TNFa and IL2 Encoding Oncolytic Adenovirus Activates Pathogen and Danger-Associated Immunological Signaling
Abstract
1. Introduction
2. Results
2.1. OAd.TNFa-IL2 Induces Immunostimulating Microenvironmental Changes Through DAMP and PAMP Release and Expression
2.2. Oncolytic Adenovirus Therapy Affects the Transcriptome Landscape of Both Cancer and Immune Cells
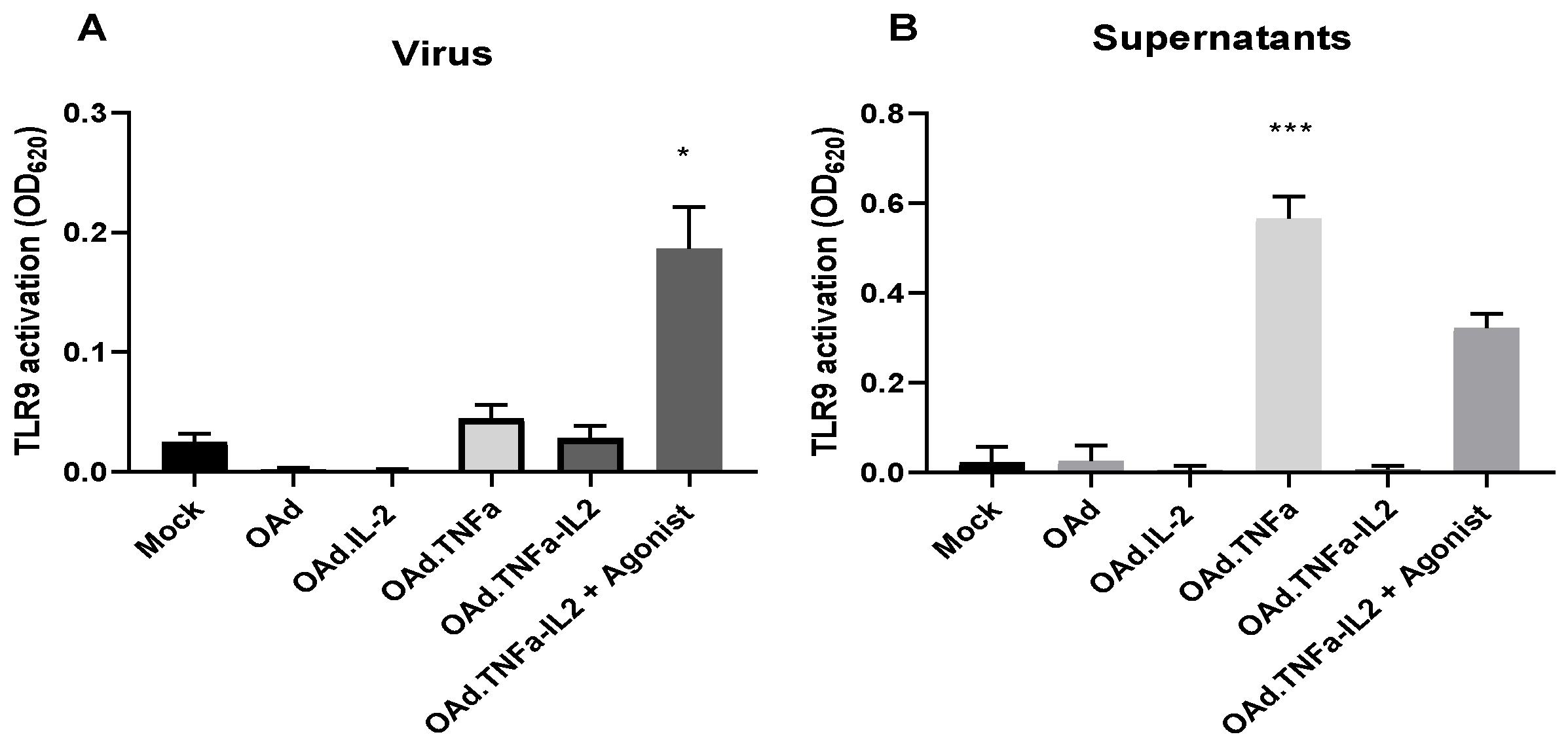
2.3. NFκB-Related Genes Are Downregulated During OAd.TNFa-IL2 Infection, But TLR9 Signaling Is Not Activated by OAd.TNFa-IL2
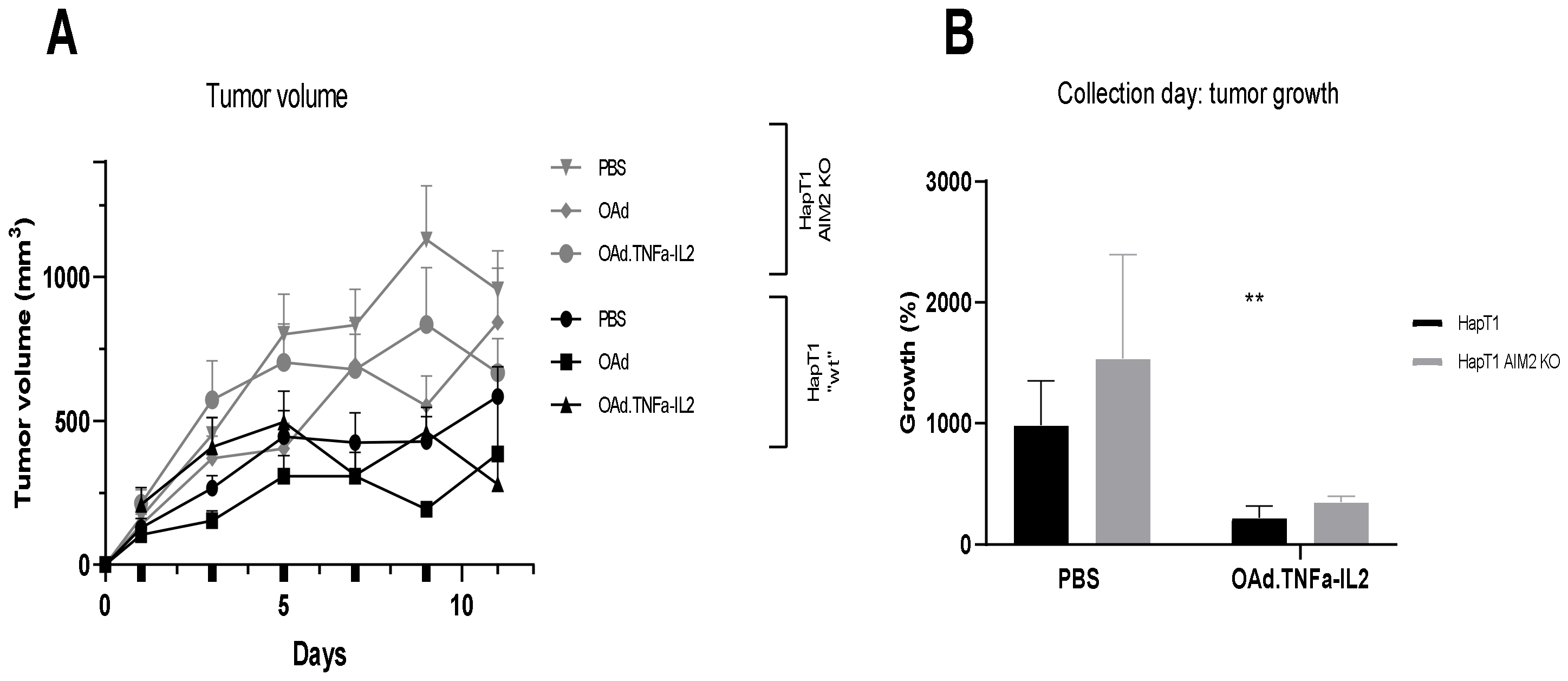
2.4. Knockout of AIM2 Diminishes the Effect of Virotherapy
2.5. Virotherapy Induces AIM2-Dependent, Pro-Inflammatory Tumor Landscape Modification In Vivo
3. Materials and Methods
3.1. Cell Lines and Viruses
3.2. DAMP and PAMP Measurement
3.3. RNA-Sequencing
3.4. NFκB Signaling Pathway Analysis
3.5. Flow Cytometry
3.6. In Vivo Analysis
3.7. Real-Time Quantitative PCR Analysis of Tumors
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Datta, M.; Coussens, L.M.; Nishikawa, H.; Hodi, F.S.; Jain, R.K. Reprogramming the Tumor Microenvironment to Improve Immunotherapy: Emerging Strategies and Combination Therapies. Am. Soc. Clin. Oncol. Educ. Book 2019, 39, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Heiniö, C.; Sorsa, S.; Siurala, M.; Grönberg-Vähä-Koskela, S.; Havunen, R.; Haavisto, E.; Koski, A.; Hemminki, O.; Zafar, S.; Cervera-Carrascon, V.; et al. Effect of Genetic Modifications on Physical and Functional Titers of Adenoviral Cancer Gene Therapy Constructs. Hum. Gene. Ther. 2019, 30, 740–752. [Google Scholar] [CrossRef]
- Corrigan, P.A.; Beaulieu, C.; Patel, R.B.; Lowe, D.K. Talimogene Laherparepvec: An Oncolytic Virus Therapy for Melanoma. Ann. Pharmacother. 2017, 51, 675–681. [Google Scholar] [CrossRef]
- Andtbacka, R.H.; Kaufman, H.L.; Collichio, F.; Amatruda, T.; Senzer, N.; Chesney, J.; Delman, K.A.; Spitler, L.E.; Puzanov, I.; Agarwala, S.S.; et al. Talimogene Laherparepvec Improves Durable Response Rate in Patients with Advanced Melanoma. J. Clin. Oncol. 2015, 33, 2780–2788. [Google Scholar] [CrossRef]
