Structural Similarity with Cholesterol Reveals Crucial Insights into Mechanisms Sustaining the Immunomodulatory Activity of the Mycotoxin Alternariol
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture Conditions and Reagents
2.2. Supernatant Analysis of Eicosanoids and Proteomics
2.3. Protein Sample Preparation
2.4. Lipids Sample Preparation
2.5. Membrane Fluidity
2.6. Live Cell Imaging: Membrane Morphology
2.7. Live Cell Imaging: Mitochondrial Superoxide
2.8. Immunofluorescence
2.9. Microscopy
2.10. Statistical Analysis
2.11. Data Analysis Secretome
2.12. Proteomics Data Analysis
2.13. UHPLC-MS/MS for Eicosanoid Measurements
2.14. In Silico Molecular Modeling
3. Results
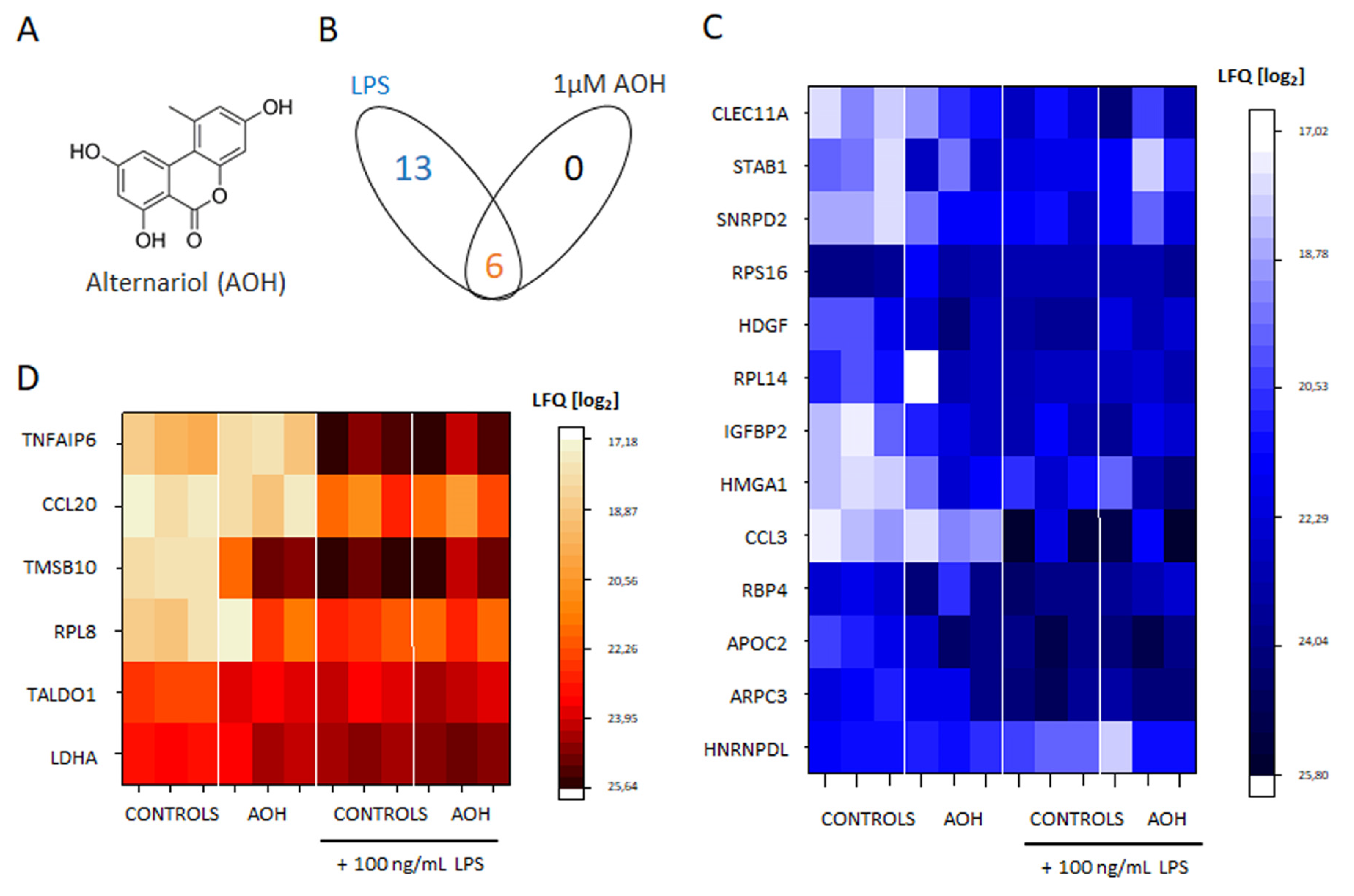
3.1. Crosstalk between AOH and LPS, with Regard to the Secretome of THP-1 Macrophages
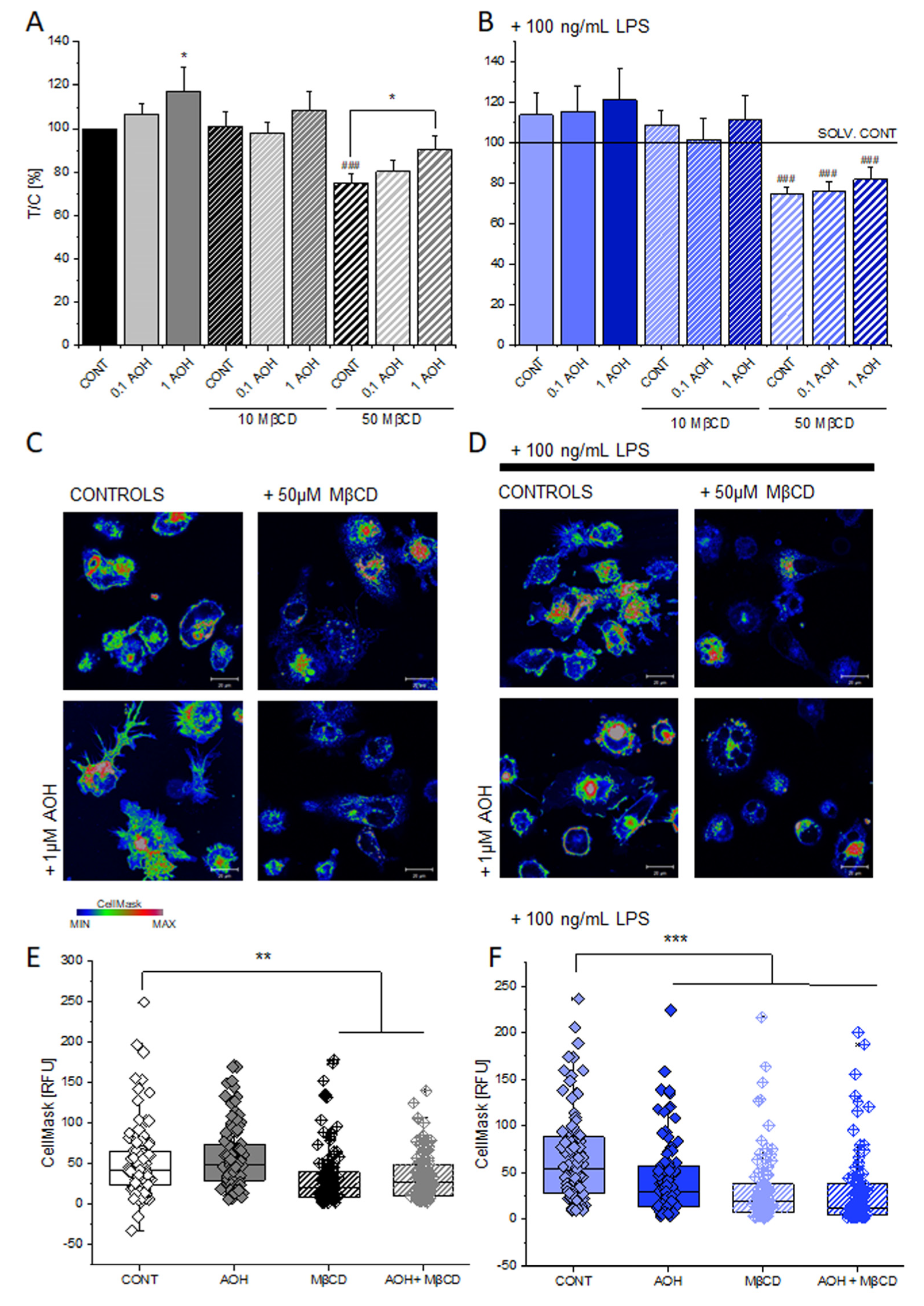
3.2. Effect of AOH on the Membrane of THP-1 Macrophages
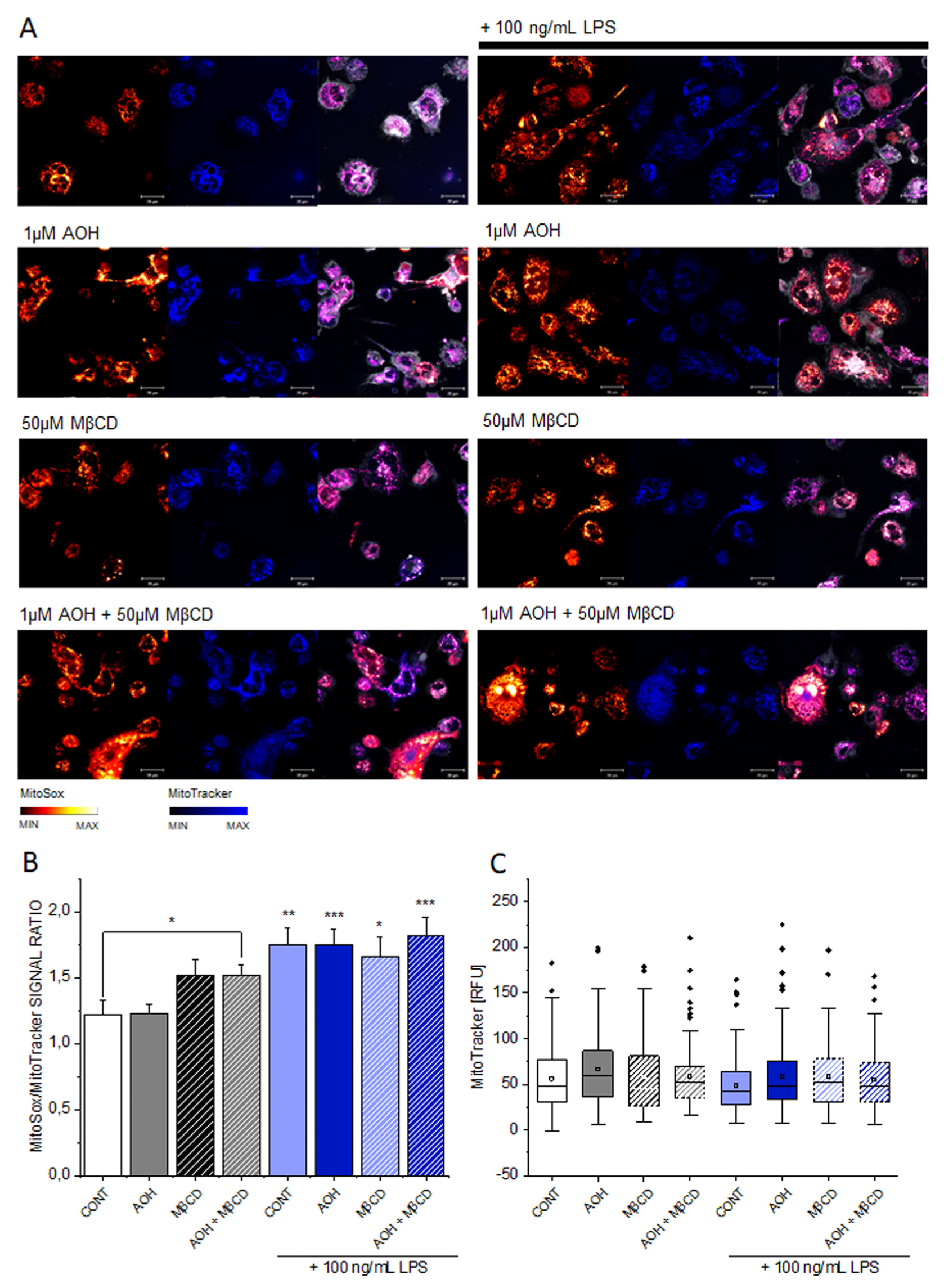
3.3. Effect of AOH on Mitochondrial Superoxide
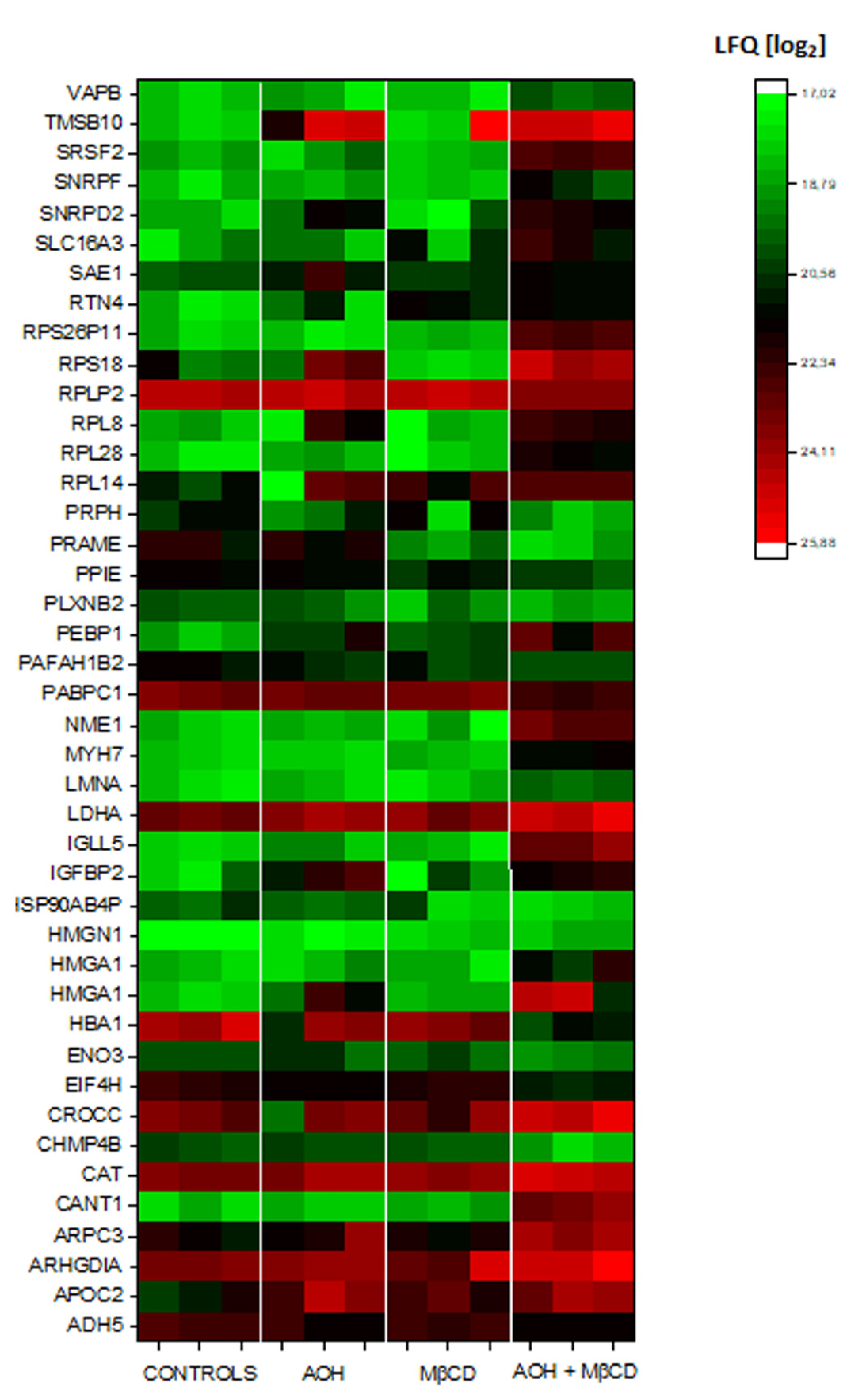
3.4. Crosstalk between AOH and MβCD on the Secretome of THP-1 Macrophages
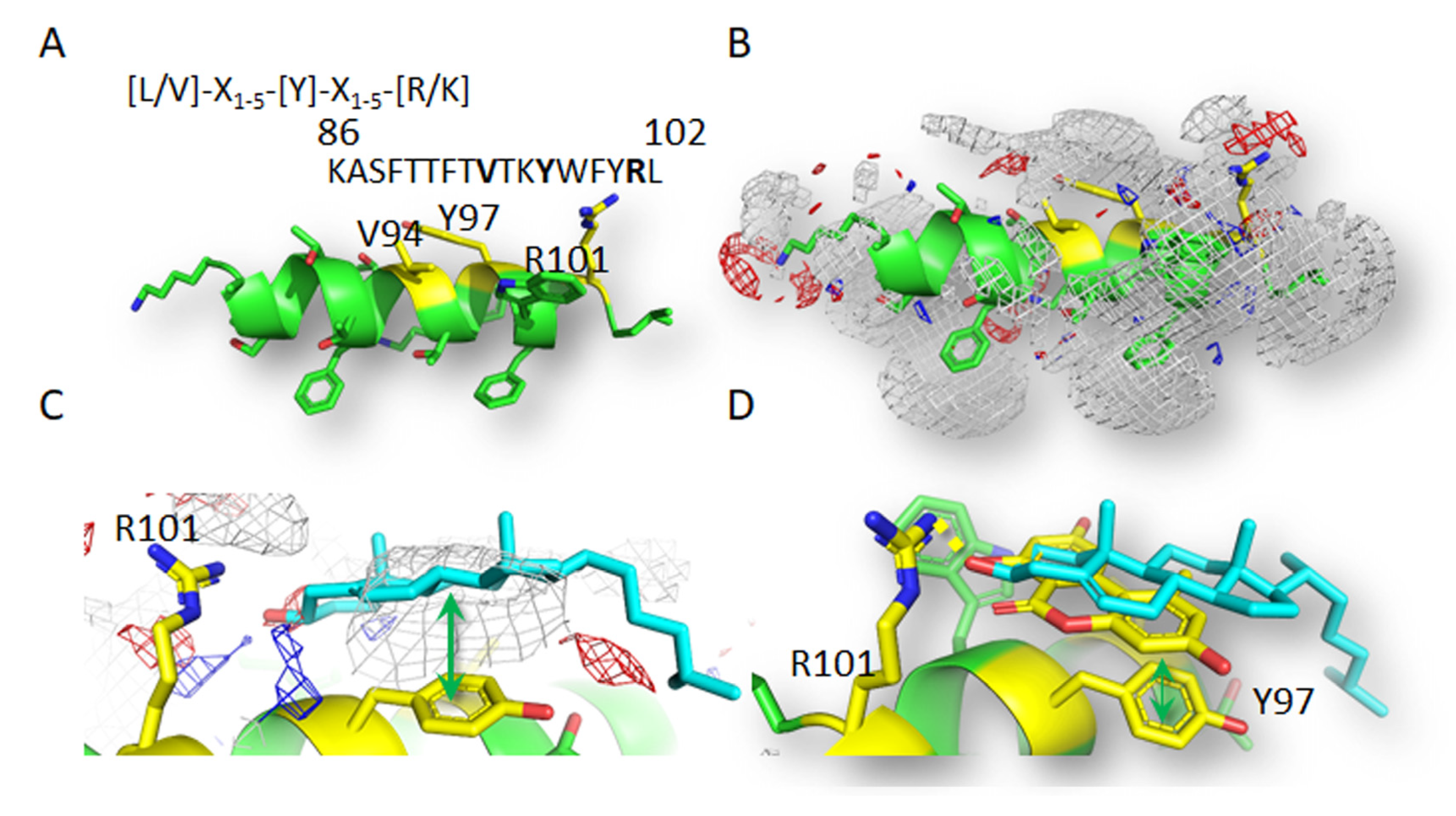
3.5. Computational Study of Molecular Interactions between AOH and Cholesterol
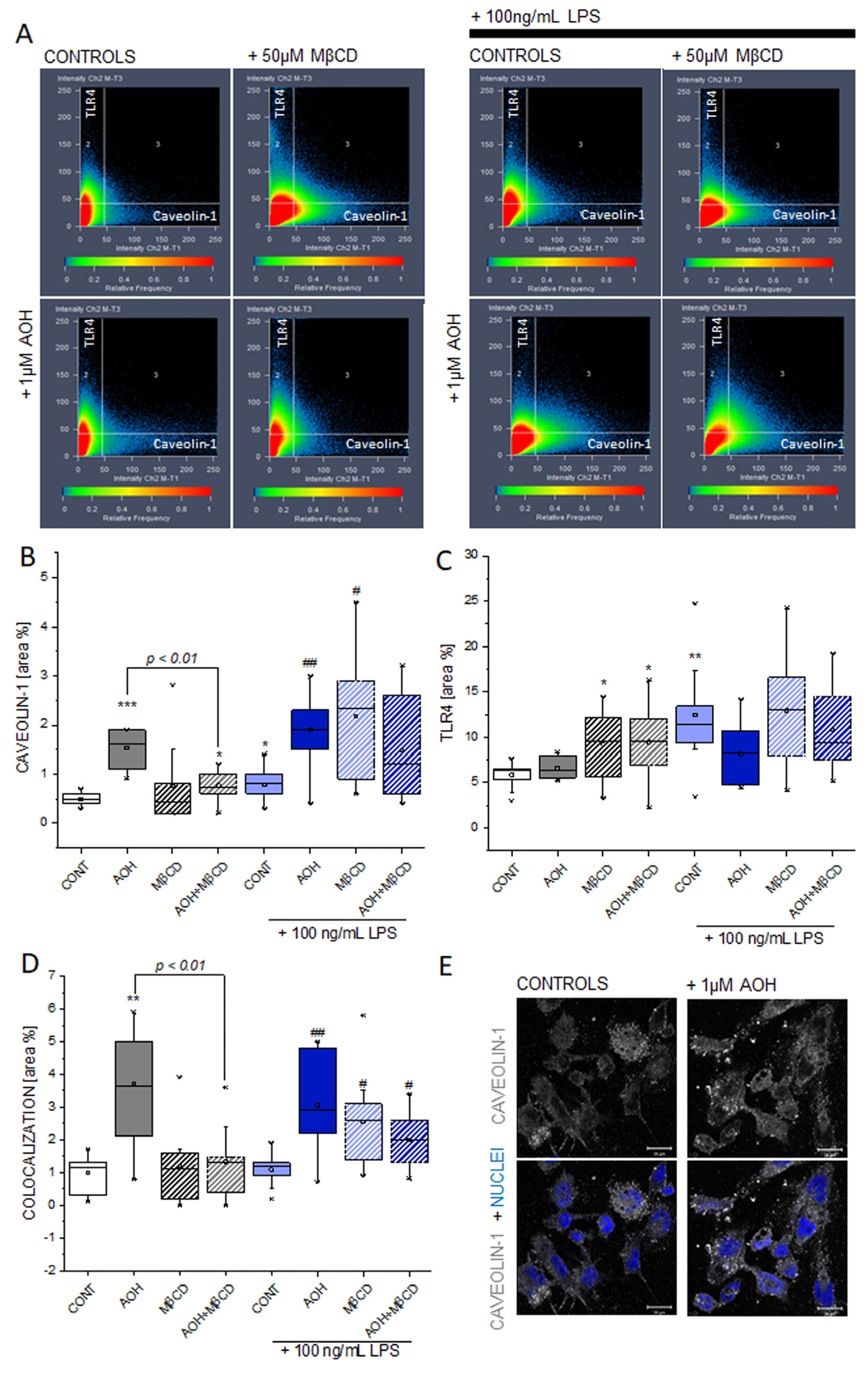
3.6. Effect of AOH and LPS on Caveolin-1 and TLR4 in THP-1 Macrophages
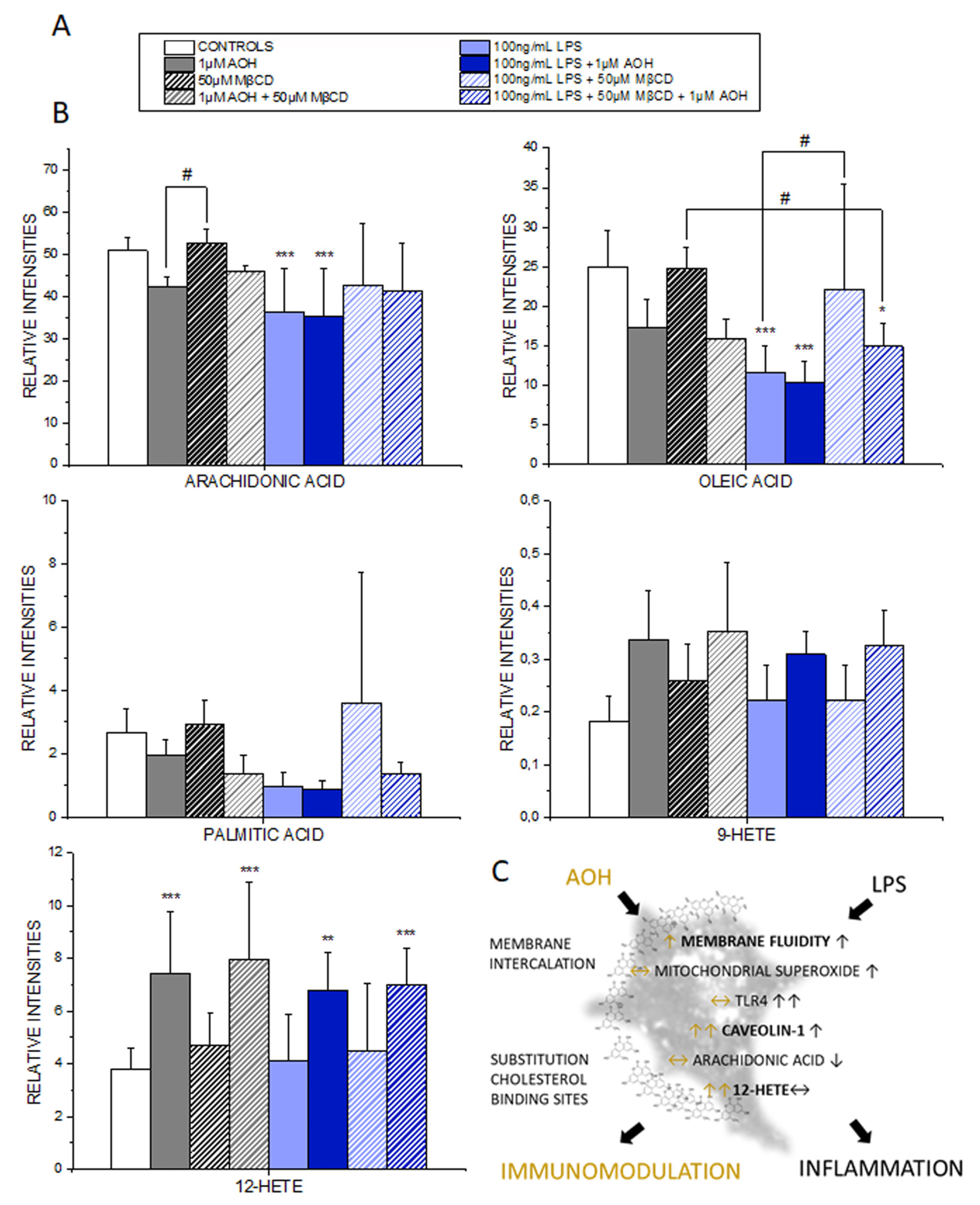
3.7. Effect of AOH on the PUFA and Oxylipin Profile of THP-1 Macrophages
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sahin, O.A.; Kececioglu, N.; Serdar, M.; Ozpinar, A. The association of residential mold exposure and adenotonsillar hypertrophy in children living in damp environments. Int. J. Pediatric Otorhinolaryngol. 2016, 88, 233–238. [Google Scholar] [CrossRef]
- Castanhinha, S.; Sherburn, R.; Walker, S.; Gupta, A.; Bossley, C.J.; Buckley, J.; Ullmann, N.; Grychtol, R.; Campbell, G.; Maglione, M.; et al. Pediatric severe asthma with fungal sensitization is mediated by steroid-resistant IL-33. J. Allergy Clin. Immunol. 2015, 136, 312–322. [Google Scholar] [CrossRef]
- Solhaug, A.; Karlsøen, L.M.; Holme, J.A.; Kristoffersen, A.B.; Eriksen, G.S. Immunomodulatory effects of individual and combined mycotoxins in the THP-1 cell line. Toxicol. Vitro (Suppl. C) 2016, 36, 120–132. [Google Scholar] [CrossRef] [PubMed]
