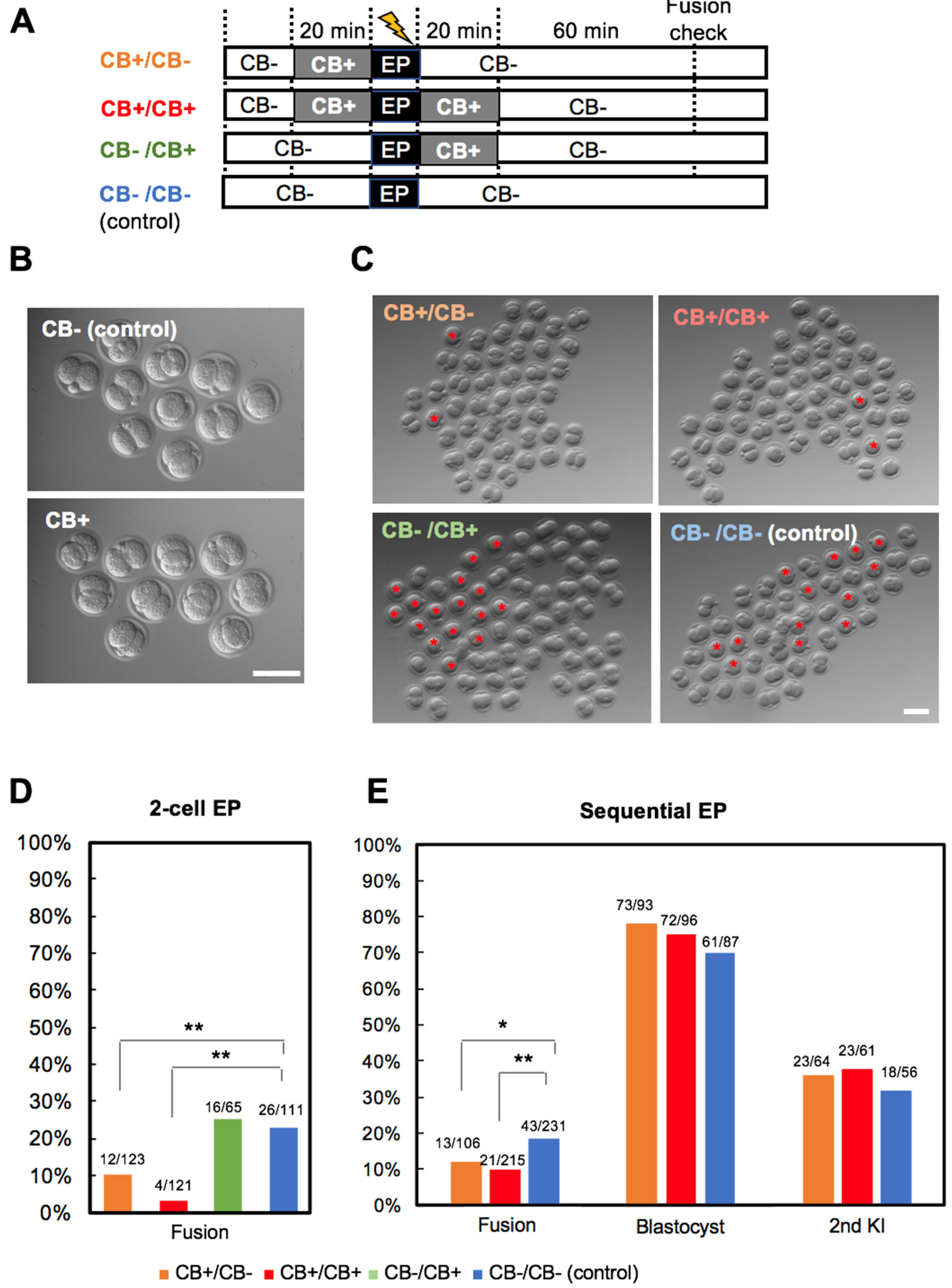
3. Results
3.1. Experiment 1: Hypertonic Treatment
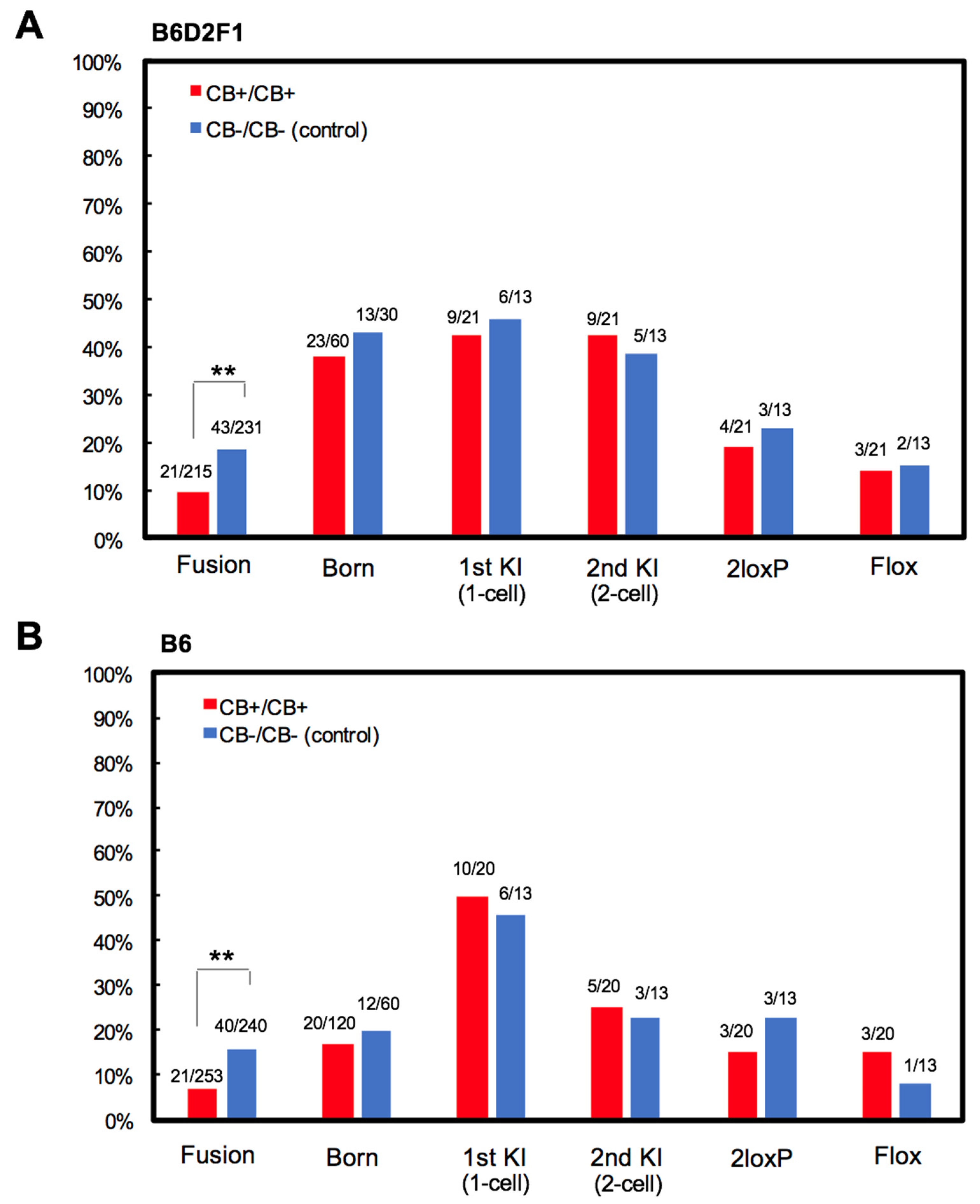
3.2. Experiment 2: Ca2+-Free Treatment
3.3. Experiment 3: Actin Polymerization Inhibition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dickinson, M.E.; Flenniken, A.M.; Ji, X.; Teboul, L.; Wong, M.D.; White, J.K.; Meehan, T.F.; Weninger, W.J.; Westerberg, H.; Adissu, H.; et al. High-throughput discovery of novel developmental phenotypes. Nature 2016, 537, 508–514. [Google Scholar] [CrossRef]
- Sauer, B.; Henderson, N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl. Acad. Sci. USA 1988, 85, 5166–5170. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, H.; Shivalila, C.S.; Dawlaty, M.M.; Cheng, A.W.; Zhang, F.; Jaenisch, R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013, 153, 910–918. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Lin, C.Y.; Gootenberg, J.S.; Konermann, S.; Trevino, A.E.; Scott, D.A.; Inoue, A.; Matoba, S.; Zhang, Y.; et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013, 154, 1380–1389. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, H.; Shivalila, C.S.; Cheng, A.W.; Shi, L.; Jaenisch, R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 2013, 154, 1370–1379. [Google Scholar] [CrossRef] [PubMed]
- Bishop, K.A.; Harrington, A.; Kouranova, E.; Weinstein, E.J.; Rosen, C.J.; Cui, X.; Liaw, L. CRISPR/Cas9-Mediated Insertion of loxP Sites in the Mouse Dock7 Gene Provides an Effective Alternative to Use of Targeted Embryonic Stem Cells. G3 2016, 6, 2051–2061. [Google Scholar] [CrossRef]
- Nakagawa, Y.; Oikawa, F.; Mizuno, S.; Ohno, H.; Yagishita, Y.; Satoh, A.; Osaki, Y.; Takei, K.; Kikuchi, T.; Han, S.I.; et al. Hyperlipidemia and hepatitis in liver-specific CREB3L3 knockout mice generated using a one-step CRISPR/Cas9 system. Sci. Rep. 2016, 6, 27857. [Google Scholar] [CrossRef]
- Ma, X.; Chen, C.; Veevers, J.; Zhou, X.; Ross, R.S.; Feng, W.; Chen, J. CRISPR/Cas9-mediated gene manipulation to create single-amino-acid-substituted and floxed mice with a cloning-free method. Sci. Rep. 2017, 7, 42244. [Google Scholar] [CrossRef]