- Havunen, R.; Siurala, M.; Sorsa, S.; Grönberg-Vähä-Koskela, S.; Behr, M.; Tähtinen, S.; Santos, J.M.; Karell, P.; Rusanen, J.; Nettelbeck, D.M.; et al. Oncolytic Adenoviruses Armed with Tumor Necrosis Factor Alpha and Interleukin-2 Enable Successful Adoptive Cell Therapy. Mol. Ther. Oncolytics 2017, 4, 77–86. [Google Scholar] [CrossRef]
- Sato, E.; Olson, S.H.; Ahn, J.; Bundy, B.; Nishikawa, H.; Qian, F.; Jungbluth, A.A.; Frosina, D.; Gnjatic, S.; Ambrosone, C.; et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc. Natl. Acad. Sci. USA 2005, 102, 18538–18543. [Google Scholar] [CrossRef]
- Dieci, M.V.; Mathieu, M.C.; Guarneri, V.; Conte, P.; Delaloge, S.; Andre, F.; Goubar, A. Prognostic and predictive value of tumor-infiltrating lymphocytes in two phase III randomized adjuvant breast cancer trials. Ann. Oncol. 2015, 26, 1698–1704. [Google Scholar] [CrossRef]
- Chen, I.F.; Ou-Yang, F.; Hung, J.Y.; Liu, J.C.; Wang, H.; Wang, S.C.; Hou, M.F.; Hortobagyi, G.N.; Hung, M.C. AIM2 suppresses human breast cancer cell proliferation in vitro and mammary tumor growth in a mouse model. Mol. Cancer Ther. 2006, 5, 1–7. [Google Scholar] [CrossRef]
- Chon, H.J.; Kim, H.; Noh, J.H.; Yang, H.; Lee, W.S.; Kong, S.J.; Lee, S.J.; Lee, Y.S.; Kim, W.R.; Kim, J.H.; et al. STING signaling is a potential immunotherapeutic target in colorectal cancer. J. Cancer 2019, 10, 4932–4938. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Z.; Yu, S. AIM2 regulates viability and apoptosis in human colorectal cancer cells via the PI3K/Akt pathway. Onco Targets Ther. 2017, 10, 811–817. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Yang, G.; Bai, P.; Gui, S.; Nyuyen, T.M.; Mercado-Uribe, I.; Yang, M.; Zou, J.; Li, Q.; Xiao, J.; et al. Inhibition of nuclear factor-kappa B enhances the tumor growth of ovarian cancer cell line derived from a low-grade papillary serous carcinoma in p53-independent pathway. BMC Cancer 2016, 16, 582. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Tan, S.; Zhou, Y.; Lin, J.; Wang, H.; Oyang, L.; Tian, Y.; Liu, L.; Su, M.; Cao, D.; et al. Role of the NFkappaB-signaling pathway in cancer. Onco Targets Ther. 2018, 11, 2063–2073. [Google Scholar] [CrossRef]
- Xia, Y.; Shen, S.; Verma, I.M. NF-κB, an active player in human cancers. Cancer Immunol. Res. 2014, 2, 823–830. [Google Scholar] [CrossRef]
