Exosomes are the Driving Force in Preparing the Soil for the Metastatic Seeds: Lessons from the Prostate Cancer
Abstract
1. Introduction
2. The Soil/Seed Analogy: Tumor Microenvironment (TME) and Tumor Cells
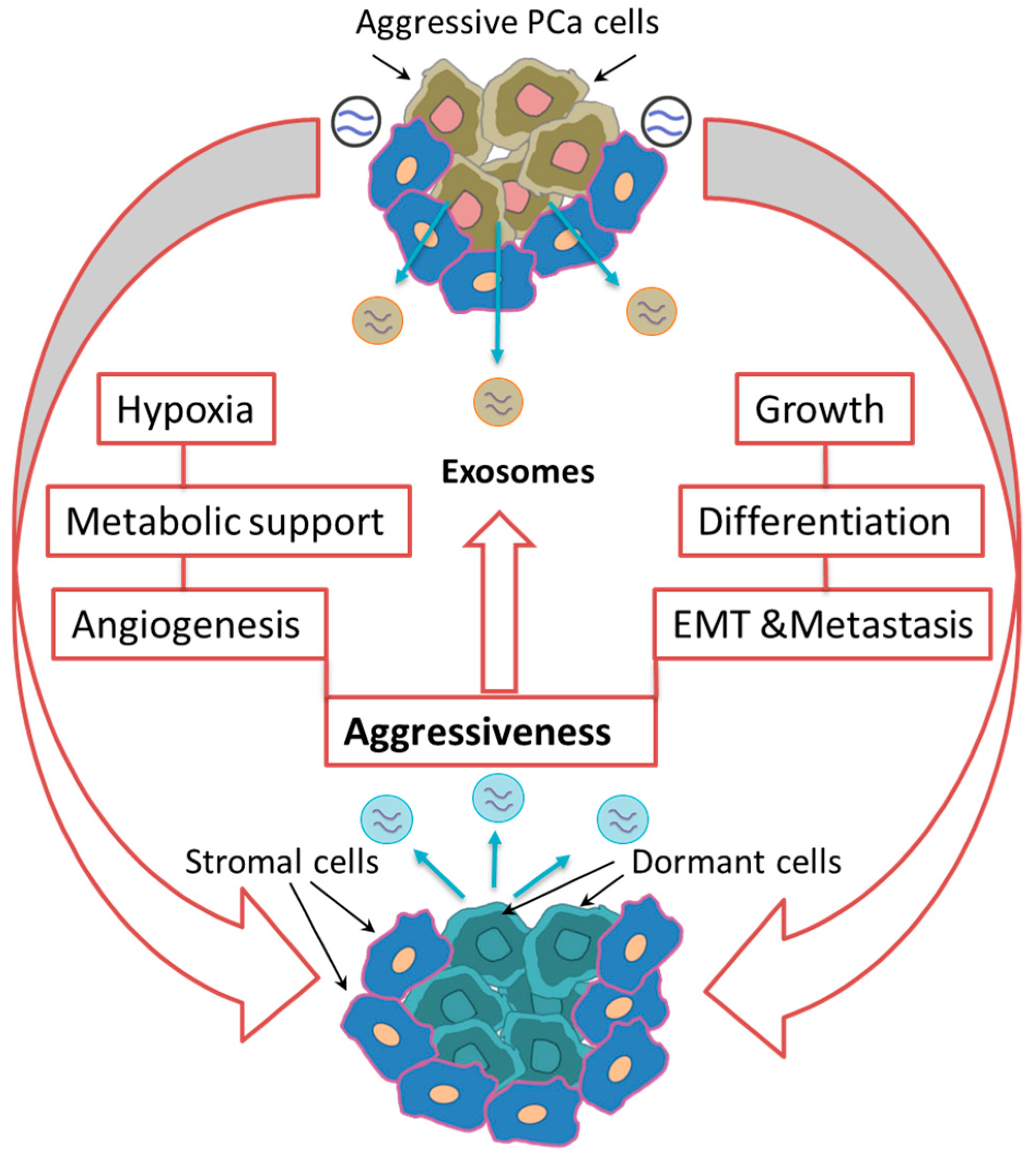
3. Tumor-Associated Exosomes Modulate the TME and Prepare the Metastatic Niche
3.1. Exosomes, Biogenesis, Trafficking, Uptake and Exosomal Cargo
3.2. Singing Together: PCa Cell-Cell Interaction via Exosomal Signals
3.3. Effect of Exosomal Cargo on the Metabolic Reprogramming of Stromal Cells in the TME
3.4. Desmoplastic Response of Stromal Cells to the PCa-Derived Exosomal Proteome
3.5. Exosomal Lipids as Messengers in PCA-Stromal Cell Crosstalk
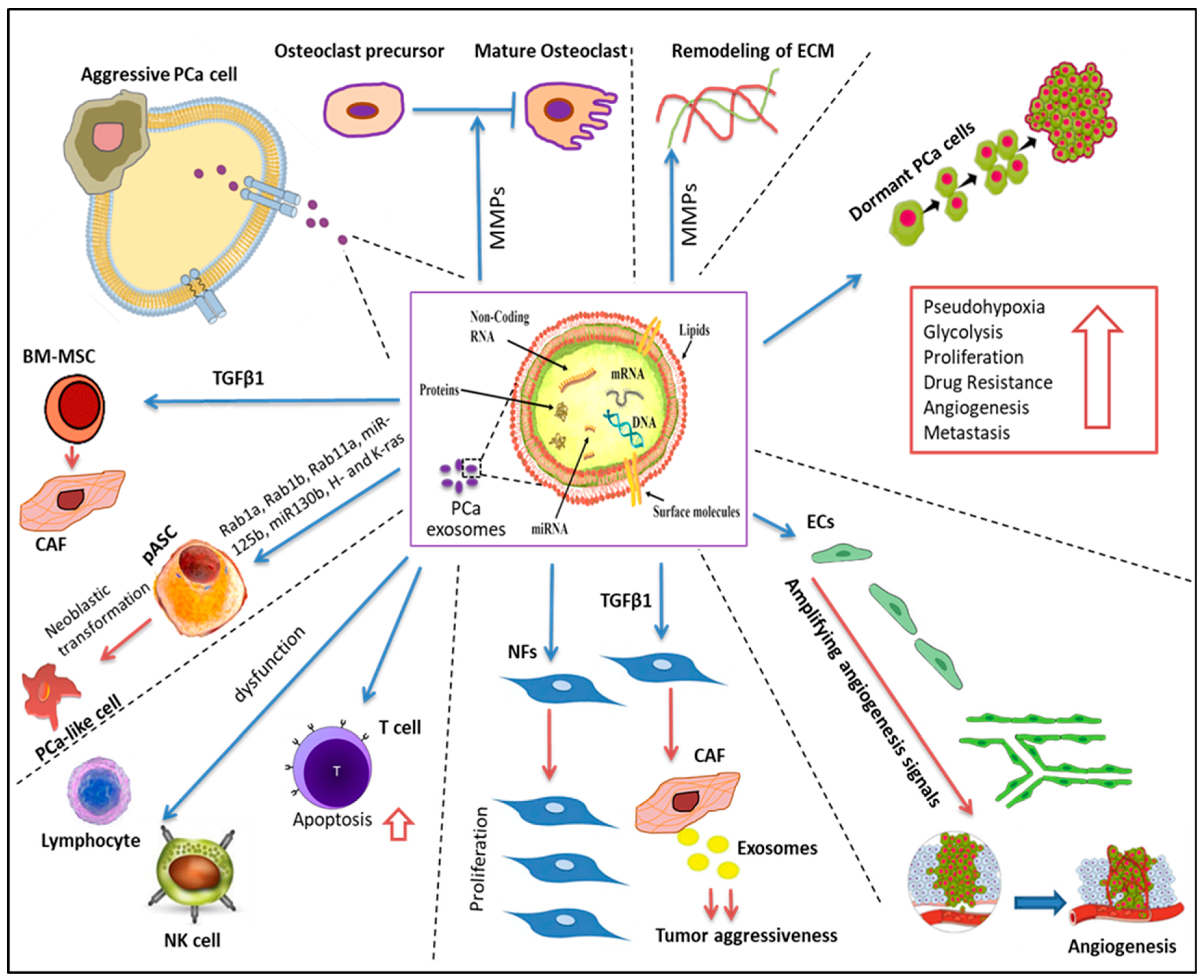
3.6. The Role of Exosomes in Shuttling Bioactive Materials between PCA and Stromal Cells
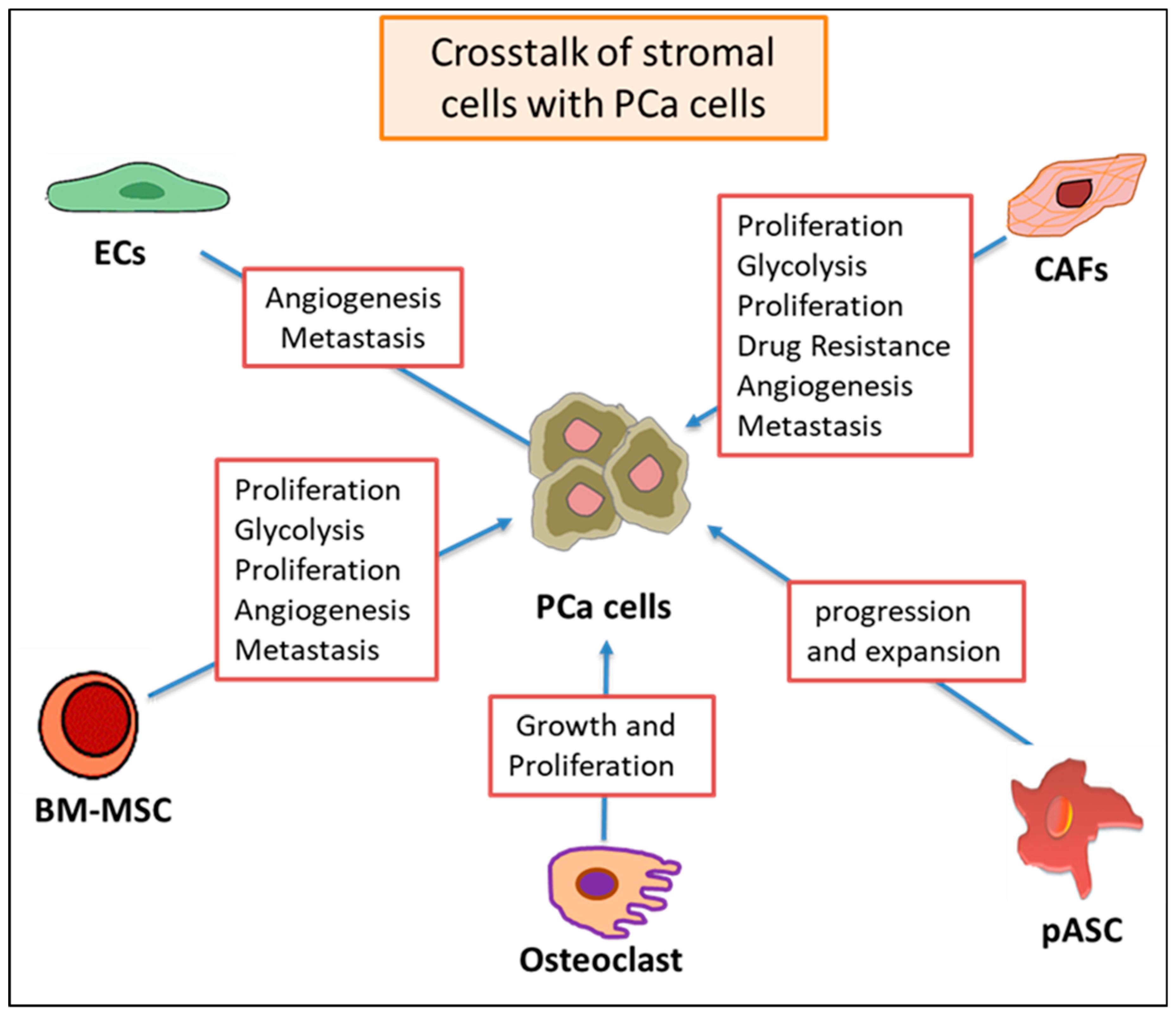
3.7. Two-Way Crosstalk between Stromal Cells and PCA Cell-Associated Exosomal Cargo
4. The Immune-Modulatory Role of Exosomes
5. Targeting the Tumor Microenvironment
6. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Minciacchi, V.; Zijlstra, A.; Rubin, M.A.; Di Vizio, D. Extracellular vesicles for liquid biopsy in prostate cancer: Where are we and where are we headed? Prostate Cancer Prostatic Dis. 2017, 20, 251. [Google Scholar] [CrossRef] [PubMed]
- Shukla, M.E.; Yu, C.; Reddy, C.A.; Stephans, K.L.; Klein, E.A.; Abdel-Wahab, M.; Ciezki, J.; Tendulkar, R.D. Evaluation of the current prostate cancer staging system based on cancer-specific mortality in the surveillance epidemiology and end results database. Clin. Genitourin Cancer 2015, 13, 17–21. [Google Scholar] [CrossRef] [PubMed]
- American Cancer Society. Cancer Facts & Figures 2019; American Cancer Society: Atlantaga, GA, USA, 2019. [Google Scholar]
- Endzeliņš, E.; Melne, V.; Kalniņa, Z.; Lietuvietis, V.; Riekstiņa, U.; Llorente, A.; Linē, A. Diagnostic, prognostic and predictive value of cell-free miRNAs in prostate cancer: A systematic review. Mol. Cancer 2016, 15, 41. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, L. Closing the controversies gap in prostate cancer? Nat. Rev. Clin. Oncol. 2014, 11, 299. [Google Scholar] [CrossRef]
- Cheville, J.C.; Tindall, D.; Boelter, C.; Jenkins, R.; Lohse, C.M.; Pankratz, V.S.; Sebo, T.J.; Davis, B.; Blute, M.L. Metastatic prostate carcinoma to bone: Clinical and pathologic features associated with cancer-specific survival. Cancer 2002, 95, 1028–1036. [Google Scholar] [CrossRef]
- Scher, H.I.; Halabi, S.; Tannock, I.; Morris, M.; Sternberg, C.N.; Carducci, M.A.; Eisenberger, M.A.; Higano, C.; Bubley, G.J.; Dreicer, R. Design and end points of clinical trials for patients with progressive prostate cancer and castrate levels of testosterone: Recommendations of the Prostate Cancer Clinical Trials Working Group. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2008, 26, 1148. [Google Scholar] [CrossRef]
- Chen, C.D.; Welsbie, D.S.; Tran, C.; Baek, S.H.; Chen, R.; Vessella, R.; Rosenfeld, M.G.; Sawyers, C.L. Molecular determinants of resistance to antiandrogen therapy. Nat. Med. 2004, 10, 33. [Google Scholar] [CrossRef]
- Debes, J.D.; Tindall, D.J. Mechanisms of androgen-refractory prostate cancer. New Engl. J. Med. 2004, 351, 1488–1490. [Google Scholar] [CrossRef]
- Hu, R.; Dunn, T.A.; Wei, S.; Isharwal, S.; Veltri, R.W.; Humphreys, E.; Han, M.; Partin, A.W.; Vessella, R.L.; Isaacs, W.B. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009, 69, 16–22. [Google Scholar] [CrossRef]
- Stanbrough, M.; Bubley, G.J.; Ross, K.; Golub, T.R.; Rubin, M.A.; Penning, T.M.; Febbo, P.G.; Balk, S.P. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res. 2006, 66, 2815–2825. [Google Scholar] [CrossRef]