- Solhaug, A.; Wisbech, C.; Christoffersen, T.E.; Hult, L.O.; Lea, T.; Eriksen, G.S.; Holme, J.A. The mycotoxin alternariol induces DNA damage and modify macrophage phenotype and inflammatory responses. Toxicol. Lett. 2015, 239, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Solhaug, A.; Holme, J.A.; Haglund, K.; Dendele, B.; Sergent, O.; Pestka, J.; Lagadic-Gossmann, D.; Eriksen, G.S. Alternariol induces abnormal nuclear morphology and cell cycle arrest in murine RAW 264.7 macrophages. Toxicol. Lett. 2013, 219, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Kollarova, J.; Cenk, E.; Schmutz, C.; Marko, D. The mycotoxin alternariol suppresses lipopolysaccharide-induced inflammation in THP-1 derived macrophages targeting the NF-kappaB signalling pathway. Arch. Toxicol. 2018, 92, 3347–3358. [Google Scholar] [CrossRef] [PubMed]
- Grover, S.; Lawrence, C.B. The Alternaria alternata Mycotoxin Alternariol Suppresses Lipopolysaccharide-Induced Inflammation. Int. J. Mol. Sci. 2017, 18, 1577. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.C.; Madec, S.; Troadec, S.; Coton, E.; Hymery, N. Effects of fusariotoxin co-exposure on THP-1 human immune cells. Cell Biol. Toxicol. 2017, 34, 191–205. [Google Scholar] [CrossRef]
- Gabriel, M.F.; Postigo, I.; Tomaz, C.T.; Martinez, J. Alternaria alternata allergens: Markers of exposure, phylogeny and risk of fungi-induced respiratory allergy. Environ. Int. 2016, 89–90, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Puntscher, H.; Hankele, S.; Tillmann, K.; Attakpah, E.; Braun, D.; Kutt, M.L.; del Favero, G.; Aichinger, G.; Pahlke, G.; Hoger, H.; et al. Warth, First insights into Alternaria multi-toxin in vivo metabolism. Toxicol. Lett. 2018, 301, 168–178. [Google Scholar] [CrossRef]
- Puntscher, H.; Kutt, M.L.; Skrinjar, P.; Mikula, H.; Podlech, J.; Frohlich, J.; Marko, D.; Warth, B. Tracking emerging mycotoxins in food: Development of an LC-MS/MS method for free and modified Alternaria toxins. Anal. Bioanal. Chem. 2018, 410, 4481–4494. [Google Scholar] [CrossRef]
- Bansal, M.; Singh, N.; Alam, S.; Pal, S.; Satyanarayana, G.N.V.; Singh, D.; Ansari, K.M. Alternariol induced proliferation in primary mouse keratinocytes and inflammation in mouse skin is regulated via PGE2/EP2/cAMP/p-CREB signaling pathway. Toxicology 2019, 412, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Solhaug, A.; Vines, L.L.; Ivanova, L.; Spilsberg, B.; Holme, J.A.; Pestka, J.; Collins, A.; Eriksen, G.S. Mechanisms involved in alternariol-induced cell cycle arrest. Mutat. Res. 2012, 738–739, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Solhaug, A.; Torgersen, M.L.; Holme, J.A.; Lagadic-Gossmann, D.; Eriksen, G.S. Autophagy and senescence, stress responses induced by the DNA-damaging mycotoxin alternariol. Toxicology 2014, 326, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, C.; Cenk, E.; Marko, D. The Alternaria Mycotoxin Alternariol Triggers the Immune Response of IL-1beta-stimulated, Differentiated Caco-2 Cells. Mol. Nutr. Food Res. 2019, 63, e1900341. [Google Scholar] [CrossRef] [PubMed]