- Wang, B.; Yin, Q. AIM2 inflammasome activation and regulation: A structural perspective. J. Struct. Biol. 2017, 200, 279–282. [Google Scholar] [CrossRef]
- Man, S.M.; Karki, R.; Kanneganti, T.D. AIM2 inflammasome in infection, cancer, and autoimmunity: Role in DNA sensing, inflammation, and innate immunity. Eur. J. Immunol. 2016, 46, 269–280. [Google Scholar] [CrossRef]
- De Koning, H.D.; van Vlijmen-Willems, I.M.; Zeeuwen, P.L.; Blokx, W.A.; Schalkwijk, J. Absent in Melanoma 2 is predominantly present in primary melanoma and primary squamous cell carcinoma, but largely absent in metastases of both tumors. J. Am. Acad. Dermatol. 2014, 71, 1012–1015. [Google Scholar] [CrossRef]
- Schierer, S.; Hesse, A.; Müller, I.; Kämpgen, E.; Curiel, D.T.; Schuler, G.; Steinkasserer, A.; Nettelbeck, D.M. Modulation of viability and maturation of human monocyte-derived dendritic cells by oncolytic adenoviruses. Int. J. Cancer 2008, 122, 219–229. [Google Scholar] [CrossRef]
- Knuefermann, P.; Chen, P.; Misra, A.; Shi, S.P.; Abdellatif, M.; Sivasubramanian, N. Myotrophin/V-1, a protein up-regulated in the failing human heart and in postnatal cerebellum, converts NFkappa B p50-p65 heterodimers to p50-p50 and p65-p65 homodimers. J. Biol. Chem. 2002, 277, 23888–23897. [Google Scholar] [CrossRef]
- Siurala, M.; Havunen, R.; Saha, D.; Lumen, D.; Airaksinen, A.J.; Tähtinen, S.; Cervera-Carrascon, V.; Bramante, S.; Parviainen, S.; Vähä-Koskela, M.; et al. Adenoviral Delivery of Tumor Necrosis Factor-α and Interleukin-2 Enables Successful Adoptive Cell Therapy of Immunosuppressive Melanoma. Mol. Ther. 2016, 24, 1435–1443. [Google Scholar] [CrossRef]
- Wang, H.T.; Lee, H.I.; Guo, J.H.; Chen, S.H.; Liao, Z.K.; Huang, K.W.; Torng, P.L.; Hwang, L.H. Calreticulin promotes tumor lymphocyte infiltration and enhances the antitumor effects of immunotherapy by up-regulating the endothelial expression of adhesion molecules. Int. J. Cancer 2012, 130, 2892–2902. [Google Scholar] [CrossRef] [PubMed]
- Vénéreau, E.; Ceriotti, C.; Bianchi, M.E. DAMPs from Cell Death to New Life. Front. Immunol. 2015, 6, 422. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.K.; Roth, J.; Oppenheim, J.J.; Tracey, K.J.; Vogl, T.; Feldmann, M.; Horwood, N.; Nanchahal, J. Alarmins: Awaiting a clinical response. J. Clin. Investig. 2012, 122, 2711–2719. [Google Scholar] [CrossRef]