- Holzbeierlein, J.; Lal, P.; LaTulippe, E.; Smith, A.; Satagopan, J.; Zhang, L.; Ryan, C.; Smith, S.; Scher, H.; Scardino, P. Gene expression analysis of human prostate carcinoma during hormonal therapy identifies androgen-responsive genes and mechanisms of therapy resistance. Am. J. Pathol. 2004, 164, 217–227. [Google Scholar] [CrossRef]
- Agarwal, N.; Hutson, T.E.; Vogelzang, N.J.; Sonpavde, G. Abiraterone acetate: A promising drug for the treatment of castration-resistant prostate cancer. Future Oncol. 2010, 6, 665–679. [Google Scholar] [CrossRef] [PubMed]
- Sonpavde, G.; Attard, G.; Bellmunt, J.; Mason, M.D.; Malavaud, B.; Tombal, B.; Sternberg, C.N. The role of abiraterone acetate in the management of prostate cancer: A critical analysis of the literature. Eur. Urol. 2011, 60, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Attard, G.; Reid, A.H.; Auchus, R.J.; Hughes, B.A.; Cassidy, A.M.; Thompson, E.; Oommen, N.B.; Folkerd, E.; Dowsett, M.; Arlt, W. Clinical and biochemical consequences of CYP17A1 inhibition with abiraterone given with and without exogenous glucocorticoids in castrate men with advanced prostate cancer. J. Clin. Endocrinol. Metab. 2012, 97, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Attard, G.; Reid, A.H.; A’Hern, R.; Parker, C.; Oommen, N.B.; Folkerd, E.; Messiou, C.; Molife, L.R.; Maier, G.; Thompson, E. Selective inhibition of CYP17 with abiraterone acetate is highly active in the treatment of castration-resistant prostate cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2009, 27, 3742. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, N.; Kaku, T.; Ojida, A.; Tanaka, T.; Hara, T.; Yamaoka, M.; Kusaka, M.; Tasaka, A. C17, 20-lyase inhibitors. Part 2: Design, synthesis and structure–activity relationships of (2-naphthylmethyl)-1H-imidazoles as novel C17, 20-lyase inhibitors. Bioorganic Med. Chem. 2004, 12, 4313–4336. [Google Scholar] [CrossRef]
- Niraula, S.; Chi, K.; Joshua, A.M. Beyond Castration—Defining Future Directions in the Hormonal Treatment of Prostate Cancer. Horm. Cancer 2012, 3, 3–13. [Google Scholar] [CrossRef]
- Taplin, M.-E.; Montgomery, R.B.; Group, A. ARMOR2: Galeterone in progressive CRPC patients who have failed oral therapy. Am. Soc. Clin. Oncol. 2014, 32, 71. [Google Scholar] [CrossRef]
- Tran, C.; Ouk, S.; Clegg, N.J.; Chen, Y.; Watson, P.A.; Arora, V.; Wongvipat, J.; Smith-Jones, P.M.; Yoo, D.; Kwon, A. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 2009, 324, 787–790. [Google Scholar] [CrossRef]
- Clegg, N.J.; Wongvipat, J.; Joseph, J.D.; Tran, C.; Ouk, S.; Dilhas, A.; Chen, Y.; Grillot, K.; Bischoff, E.D.; Cai, L. ARN-509: A novel antiandrogen for prostate cancer treatment. Cancer Res. 2012, 72, 1494–1503. [Google Scholar] [CrossRef]
- Kirby, M.; Hirst, C.; Crawford, E. Characterising the castration-resistant prostate cancer population: A systematic review. Int. J. Clin. Pract. 2011, 65, 1180–1192. [Google Scholar] [CrossRef] [PubMed]
- Giannakakou, P.; Nakano, M.; Nicolaou, K.C.; O’Brate, A.; Yu, J.; Blagosklonny, M.V.; Greber, U.F.; Fojo, T. Enhanced microtubule-dependent trafficking and p53 nuclear accumulation by suppression of microtubule dynamics. Proc. Natl. Acad. Sci. USA 2002, 99, 10855–10860. [Google Scholar] [CrossRef] [PubMed]
- Tannock, I.F.; De Wit, R.; Berry, W.R.; Horti, J.; Pluzanska, A.; Chi, K.N.; Oudard, S.; Théodore, C.; James, N.D.; Turesson, I. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. New Engl. J. Med. 2004, 351, 1502–1512. [Google Scholar] [CrossRef] [PubMed]