- Köberlin, M.S.; Heinz, L.X.; Superti-Furga, G. Functional crosstalk between membrane lipids and TLR biology. Curr. Opin. Cell Biol. 2016, 39, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Hoop, C.L.; Sivanandam, V.N.; Kodali, R.; Srnec, M.N.; van der Wel, P.C. Structural characterization of the caveolin scaffolding domain in association with cholesterol-rich membranes. Biochemistry 2012, 51, 90–99. [Google Scholar] [CrossRef]
- Agache, I.; Annesi-Maesano, I.; Bonertz, A.; Branca, F.; Cant, A.; Fras, Z.; Ingenrieth, F.; Namazova-Baranova, L.; Odemyr, M.; Spanevello, A.; et al. Prioritising research challenges and funding for allergy and asthma and the need for translational research—The European Strategic Forum on Allergic Diseases. Allergy 2019, 74, 2064–2076. [Google Scholar] [CrossRef]
- Kandasamy, P.; Numata, M.; Berry, K.Z.; Fickes, R.; Leslie, C.C.; Murphy, R.C.; Voelker, D.R. Structural analogs of pulmonary surfactant phosphatidylglycerol inhibit toll-like receptor 2 and 4 signaling. J. Lipid Res. 2016, 57, 993–1005. [Google Scholar] [CrossRef]
- Pande, A.H.; Qin, S.; Tatulian, S.A. Membrane fluidity is a key modulator of membrane binding, insertion, and activity of 5-lipoxygenase. Biophys. J. 2005, 88, 4084–4094. [Google Scholar] [CrossRef]
- De la Haba, C.; Morros, A.; Martinez, P.; Palacio, J.R. LPS-Induced Macrophage Activation and Plasma Membrane Fluidity Changes are Inhibited Under Oxidative Stress. J. Membr. Biol. 2016, 249, 789–800. [Google Scholar] [CrossRef] [PubMed]
- De la Haba, C.; Palacio, J.R.; Martinez, P.; Morros, A. Effect of oxidative stress on plasma membrane fluidity of THP-1 induced macrophages. Biochim. Biophys. Acta 2013, 1828, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Del Favero, G.; Hohenbichler, J.; Mayer, R.M.; Rychlik, M.; Marko, D. Mycotoxin Altertoxin II Induces Lipid Peroxidation Connecting Mitochondrial Stress Response to NF-kappaB Inhibition in THP-1 Macrophages. Chem. Res. Toxicol. 2020, 33, 492–504. [Google Scholar] [CrossRef] [PubMed]
- Chanput, W.; Mes, J.J.; Wichers, H.J. THP-1 cell line: An in vitro cell model for immune modulation approach. Int. Immunopharmacol. 2014, 23, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Mayer, R.L.; Schwarzmeier, J.D.; Gerner, M.C.; Bileck, A.; Mader, J.C.; Meier-Menches, S.M.; Gerner, S.M.; Schmetterer, K.G.; Pukrop, T.; Reichle, A.; et al. Proteomics and metabolomics identify molecular mechanisms of aging potentially predisposing for chronic lymphocytic leukemia. Mol. Cell. Proteom. MCP 2018, 17, 290–303. [Google Scholar] [CrossRef]