- Horii, T.; Morita, S.; Kimura, M.; Terawaki, N.; Shibutani, M.; Hatada, I. Efficient generation of conditional knockout mice via sequential introduction of lox sites. Sci. Rep. 2017, 7, 7891. [Google Scholar] [CrossRef]
- Gurumurthy, C.B.; O’Brien, A.R.; Quadros, R.M.; Adams, J., Jr.; Alcaide, P.; Ayabe, S.; Ballard, J.; Batra, S.K.; Beauchamp, M.C.; Becker, K.A.; et al. Reproducibility of CRISPR-Cas9 methods for generation of conditional mouse alleles: A multi-center evaluation. Genome Biol. 2019, 20, 171. [Google Scholar] [CrossRef] [PubMed]
- Horii, T.; Yamamoto, M.; Morita, S.; Kimura, M.; Nagao, Y.; Hatada, I. p53 suppresses tetraploid development in mice. Sci. Rep. 2015, 5, 8907. [Google Scholar] [CrossRef]
- Hogan, B.; Constantini, F.; Lacy, E. Manipulating the Mouse Embryo: A Laboratory Manual, 1st ed.; Cold Spring Harbor Laboratory: Huntington, NY, USA, 1986; pp. 252–253. [Google Scholar]
- Schmitt, J.J.; Zimmermann, U. Enhanced hybridoma production by electrofusion in strongly hypo-osmolar solutions. Biochim. Biophys. Acta 1989, 983, 42–50. [Google Scholar] [CrossRef]
- Usaj, M.; Kanduser, M. The systematic study of the electroporation and electrofusion of B16-F1 and CHO cells in isotonic and hypotonic buffer. J. Membr. Biol. 2012, 245, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Takeichi, M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science 1991, 251, 1451–1455. [Google Scholar] [CrossRef] [PubMed]
- Larue, L.; Ohsugi, M.; Hirchenhain, J.; Kemler, R. E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc. Natl. Acad. Sci. USA 1994, 91, 8263–8267. [Google Scholar] [CrossRef]
- Riethmacher, D.; Brinkmann, V.; Birchmeier, C. A targeted mutation in the mouse E-cadherin gene results in defective preimplantation development. Proc. Natl. Acad. Sci. USA 1995, 92, 855–859. [Google Scholar] [CrossRef]