- Mita, A.C.; Denis, L.J.; Rowinsky, E.K.; DeBono, J.S.; Goetz, A.D.; Ochoa, L.; Forouzesh, B.; Beeram, M.; Patnaik, A.; Molpus, K. Phase I and pharmacokinetic study of XRP6258 (RPR 116258A), a novel taxane, administered as a 1-h infusion every 3 weeks in patients with advanced solid tumors. Clin. Cancer Res. 2009, 15, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Taylor, B.; Varambally, S.; Chinnaiyan, A. Differential proteomic alterations between localised and metastatic prostate cancer. Br. J. Cancer 2006, 95, 425. [Google Scholar] [CrossRef]
- Joniau, S.; Abrahamsson, P.-A.; Bellmunt, J.; Figdor, C.; Hamdy, F.; Verhagen, P.; Vogelzang, N.J.; Wirth, M.; Van Poppel, H.; Osanto, S. Current vaccination strategies for prostate cancer. Eur. Urol. 2012, 61, 290–306. [Google Scholar] [CrossRef]
- Bissell, M.J.; Hines, W.C. Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat. Med. 2011, 17, 320. [Google Scholar] [CrossRef]
- Wiseman, B.S.; Werb, Z. Stromal effects on mammary gland development and breast cancer. Science 2002, 296, 1046–1049. [Google Scholar] [CrossRef]
- Egeblad, M.; Nakasone, E.S.; Werb, Z. Tumors as organs: Complex tissues that interface with the entire organism. Dev. Cell 2010, 18, 884–901. [Google Scholar] [CrossRef]
- Shephard, A.P.; Yeung, V.; Clayton, A.; Webber, J.P. Prostate cancer exosomes as modulators of the tumor microenvironment. J. Cancer Metastasis Treat. 2017, 3, 288–301. [Google Scholar] [CrossRef]
- Saleem, S.N.; Abdel-Mageed, A.B. Tumor-derived exosomes in oncogenic reprogramming and cancer progression. Cell Mol. Life Sci 2015, 72, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Akers, J.C.; Gonda, D.; Kim, R.; Carter, B.S.; Chen, C.C. Biogenesis of extracellular vesicles (EV): Exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J. Neuro-Oncol. 2013, 113, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Riches, A.; Campbell, E.; Borger, E.; Powis, S. Regulation of exosome release from mammary epithelial and breast cancer cells–a new regulatory pathway. Eur. J. Cancer 2014, 50, 1025–1034. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Wang, D.; Jin, F.; Bian, Z.; Li, L.; Liang, H.; Li, M.; Shi, L.; Pan, C.; Zhu, D. Pyruvate kinase type M2 promotes tumour cell exosome release via phosphorylating synaptosome-associated protein 23. Nat. Commun. 2017, 8, 14041. [Google Scholar] [CrossRef]
- Liu, C.; Su, C. Design strategies and application progress of therapeutic exosomes. Theranostics 2019, 9, 1015–1028. [Google Scholar] [CrossRef]
- Mashouri, L.; Yousefi, H.; Aref, A.R.; Ahadi, A.M.; Molaei, F.; Alahari, S.K. Exosomes: Composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol. Cancer 2019, 18, 75. [Google Scholar] [CrossRef]
- Robbins, P.D.; Morelli, A.E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 2014, 14, 195. [Google Scholar] [CrossRef]
- Koliopanos, A.; Friess, H.; Kleeff, J.; Shi, X.; Liao, Q.; Pecker, I.; Vlodavsky, I.; Zimmermann, A.; Büchler, M.W. Heparanase expression in primary and metastatic pancreatic cancer. Cancer Res. 2001, 61, 4655–4659. [Google Scholar]
- Koo, T.H.; Lee, J.-J.; Kim, E.-M.; Kim, K.-W.; Do Kim, H.; Lee, J.-H. Syntenin is overexpressed and promotes cell migration in metastatic human breast and gastric cancer cell lines. Oncogene 2002, 21, 4080. [Google Scholar] [CrossRef]
- Liu, R.-T.; Huang, C.-C.; You, H.-L.; Chou, F.-F.; Hu, C.-C.A.; Chao, F.-P.; Chen, C.-M.; Cheng, J.-T. Overexpression of tumor susceptibility gene TSG101 in human papillary thyroid carcinomas. Oncogene 2002, 21, 4830. [Google Scholar] [CrossRef]
- Oh, K.; Stanton, M.; West, W.; Todd, G.; Wagner, K. Tsg101 is upregulated in a subset of invasive human breast cancers and its targeted overexpression in transgenic mice reveals weak oncogenic properties for mammary cancer initiation. Oncogene 2007, 26, 5950. [Google Scholar] [CrossRef] [PubMed]