- Jarolim, K.; del Favero, G.; Pahlke, G.; Dostal, V.; Zimmermann, K.; Heiss, E.; Ellmer, D.; Stark, T.D.; Hofmann, T.; Marko, D. Activation of the Nrf2-ARE pathway by the Alternaria alternata mycotoxins altertoxin I and II. Arch. Toxicol. 2017, 91, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Carlson, A.; Sinitcyn, P.; Mann, M.; Cox, J. Visualization of LC-MS/MS proteomics data in MaxQuant. Proteomics 2015, 15, 1453–1456. [Google Scholar] [CrossRef]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. 1D and 2D annotation enrichment: A statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinform. 2012, 13, S12. [Google Scholar] [CrossRef] [PubMed]
- Fantini, J.; Barrantes, F.J. How cholesterol interacts with membrane proteins: An exploration of cholesterol-binding sites including CRAC, CARC, and tilted domains. Front. Physiol. 2013, 4, 31. [Google Scholar] [CrossRef] [PubMed]
- Baier, C.J.; Fantini, J.; Barrantes, F.J. Disclosure of cholesterol recognition motifs in transmembrane domains of the human nicotinic acetylcholine receptor. Sci. Rep. 2011, 1, 69. [Google Scholar] [CrossRef] [PubMed]
- Le Lan, C.; Gallay, J.; Vincent, M.; Neumann, J.M.; de Foresta, B.; Jamin, N. Structural and dynamic properties of juxta-membrane segments of caveolin-1 and caveolin-2 at the membrane interface. Eur. Biophys. J. EBJ 2010, 39, 307–325. [Google Scholar] [CrossRef]
- Epand, R.M. Proteins and cholesterol-rich domains. Biochim. Biophys. Acta 2008, 1778, 1576–1582. [Google Scholar] [CrossRef]
- Baroni, M.; Cruciani, G.; Sciabola, S.; Perruccio, F.; Mason, J.S. A common reference framework for analyzing/comparing proteins and ligands. Fingerprints for Ligands and Proteins (FLAP): Theory and application. J. Chem. Inf. Model. 2007, 47, 279–294. [Google Scholar] [CrossRef]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef]
- Dellafiora, L.; Galaverna, G.; Dall’Asta, C. Mechanisms of Fumonisin B1 Toxicity: A Computational Perspective beyond the Ceramide Synthases Inhibition. Chem. Res. Toxicol. 2018, 31, 1203–1212. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.; Mittal, J.; Feig, M.; Mackerell, A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef]
- Dellafiora, L.; Galaverna, G.; Cruciani, G.; Dall’Asta, C. A computational study toward the “personalized” activity of alternariol - Does it matter for safe food at individual level? Food Chem. Toxicol. 2019, 130, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Kanzler, H.; Barrat, F.J.; Hessel, E.M.; Coffman, R.L. Therapeutic targeting of innate immunity with Toll-like receptor agonists and antagonists. Nat. Med. 2007, 13, 552. [Google Scholar] [CrossRef] [PubMed]