- De Vries, W.N.; Evsikov, A.V.; Haac, B.E.; Fancher, K.S.; Holbrook, A.E.; Kemler, R.; Solter, D.; Knowles, B.B. Maternal beta-catenin and E-cadherin in mouse development. Development 2004, 131, 4435–4445. [Google Scholar] [CrossRef]
- Johnson, M.H.; Maro, B.; Takeichi, M. The role of cell adhesion in the synchronization and orientation of polarization in 8-cell mouse blastomeres. J. Embryol. Exp. Morphol. 1986, 93, 239–255. [Google Scholar]
- Cooper, J.A. Effects of cytochalasin and phalloidin on actin. J. Cell Biol. 1987, 105, 1473–1478. [Google Scholar] [CrossRef]
- Kimber, S.J.; Surani, M.A. Morphogenetic analysis of changing cell associations following release of 2-cell and 4-cell mouse embryos from cleavage arrest. J. Embryol. Exp. Morphol. 1981, 61, 331–345. [Google Scholar] [PubMed]
- Pey, R.; Vial, C.; Schatten, G.; Hafner, M. Increase of intracellular Ca2+ and relocation of E-cadherin during experimental decompaction of mouse embryos. Proc. Natl. Acad. Sci. USA 1998, 95, 12977–12982. [Google Scholar] [CrossRef] [PubMed]
- Lehtonen, E.; Badley, R.A. Localization of cytoskeletal proteins in preimplantation mouse embryos. J. Embryol. Exp. Morphol. 1980, 55, 211–225. [Google Scholar]
- Duan, X.; Zhang, H.L.; Wu, L.L.; Liu, M.Y.; Pan, M.H.; Ou, X.H.; Sun, S.C. Involvement of LIMK1/2 in actin assembly during mouse embryo development. Cell Cycle 2018, 17, 1381–1389. [Google Scholar] [CrossRef]
- Quadros, R.M.; Miura, H.; Harms, D.W.; Akatsuka, H.; Sato, T.; Aida, T.; Redder, R.; Richardson, G.P.; Inagaki, Y.; Sakai, D.; et al. Easi-CRISPR: A robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol. 2017, 18, 92. [Google Scholar] [CrossRef]
- Miyasaka, Y.; Uno, Y.; Yoshimi, K.; Kunihiro, Y.; Yoshimura, T.; Tanaka, T.; Ishikubo, H.; Hiraoka, Y.; Takemoto, N.; Tanaka, T.; et al. CLICK: One-step generation of conditional knockout mice. BMC Genom. 2018, 19, 318. [Google Scholar] [CrossRef]
- Yao, X.; Zhang, M.; Wang, X.; Ying, W.; Hu, X.; Dai, P.; Meng, F.; Shi, L.; Sun, Y.; Yao, N.; et al. Tild-CRISPR Allows for Efficient and Precise Gene Knockin in Mouse and Human Cells. Dev. Cell 2018, 45, 526–536. [Google Scholar] [CrossRef]