- Toyoshima, M.; Tanaka, N.; Aoki, J.; Tanaka, Y.; Murata, K.; Kyuuma, M.; Kobayashi, H.; Ishii, N.; Yaegashi, N.; Sugamura, K. Inhibition of tumor growth and metastasis by depletion of vesicular sorting protein Hrs: Its regulatory role on E-cadherin and β-catenin. Cancer Res. 2007, 67, 5162–5171. [Google Scholar] [CrossRef] [PubMed]
- Morgan-Fisher, M.; Wewer, U.M.; Yoneda, A. Regulation of ROCK activity in cancer. J. Histochem. Cytochem. 2013, 61, 185–198. [Google Scholar] [CrossRef]
- Ruiz-Martinez, M.; Navarro, A.; Marrades, R.M.; Viñolas, N.; Santasusagna, S.; Muñoz, C.; Ramírez, J.; Molins, L.; Monzo, M. YKT6 expression, exosome release, and survival in non-small cell lung cancer. Oncotarget 2016, 7, 51515. [Google Scholar] [CrossRef] [PubMed]
- Al-Nedawi, K.; Meehan, B.; Micallef, J.; Lhotak, V.; May, L.; Guha, A.; Rak, J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008, 10, 619. [Google Scholar] [CrossRef] [PubMed]
- Takasugi, M.; Okada, R.; Takahashi, A.; Chen, D.V.; Watanabe, S.; Hara, E. Small extracellular vesicles secreted from senescent cells promote cancer cell proliferation through EphA2. Nat. Commun. 2017, 8, 15729. [Google Scholar] [CrossRef] [PubMed]
- Imjeti, N.S.; Menck, K.; Egea-Jimenez, A.L.; Lecointre, C.; Lembo, F.; Bouguenina, H.; Badache, A.; Ghossoub, R.; David, G.; Roche, S. Syntenin mediates SRC function in exosomal cell-to-cell communication. Proc. Natl. Acad. Sci. USA 2017, 114, 12495–12500. [Google Scholar] [CrossRef]
- Parolini, I.; Federici, C.; Raggi, C.; Lugini, L.; Palleschi, S.; De Milito, A.; Coscia, C.; Iessi, E.; Logozzi, M.; Molinari, A. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J. Biol. Chem. 2009, 284, 34211–34222. [Google Scholar] [CrossRef]
- King, H.W.; Michael, M.Z.; Gleadle, J.M. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer 2012, 12, 421. [Google Scholar] [CrossRef]
- Li, L.; Li, C.; Wang, S.; Wang, Z.; Jiang, J.; Wang, W.; Li, X.; Chen, J.; Liu, K.; Li, C. Exosomes derived from hypoxic oral squamous cell carcinoma cells deliver miR-21 to normoxic cells to elicit a prometastatic phenotype. Cancer Res. 2016, 76, 1770–1780. [Google Scholar] [CrossRef]
- Wang, T.; Gilkes, D.M.; Takano, N.; Xiang, L.; Luo, W.; Bishop, C.J.; Chaturvedi, P.; Green, J.J.; Semenza, G.L. Hypoxia-inducible factors and RAB22A mediate formation of microvesicles that stimulate breast cancer invasion and metastasis. Proc. Natl. Acad. Sci. USA 2014, 111, E3234–E3242. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Harris, S.L.; Levine, A.J. The regulation of exosome secretion: A novel function of the p53 protein. Cancer Res. 2006, 66, 4795–4801. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Iguchi, H.; Yoshioka, Y.; Takeshita, F.; Matsuki, Y.; Ochiya, T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 2010, 285, 17442–17452. [Google Scholar] [CrossRef] [PubMed]
- Mittelbrunn, M.; Gutiérrez-Vázquez, C.; Villarroya-Beltri, C.; González, S.; Sánchez-Cabo, F.; González, M.Á.; Bernad, A.; Sánchez-Madrid, F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2011, 2, 282. [Google Scholar] [CrossRef]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brugger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Baixauli, F.; Gutiérrez-Vázquez, C.; Sánchez-Madrid, F.; Mittelbrunn, M. Sorting it out: Regulation of exosome loading. Semin. Cancer Biol. 2014, 28, 3–13. [Google Scholar] [CrossRef]
- Shen, B.; Wu, N.; Yang, J.-M.; Gould, S.J. Protein targeting to exosomes/microvesicles by plasma membrane anchors. J. Biol. Chem. 2011, 286, 14383–14395. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Gutiérrez-Vázquez, C.; Sánchez-Cabo, F.; Pérez-Hernández, D.; Vázquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sánchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef]