- Lopez, C.A.; de Vries, A.H.; Marrink, S.J. Computational microscopy of cyclodextrin mediated cholesterol extraction from lipid model membranes. Sci. Rep. 2013, 3, 2071. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, M.; Ogura, K.; Mizoguchi, I.; Chiba, Y.; Higuchi, K.; Ohtsuka, H.; Mizuguchi, J.; Yoshimoto, T. IL-27 promotes nitric oxide production induced by LPS through STAT1, NF-kappaB and MAPKs. Immunobiology 2013, 218, 628–634. [Google Scholar] [CrossRef]
- Yusupov, M.; Wende, K.; Kupsch, S.; Neyts, E.C.; Reuter, S.; Bogaerts, A. Effect of head group and lipid tail oxidation in the cell membrane revealed through integrated simulations and experiments. Sci. Rep. 2017, 7, 5761. [Google Scholar] [CrossRef]
- Morgan, M.J.; Liu, Z.-G. Crosstalk of reactive oxygen species and NF-κB signaling. Cell Res. 2011, 21, 103–115. [Google Scholar] [CrossRef]
- Gerner, M.C.; Niederstaetter, L.; Ziegler, L.; Bileck, A.; Slany, A.; Janker, L.; Schmidt, R.L.J.; Gerner, C.; del Favero, G.; Schmetterer, K.G. Proteome Analysis Reveals Distinct Mitochondrial Functions Linked to Interferon Response Patterns in Activated CD4+ and CD8+ T Cells. Front. Pharmacol. 2019, 10, 727. [Google Scholar] [CrossRef]
- Attwood, P.V.; Muimo, R. The actions of NME1/NDPK-A and NME2/NDPK-B as protein kinases. Lab. Invest. 2018, 98, 283–290. [Google Scholar]
- Boulter, E.; Garcia-Mata, R.; Guilluy, C.; Dubash, A.; Rossi, G.; Brennwald, P.J.; Burridge, K. Regulation of Rho GTPase crosstalk, degradation and activity by RhoGDI1. Nat. Cell Biol. 2010, 12, 477–483. [Google Scholar] [CrossRef]
- Bozza, W.P.; Zhang, Y.; Hallett, K.; Rosado, L.A.R.; Zhang, B. RhoGDI deficiency induces constitutive activation of Rho GTPases and COX-2 pathways in association with breast cancer progression. Oncotarget 2015, 6, 32723–32736. [Google Scholar] [CrossRef][Green Version]
- Otto, A.; Müller, C.; Huff, T.; Hannappel, E. Chemotherapeutic drugs change actin skeleton organization and the expression of beta-thymosins in human breast cancer cells. J. Cancer Res. Clin. Oncol. 2002, 128, 247–256. [Google Scholar]
- Vlijm, R.; Li, X.; Panic, M.; Rüthnick, D.; Hata, S.; Herrmannsdörfer, F.; Kuner, T.; Heilemann, M.; Engelhardt, J.; Hell, S.W.; et al. STED nanoscopy of the centrosome linker reveals a CEP68-organized, periodic rootletin network anchored to a C-Nap1 ring at centrioles. Proc. Natl. Acad. Sci. USA 2018, 115, E2246–E2253. [Google Scholar] [CrossRef]
- Baietti, M.F.; Zhang, Z.; Mortier, E.; Melchior, A.; Degeest, G.; Geeraerts, A.; Ivarsson, Y.; Depoortere, F.; Coomans, C.; Vermeiren, E.; et al. Syndecan–syntenin–ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012, 14, 677–685. [Google Scholar] [CrossRef]
- Frank, P.G.; Marcel, Y.L.; Connelly, M.A.; Lublin, D.M.; Franklin, V.; Williams, D.L.; Lisanti, M.P. Stabilization of caveolin-1 by cellular cholesterol and scavenger receptor class B type I. Biochemistry 2002, 41, 11931–11940. [Google Scholar] [CrossRef] [PubMed]