- Sato, M.; Miyagasako, R.; Takabayashi, S.; Ohtsuka, M.; Hatada, I.; Horii, T. Sequential i-GONAD: An Improved In Vivo Technique for CRISPR/Cas9-Based Genetic Manipulations in Mice. Cells 2020, 9, 546. [Google Scholar] [CrossRef]



| Locus | crRNA | ssODN |
|---|---|---|
| Mecp2 intron 2 (Left) | Mecp2-L2 (5′-CCCAAGGATACAGTATCCTA-3′) | Mecp2-L2-lox66 (5′-ccagcaacctaaagctgttaagaaatctttgggccccagcttgacccaaggatacagtatgctagcTACCGTTCGTATAATGTATGCTATACGAAGTTATCCTAGGgaagttaccaaaatcagagatagtatgcagcagccaggggtctcatgtgtggca-3′) |
| Mecp2 intron 3 (Right) | Mecp2-R1 (5′-AGGAGTGAGGTCTAGTACTT-3′) | Mecp2-R1-lox71 (5′-ccactcctctgtactccctggcttttccacaatccttaaactgaaggagtgaggtctagtTACCGTTCGTATAGCATACATTATACGAAGTTATGAATTCacttgggggtcattgggctagactgaata tctttggttggtacccagacctaatccacca-3′) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Horii, T.; Kobayashi, R.; Kimura, M.; Morita, S.; Hatada, I. Calcium-Free and Cytochalasin B Treatment Inhibits Blastomere Fusion in 2-Cell Stage Embryos for the Generation of Floxed Mice via Sequential Electroporation. Cells 2020, 9, 1088. https://doi.org/10.3390/cells9051088
Horii T, Kobayashi R, Kimura M, Morita S, Hatada I. Calcium-Free and Cytochalasin B Treatment Inhibits Blastomere Fusion in 2-Cell Stage Embryos for the Generation of Floxed Mice via Sequential Electroporation. Cells. 2020; 9(5):1088. https://doi.org/10.3390/cells9051088
Chicago/Turabian StyleHorii, Takuro, Ryosuke Kobayashi, Mika Kimura, Sumiyo Morita, and Izuho Hatada. 2020. "Calcium-Free and Cytochalasin B Treatment Inhibits Blastomere Fusion in 2-Cell Stage Embryos for the Generation of Floxed Mice via Sequential Electroporation" Cells 9, no. 5: 1088. https://doi.org/10.3390/cells9051088
APA StyleHorii, T., Kobayashi, R., Kimura, M., Morita, S., & Hatada, I. (2020). Calcium-Free and Cytochalasin B Treatment Inhibits Blastomere Fusion in 2-Cell Stage Embryos for the Generation of Floxed Mice via Sequential Electroporation. Cells, 9(5), 1088. https://doi.org/10.3390/cells9051088