- Guduric-Fuchs, J.; O’Connor, A.; Camp, B.; O’Neill, C.L.; Medina, R.J.; Simpson, D.A. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. Bmc Genom. 2012, 13, 357. [Google Scholar] [CrossRef]
- Squadrito, M.L.; Baer, C.; Burdet, F.; Maderna, C.; Gilfillan, G.D.; Lyle, R.; Ibberson, M.; De Palma, M. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014, 8, 1432–1446. [Google Scholar] [CrossRef]
- Rajagopal, C.; Harikumar, K. The origin and functions of exosomes in cancer. Front. Oncol. 2018, 8, 66. [Google Scholar] [CrossRef] [PubMed]
- Jansen, F.H.; Krijgsveld, J.; van Rijswijk, A.; van den Bemd, G.-J.; van den Berg, M.S.; van Weerden, W.M.; Willemsen, R.; Dekker, L.J.; Luider, T.M.; Jenster, G. Exosomal secretion of cytoplasmic prostate cancer xenograft-derived proteins. Mol. Cell. Proteom. 2009, 8, 1192–1205. [Google Scholar] [CrossRef]
- Nilsson, J.; Skog, J.; Nordstrand, A.; Baranov, V.; Mincheva-Nilsson, L.; Breakefield, X.; Widmark, A. Prostate cancer-derived urine exosomes: A novel approach to biomarkers for prostate cancer. Br. J. Cancer 2009, 100, 1603. [Google Scholar] [CrossRef] [PubMed]
- Duijvesz, D.; Luider, T.; Bangma, C.H.; Jenster, G. Exosomes as biomarker treasure chests for prostate cancer. Eur. Urol. 2011, 59, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654. [Google Scholar] [CrossRef]
- Hosseini-Beheshti, E.; Pham, S.; Adomat, H.; Li, N.; Tomlinson Guns, E.S. Exosomes as biomarker enriched microvesicles: Characterization of exosomal proteins derived from a panel of prostate cell lines with distinct AR phenotypes. Mol. Cell Proteom. 2012, 11, 863–885. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Bahassi, E.M. Biofluid-based circulating tumor molecules as diagnostic tools for use in personalized medicine. J. Mol. Biomark. Diagn. 2013, 5, 157–163. [Google Scholar]
- Perkins, G.L.; Slater, E.D.; Sanders, G.K.; Prichard, J.G. Serum tumor markers. Am. Fam. Physician 2003, 68, 1075–1088. [Google Scholar]
- Kharmate, G.; Hosseini-Beheshti, E.; Caradec, J.; Chin, M.Y.; Guns, E.S.T. Epidermal growth factor receptor in prostate cancer derived exosomes. PLoS ONE 2016, 11. [Google Scholar]
- Khan, S.; Jutzy, J.M.; Valenzuela, M.M.A.; Turay, D.; Aspe, J.R.; Ashok, A.; Mirshahidi, S.; Mercola, D.; Lilly, M.B.; Wall, N.R. Plasma-derived exosomal survivin, a plausible biomarker for early detection of prostate cancer. PLoS ONE 2012, 7, e46737. [Google Scholar] [CrossRef]
- Panigrahi, G.K.; Praharaj, P.P.; Kittaka, H.; Mridha, A.R.; Black, O.M.; Singh, R.; Mercer, R.; van Bokhoven, A.; Torkko, K.C.; Agarwal, C. Exosome proteomic analyses identify inflammatory phenotype and novel biomarkers in African American prostate cancer patients. Cancer Med. 2019, 8, 1110–1123. [Google Scholar] [CrossRef] [PubMed]
- McKiernan, J.; Donovan, M.J.; O’Neill, V.; Bentink, S.; Noerholm, M.; Belzer, S.; Skog, J.; Kattan, M.W.; Partin, A.; Andriole, G. A novel urine exosome gene expression assay to predict high-grade prostate cancer at initial biopsy. Jama Oncol. 2016, 2, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yuan, T.; Liang, M.; Du, M.; Xia, S.; Dittmar, R.; Wang, D.; See, W.; Costello, B.A.; Quevedo, F. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur. Urol. 2015, 67, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, C.; Rani, S.; O’driscoll, L. miR-34a is an intracellular and exosomal predictive biomarker for response to docetaxel with clinical relevance to prostate cancer progression. Prostate 2014, 74, 1320–1334. [Google Scholar] [CrossRef] [PubMed]