- Jiao, H.; Zhang, Y.; Yan, Z.; Wang, Z.G.; Liu, G.; Minshall, R.D.; Malik, A.B.; Hu, G. Caveolin-1 Tyr14 phosphorylation induces interaction with TLR4 in endothelial cells and mediates MyD88-dependent signaling and sepsis-induced lung inflammation. J. Immunol. 1950 2013, 191, 6191–6199. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Hirabayashi, Y. Caveolin-1 prevents palmitate-induced NF-kappaB signaling by inhibiting GPRC5B-phosphorylation. Biochem. Biophys. Res. Commun. 2018, 503, 2673–2677. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.Y.; Lai, T.Y.; Tsai, M.K.; Chang, Y.C.; Ho, Y.H.; Yu, I.S.; Yeh, T.W.; Chou, C.C.; Lin, Y.S.; Lawrence, T.; et al. The ubiquitin ligase ZNRF1 promotes caveolin-1 ubiquitination and degradation to modulate inflammation. Nat. Commun. 2017, 8, 15502. [Google Scholar] [CrossRef]
- De Almeida, C.J.G. Caveolin-1 and Caveolin-2 Can Be Antagonistic Partners in Inflammation and Beyond. Front. Immunol. 2017, 8, 1530. [Google Scholar] [CrossRef]
- Yeh, Y.C.; Ling, J.Y.; Chen, W.C.; Lin, H.H.; Tang, M.J. Mechanotransduction of matrix stiffness in regulation of focal adhesion size and number: Reciprocal regulation of caveolin-1 and beta1 integrin. Sci. Rep. 2017, 7, 15008. [Google Scholar] [CrossRef]
- Wilde, L.; Roche, M.; Domingo-Vidal, M.; Tanson, K.; Philp, N.; Curry, J.; Martinez-Outschoorn, U. Metabolic coupling and the Reverse Warburg Effect in cancer: Implications for novel biomarker and anticancer agent development. Semin. Oncol. 2017, 44, 198–203. [Google Scholar] [CrossRef]
- Busija, A.R.; Patel, H.H.; Insel, P.A. Caveolins and cavins in the trafficking, maturation, and degradation of caveolae: Implications for cell physiology. Am. J. Physiol. Cell Physiol. 2017, 312, C459–C477. [Google Scholar] [CrossRef] [PubMed]
- Dellafiora, L.; Galaverna, G.; Dall’Asta, C.; Cozzini, P. Hazard identification of cis/trans-zearalenone through the looking-glass. Food Chem. Toxicol. 2015, 86, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Tao, X.; Shen, B.; Horng, T.; Medzhitov, R.; Manley, J.L.; Tong, L. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature 2000, 408, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Kain, V.; Halade, G.V. Immune responsive resolvin D1 programs peritoneal macrophages and cardiac fibroblast phenotypes in diversified metabolic microenvironment. J. Cell. Physiol. 2018, 234, 3910–3920. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Lee, M.Y.; Jeon, Y.J. Silymarin Inhibits Morphological Changes in LPS-Stimulated Macrophages by Blocking NF-kappaB Pathway. Korean J. Physiol. Pharmacol. 2015, 19, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Graziani, A.; Bricko, V.; Carmignani, M.; Graier, W.F.; Groschner, K. Cholesterol- and caveolin-rich membrane domains are essential for phospholipase A2-dependent EDHF formation. Cardiovasc. Res. 2004, 64, 234–242. [Google Scholar] [CrossRef]