- Krysko, O.; Løve Aaes, T.; Bachert, C.; Vandenabeele, P.; Krysko, D.V. Many faces of DAMPs in cancer therapy. Cell Death Dis. 2013, 4, e631. [Google Scholar] [CrossRef] [PubMed]
- La Sala, A.; Ferrari, D.; Corinti, S.; Cavani, A.; Di Virgilio, F.; Girolomoni, G. Extracellular ATP induces a distorted maturation of dendritic cells and inhibits their capacity to initiate Th1 responses. J. Immunol. 2001, 166, 1611–1617. [Google Scholar] [CrossRef] [PubMed]
- Cerullo, V.; Seiler, M.P.; Mane, V.; Brunetti-Pierri, N.; Clarke, C.; Bertin, T.K.; Rodgers, J.R.; Lee, B. Toll-like receptor 9 triggers an innate immune response to helper-dependent adenoviral vectors. Mol. Ther. 2007, 15, 378–385. [Google Scholar] [CrossRef]
- Kaczanowska, S.; Joseph, A.M.; Davila, E. TLR agonists: Our best frenemy in cancer immunotherapy. J. Leukoc. Biol. 2013, 93, 847–863. [Google Scholar] [CrossRef]
- Chen, M.F.; Chen, P.T.; Lu, M.S.; Lin, P.Y.; Chen, W.C.; Lee, K.D. IL-6 expression predicts treatment response and outcome in squamous cell carcinoma of the esophagus. Mol. Cancer 2013, 12, 26. [Google Scholar] [CrossRef]
- Nish, S.A.; Schenten, D.; Wunderlich, F.T.; Pope, S.D.; Gao, Y.; Hoshi, N.; Yu, S.; Yan, X.; Lee, H.K.; Pasman, L.; et al. T cell-intrinsic role of IL-6 signaling in primary and memory responses. eLife 2014, 3, e01949. [Google Scholar] [CrossRef]
- Ross, S.H.; Cantrell, D.A. Signaling and Function of Interleukin-2 in T Lymphocytes. Annu. Rev. Immunol. 2018, 36, 411–433. [Google Scholar] [CrossRef]
- Parameswaran, N.; Patial, S. Tumor necrosis factor-α signaling in macrophages. Crit. Rev. Eukaryot. Gene Expr. 2010, 20, 87–103. [Google Scholar] [CrossRef] [PubMed]



| SK-MEL-28 | ||||
|---|---|---|---|---|
| Gene | Abbreviation | Log2 Ratio (Infected versus Uninfected Cancer Cells) | Up- or Downregulation | p-Value |
| Interleukin 2 | IL-2 | 11.620 | Up | 0 |
| Tumor necrosis factor alfa | TNFa | 11.388 | Up | 0 |
| Ubiquitin | UBD | 9.958 | Up | 1.284006 × 10−182 |
| Histone H2A type 1 | HIST1H2AI | 9.935 | Up | 1.307986 × 10−08 |
| Chemokine (C-C motif) ligand 2 | CCL2 | 9.527 | Up | 6.97892 × 10 −71 |