- Shedden, K.; Xie, X.T.; Chandaroy, P.; Chang, Y.T.; Rosania, G.R. Expulsion of small molecules in vesicles shed by cancer cells: Association with gene expression and chemosensitivity profiles. Cancer Res. 2003, 63, 4331–4337. [Google Scholar] [PubMed]
- Safaei, R.; Larson, B.J.; Cheng, T.C.; Gibson, M.A.; Otani, S.; Naerdemann, W.; Howell, S.B. Abnormal lysosomal trafficking and enhanced exosomal export of cisplatin in drug-resistant human ovarian carcinoma cells. Mol. Cancer Ther. 2005, 4, 1595–1604. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.; Zahid, S.; Wan, Y.-J.Y.; Forster, J.; Karim, A.-B.A.; Nawabi, A.M.; Azhar, A.; Rahman, M.A.; Ahmed, N. Protein expression profiling of nuclear membrane protein reveals potential biomarker of human hepatocellular carcinoma. Clin. Proteom. 2013, 10, 6. [Google Scholar] [CrossRef]
- Aung, T.; Chapuy, B.; Vogel, D.; Wenzel, D.; Oppermann, M.; Lahmann, M.; Weinhage, T.; Menck, K.; Hupfeld, T.; Koch, R. Exosomal evasion of humoral immunotherapy in aggressive B-cell lymphoma modulated by ATP-binding cassette transporter A3. Proc. Natl. Acad. Sci. USA 2011, 108, 15336–15341. [Google Scholar] [CrossRef]
- Ciravolo, V.; Huber, V.; Ghedini, G.C.; Venturelli, E.; Bianchi, F.; Campiglio, M.; Morelli, D.; Villa, A.; Mina, P.D.; Menard, S. Potential role of HER2-overexpressing exosomes in countering trastuzumab-based therapy. J. Cell. Physiol. 2012, 227, 658–667. [Google Scholar] [CrossRef]
- Zhang, H.-G.; Grizzle, W.E. Exosomes and cancer: A newly described pathway of immune suppression. Clin. Cancer Res. 2011, 17, 959–964. [Google Scholar] [CrossRef]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S.-i.; Ishikawa, A.; Kuroda, M. Roles of exosomes and microvesicles in disease pathogenesis. Adv. Drug Deliv. Rev. 2013, 65, 398–401. [Google Scholar] [CrossRef] [PubMed]
- Camussi, G.; Deregibus, M.C.; Bruno, S.; Cantaluppi, V.; Biancone, L. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int. 2010, 78, 838–848. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, S.; Palanisamy, V. Horizontal transfer of RNAs: Exosomes as mediators of intercellular communication. Wiley Interdiscip. Rev. 2012, 3, 286–293. [Google Scholar] [CrossRef]
- Han, L.; Xu, J.; Xu, Q.; Zhang, B.; Lam, E.F.; Sun, Y. Extracellular vesicles in the tumor microenvironment: Therapeutic resistance, clinical biomarkers, and targeting strategies. Med. Res. Rev. 2017, 37, 1318–1349. [Google Scholar] [CrossRef]
- Soekmadji, C.; Riches, J.D.; Russell, P.J.; Ruelcke, J.E.; McPherson, S.; Wang, C.; Hovens, C.M.; Corcoran, N.M.; BioResource, T.A.P.C.C.; Hill, M.M. Modulation of paracrine signaling by CD9 positive small extracellular vesicles mediates cellular growth of androgen deprived prostate cancer. Oncotarget 2017, 8, 52237. [Google Scholar] [CrossRef]
- Souza, A.G.; B Silva, I.B.; Campos-Fernández, E.; Marangoni, K.; F Bastos, V.A.; Alves, P.T.; Goulart, L.R.; Alonso-Goulart, V. Extracellular vesicles as drivers of epithelial-mesenchymal transition and carcinogenic characteristics in normal prostate cells. Mol. Carcinog. 2018, 57, 503–511. [Google Scholar] [CrossRef]
- Hosseini-Beheshti, E.; Choi, W.; Weiswald, L.-B.; Kharmate, G.; Ghaffari, M.; Roshan-Moniri, M.; Hassona, M.D.; Chan, L.; Chin, M.Y.; Tai, I.T. Exosomes confer pro-survival signals to alter the phenotype of prostate cells in their surrounding environment. Oncotarget 2016, 7, 14639. [Google Scholar] [CrossRef]
- Waugh, D.J.; Wilson, C. The interleukin-8 pathway in cancer. Clin. Cancer Res. 2008, 14, 6735–6741. [Google Scholar] [CrossRef]
- Ramteke, A.; Ting, H.; Agarwal, C.; Mateen, S.; Somasagara, R.; Hussain, A.; Graner, M.; Frederick, B.; Agarwal, R.; Deep, G. Exosomes secreted under hypoxia enhance invasiveness and stemness of prostate cancer cells by targeting adherens junction molecules. Mol. Carcinog. 2015, 54, 554–565. [Google Scholar] [CrossRef]