- Jagielska, J.; Kapopara, P.R.; Salguero, G.; Scherr, M.; Schutt, H.; Grote, K.; Schieffer, B.; Bavendiek, U. Interleukin-1 assembles a proangiogenic signaling module consisting of caveolin-1, tumor necrosis factor receptor-associated factor 6, p38-mitogen-activated protein kinase (MAPK), and MAPK-activated protein kinase 2 in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 1280–1288. [Google Scholar] [CrossRef]
- Zheng, Y.Z.; Boscher, C.; Inder, K.L.; Fairbank, M.; Loo, D.; Hill, M.M.; Nabi, I.R.; Foster, L.J. Differential impact of caveolae and caveolin-1 scaffolds on the membrane raft proteome. Mol. Cell. Proteomics 2011, 10, M110.007146. [Google Scholar] [CrossRef]
- Martins, D.; Beca, F.F.; Sousa, B.; Baltazar, F.; Paredes, J.; Schmitt, F. Loss of caveolin-1 and gain of MCT4 expression in the tumor stroma: Key events in the progression from an in situ to an invasive breast carcinoma. Cell Cycle 2013, 12, 2684–2890. [Google Scholar] [CrossRef]
- Hunter, I.; Nixon, G.F. Spatial compartmentalization of tumor necrosis factor (TNF) receptor 1-dependent signaling pathways in human airway smooth muscle cells. Lipid rafts are essential for TNF-alpha-mediated activation of RhoA but dispensable for the activation of the NF-kappaB and MAPK pathways. J. Biol. Chem. 2006, 281, 34705–34715. [Google Scholar] [PubMed]
- Kagawa, Y.; Yasumoto, Y.; Sharifi, K.; Ebrahimi, M.; Islam, A.; Miyazaki, H.; Yamamoto, Y.; Sawada, T.; Kishi, H.; Kobayashi, S.; et al. Fatty acid-binding protein 7 regulates function of caveolae in astrocytes through expression of caveolin-1. Glia 2015, 63, 780–794. [Google Scholar] [CrossRef] [PubMed]
- Pohl, J.; Ring, A.; Stremmel, W. Uptake of long-chain fatty acids in HepG2 cells involves caveolae: Analysis of a novel pathway. J. Lipid Res. 2002, 43, 1390–1399. [Google Scholar] [CrossRef]
- Astudillo, A.M.; Perez-Chacon, G.; Meana, C.; Balgoma, D.; Pol, A.; del Pozo, M.A.; Balboa, M.A.; Balsinde, J. Altered arachidonate distribution in macrophages from caveolin-1 null mice leading to reduced eicosanoid synthesis. J. Biol. Chem. 2011, 286, 35299–35307. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Guo, L.; Gao, H.; Li, X.-A. Caveolar Fatty Acids and Acylation of Caveolin-1. PLoS ONE 2013, 8, e60884. [Google Scholar] [CrossRef]