| Abhydrolase domain containing 14A and aminoacylase 1 (read-through transcription) | ABHD14A-ACY1 | 8.991 | Up | 1.422174 × 1018 |
| Myotrophin | MTPN | 8.969 | Up | 1.681326 × 10−38 |
| Interleukin 32 | IL32 | 8.394 | Up | 9.93188 × 10−74 |
| DC | ||||
| Gene | Abbreviation | Log2 Ratio (Infected versus Uninfected DC) | Up or Down-Regulation | p-Value |
| Myothrophin | MTPN | 9.629 | Up | 3.8971 × 10−65 |
| Locus101929802 | LOC101929802 | 6.931 | Up | 9.91084 × 10−14 |
| Locus102724994 | LOC102724994 | 6.919 | Up | 8.93212 × 10−07 |
| Zink finger816-Zink finger321P | ZNF816-ZNF321P | 6.443 | Up | 1.447082 × 10−05 |
| Heat shock 10kDa protein 1 and MOB family member 4, phocein (read-through transcription) | HSPE1-MOB4 | 5.807 | Up | 1.447082 × 10−05 |
| Nuclear Pore Complex Interacting Protein Family Member A3 | NPIPA3 | 4.518 | Up | 1.870752 × 10−16 |
| Chemokine (C-X-C motif) ligand 9 | CXCL9 | 3.0 | Up | 2.73386 × 10−10 |
| Platelet glycoprotein Ib alpha chain | GP1BA | 2.269 | Up | 7.3774 × 10−106 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heiniö, C.; Havunen, R.; Santos, J.; de Lint, K.; Cervera-Carrascon, V.; Kanerva, A.; Hemminki, A. TNFa and IL2 Encoding Oncolytic Adenovirus Activates Pathogen and Danger-Associated Immunological Signaling. Cells 2020, 9, 798. https://doi.org/10.3390/cells9040798
Heiniö C, Havunen R, Santos J, de Lint K, Cervera-Carrascon V, Kanerva A, Hemminki A. TNFa and IL2 Encoding Oncolytic Adenovirus Activates Pathogen and Danger-Associated Immunological Signaling. Cells. 2020; 9(4):798. https://doi.org/10.3390/cells9040798
Chicago/Turabian StyleHeiniö, Camilla, Riikka Havunen, Joao Santos, Klaas de Lint, Victor Cervera-Carrascon, Anna Kanerva, and Akseli Hemminki. 2020. "TNFa and IL2 Encoding Oncolytic Adenovirus Activates Pathogen and Danger-Associated Immunological Signaling" Cells 9, no. 4: 798. https://doi.org/10.3390/cells9040798
APA StyleHeiniö, C., Havunen, R., Santos, J., de Lint, K., Cervera-Carrascon, V., Kanerva, A., & Hemminki, A. (2020). TNFa and IL2 Encoding Oncolytic Adenovirus Activates Pathogen and Danger-Associated Immunological Signaling. Cells, 9(4), 798. https://doi.org/10.3390/cells9040798