- Fedele, C.; Singh, A.; Zerlanko, B.J.; Iozzo, R.V.; Languino, L.R. The αvβ6 integrin is transferred intercellularly via exosomes. J. Biol. Chem. 2015, 290, 4545–4551. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Fedele, C.; Lu, H.; Nevalainen, M.T.; Keen, J.H.; Languino, L.R. Exosome-mediated transfer of αvβ3 integrin from tumorigenic to nontumorigenic cells promotes a migratory phenotype. Mol. Cancer Res. 2016, 14, 1136–1146. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, C.; Rani, S.; O’Brien, K.; O’Neill, A.; Prencipe, M.; Sheikh, R.; Webb, G.; McDermott, R.; Watson, W.; Crown, J. Docetaxel-resistance in prostate cancer: Evaluating associated phenotypic changes and potential for resistance transfer via exosomes. PLoS ONE 2012, 7, e50999. [Google Scholar] [CrossRef] [PubMed]
- Panagopoulos, K.; Cross-Knorr, S.; Dillard, C.; Pantazatos, D.; Del Tatto, M.; Mills, D.; Goldstein, L.; Renzulli, J.; Quesenberry, P.; Chatterjee, D. Reversal of chemosensitivity and induction of cell malignancy of a non-malignant prostate cancer cell line upon extracellular vesicle exposure. Mol. Cancer 2013, 12, 118. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Q.; DeChalus, A.; Chatterjee, D.N.; Keller, E.T.; Mizokami, A.; Camussi, G.; Mendelsohn, A.R.; Renzulli, J.F. Extracellular vesicle-mediated reversal of paclitaxel resistance in prostate cancer. Crit. Rev. Oncog. 2015, 20, 407. [Google Scholar] [CrossRef]
- Tomasetti, M.; Nocchi, L.; Staffolani, S.; Manzella, N.; Amati, M.; Goodwin, J.; Kluckova, K.; Nguyen, M.; Strafella, E.; Bajzikova, M. MicroRNA-126 suppresses mesothelioma malignancy by targeting IRS1 and interfering with the mitochondrial function. Antioxid. Redox Signal. 2014, 21, 2109–2125. [Google Scholar] [CrossRef]
- Tomasetti, M.; Monaco, F.; Manzella, N.; Rohlena, J.; Rohlenova, K.; Staffolani, S.; Gaetani, S.; Ciarapica, V.; Amati, M.; Bracci, M. MicroRNA-126 induces autophagy by altering cell metabolism in malignant mesothelioma. Oncotarget 2016, 7, 36338. [Google Scholar] [CrossRef]
- Chang, Q.; Li, Y.; White, M.F.; Fletcher, J.A.; Xiao, S. Constitutive activation of insulin receptor substrate 1 is a frequent event in human tumors: Therapeutic implications. Cancer Res. 2002, 62, 6035–6038. [Google Scholar]
- Huang, T.; Chu, T. Repression of miR-126 and upregulation of adrenomedullin in the stromal endothelium by cancer-stromal cross talks confers angiogenesis of cervical cancer. Oncogene 2014, 33, 3636. [Google Scholar] [CrossRef]
- Sun, X.; Wang, Z.-M.; Song, Y.; Tai, X.-H.; Ji, W.-Y.; Gu, H. MicroRNA-126 modulates the tumor microenvironment by targeting calmodulin-regulated spectrin-associated protein 1 (Camsap1). Int. J. Oncol. 2014, 44, 1678–1684. [Google Scholar] [CrossRef]
- Garcia, N.A.; Ontoria-Oviedo, I.; González-King, H.; Diez-Juan, A.; Sepúlveda, P. Glucose starvation in cardiomyocytes enhances exosome secretion and promotes angiogenesis in endothelial cells. PLoS ONE 2015, 10, e0138849. [Google Scholar] [CrossRef] [PubMed]
- Garcia, N.A.; Moncayo-Arlandi, J.; Sepulveda, P.; Diez-Juan, A. Cardiomyocyte exosomes regulate glycolytic flux in endothelium by direct transfer of GLUT transporters and glycolytic enzymes. Cardiovasc. Res. 2015, 109, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Valis, K.; Prochazka, L.; Boura, E.; Chladova, J.; Obsil, T.; Rohlena, J.; Truksa, J.; Dong, L.-F.; Ralph, S.J.; Neuzil, J. Hippo/Mst1 stimulates transcription of the proapoptotic mediator NOXA in a FoxO1-dependent manner. Cancer Res. 2011, 71, 946–954. [Google Scholar] [CrossRef] [PubMed]