- Dellafiora, L.; Warth, B.; Schmidt, V.; del Favero, G.; Mikula, H.; Frohlich, J.; Marko, D. An integrated in silico/in vitro approach to assess the xenoestrogenic potential of Alternaria mycotoxins and metabolites. Food Chem. 2018, 248, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Dainese, E.; Sabatucci, A.; Angelucci, C.B.; Barsacchi, D.; Chiarini, M.; Maccarrone, M. Impact of embedded endocannabinoids and their oxygenation by lipoxygenase on membrane properties. ACS Chem. Neurosci. 2012, 3, 386–392. [Google Scholar] [CrossRef]
- Pereira-Leite, C.; Nunes, C.; Reis, S. Interaction of nonsteroidal anti-inflammatory drugs with membranes: In vitro assessment and relevance for their biological actions. Prog. Lipid Res. 2013, 52, 571–584. [Google Scholar] [CrossRef]
- Sousa, C.; Nunes, C.; Lucio, M.; Ferreira, H.; Lima, J.L.; Tavares, J.; Cordeiro-da-Silva, A.; Reis, S. Effect of nonsteroidal anti-inflammatory drugs on the cellular membrane fluidity. J. Pharm. Sci. 2008, 97, 3195–3206. [Google Scholar] [CrossRef]
- De la Roche, M.; Hamilton, C.; Mortensen, R.; Jeyaprakash, A.A.; Ghosh, S.; Anand, P.K. Trafficking of cholesterol to the ER is required for NLRP3 inflammasome activation. J. Cell Biol. 2018, 217, 3560–3576. [Google Scholar] [CrossRef]
- Lehmann, L.; Wagner, J.; Metzler, M. Estrogenic and clastogenic potential of the mycotoxin alternariol in cultured mammalian cells. Food Chem. Toxicol. 2006, 44, 398–408. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Del Favero, G.; Mayer, R.M.; Dellafiora, L.; Janker, L.; Niederstaetter, L.; Dall’Asta, C.; Gerner, C.; Marko, D. Structural Similarity with Cholesterol Reveals Crucial Insights into Mechanisms Sustaining the Immunomodulatory Activity of the Mycotoxin Alternariol. Cells 2020, 9, 847. https://doi.org/10.3390/cells9040847
Del Favero G, Mayer RM, Dellafiora L, Janker L, Niederstaetter L, Dall’Asta C, Gerner C, Marko D. Structural Similarity with Cholesterol Reveals Crucial Insights into Mechanisms Sustaining the Immunomodulatory Activity of the Mycotoxin Alternariol. Cells. 2020; 9(4):847. https://doi.org/10.3390/cells9040847
Chicago/Turabian StyleDel Favero, Giorgia, Raphaela M. Mayer, Luca Dellafiora, Lukas Janker, Laura Niederstaetter, Chiara Dall’Asta, Christopher Gerner, and Doris Marko. 2020. "Structural Similarity with Cholesterol Reveals Crucial Insights into Mechanisms Sustaining the Immunomodulatory Activity of the Mycotoxin Alternariol" Cells 9, no. 4: 847. https://doi.org/10.3390/cells9040847
APA StyleDel Favero, G., Mayer, R. M., Dellafiora, L., Janker, L., Niederstaetter, L., Dall’Asta, C., Gerner, C., & Marko, D. (2020). Structural Similarity with Cholesterol Reveals Crucial Insights into Mechanisms Sustaining the Immunomodulatory Activity of the Mycotoxin Alternariol. Cells, 9(4), 847. https://doi.org/10.3390/cells9040847