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.F.; Li, S.; Chin, A.R. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 2015, 17, 183. [Google Scholar] [CrossRef]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Tomasetti, M.; Lee, W.; Santarelli, L.; Neuzil, J. Exosome-derived microRNAs in cancer metabolism: Possible implications in cancer diagnostics and therapy. Exp. Mol. Med. 2017, 49, e285. [Google Scholar] [CrossRef]
- Castellana, D.; Zobairi, F.; Martinez, M.C.; Panaro, M.A.; Mitolo, V.; Freyssinet, J.-M.; Kunzelmann, C. Membrane microvesicles as actors in the establishment of a favorable prostatic tumoral niche: A role for activated fibroblasts and CX3CL1-CX3CR1 axis. Cancer Res. 2009, 69, 785–793. [Google Scholar] [CrossRef]
- Martinez-Outschoorn, U.E.; Lisanti, M.P.; Sotgia, F. Catabolic cancer-associated fibroblasts transfer energy and biomass to anabolic cancer cells, fueling tumor growth. Semin. Cancer Biol. 2014, 25, 47–60. [Google Scholar] [CrossRef]
- Shiga, K.; Hara, M.; Nagasaki, T.; Sato, T.; Takahashi, H.; Takeyama, H. Cancer-associated fibroblasts: Their characteristics and their roles in tumor growth. Cancers 2015, 7, 2443–2458. [Google Scholar] [CrossRef]
- Cirri, P.; Chiarugi, P. Cancer-associated-fibroblasts and tumour cells: A diabolic liaison driving cancer progression. Cancer Metastasis Rev. 2012, 31, 195–208. [Google Scholar] [CrossRef]
- Flaberg, E.; Markasz, L.; Petranyi, G.; Stuber, G.; Dicső, F.; Alchihabi, N.; Oláh, È.; Csízy, I.; Józsa, T.; Andrén, O. High-throughput live-cell imaging reveals differential inhibition of tumor cell proliferation by human fibroblasts. Int. J. Cancer 2011, 128, 2793–2802. [Google Scholar] [CrossRef] [PubMed]
- Öhlund, D.; Elyada, E.; Tuveson, D. Fibroblast heterogeneity in the cancer wound. J. Exp. Med. 2014, 211, 1503–1523. [Google Scholar] [CrossRef] [PubMed]
- Fiaschi, T.; Marini, A.; Giannoni, E.; Taddei, M.L.; Gandellini, P.; De Donatis, A.; Lanciotti, M.; Serni, S.; Cirri, P.; Chiarugi, P. Reciprocal metabolic reprogramming through lactate shuttle coordinately influences tumor-stroma interplay. Cancer Res. 2012, 72, 5130–5140. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Zhao, H.; Yang, L.; Baddour, J.; Achreja, A.; Bernard, V.; Moss, T.; Marini, J.C.; Tudawe, T.; Seviour, E.G.; San Lucas, F.A. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife 2016, 5, e10250. [Google Scholar] [CrossRef]
- Paggetti, J.; Haderk, F.; Seiffert, M.; Janji, B.; Distler, U.; Ammerlaan, W.; Kim, Y.J.; Adam, J.; Lichter, P.; Solary, E. Exosomes released by chronic lymphocytic leukemia cells induce the transition of stromal cells into cancer-associated fibroblasts. Blood 2015, 126, 1106–1117. [Google Scholar] [CrossRef]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423. [Google Scholar] [CrossRef]
- Duijvesz, D.; Burnum-Johnson, K.E.; Gritsenko, M.A.; Hoogland, A.M.; Vredenbregt-van den Berg, M.S.; Willemsen, R.; Luider, T.; Paša-Tolić, L.; Jenster, G. Proteomic profiling of exosomes leads to the identification of novel biomarkers for prostate cancer. PLoS ONE 2013, 8, e82589. [Google Scholar] [CrossRef]
- Ronquist, K.G.; Ronquist, G.; Larsson, A.; Carlsson, L. Proteomic analysis of prostate cancer metastasis-derived prostasomes. Anticancer Res. 2010, 30, 285–290. [Google Scholar]
- Bang, C.; Thum, T. Exosomes: New players in cell–cell communication. Int. J. Biochem. Cell Biol. 2012, 44, 2060–2064. [Google Scholar] [CrossRef]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intercellular communication. J. Proteom. 2010, 73, 1907–1920. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Beach, A.; Zhang, H.-G.; Ratajczak, M.Z.; Kakar, S.S. Exosomes: An overview of biogenesis, composition and role in ovarian cancer. J. Ovarian Res. 2014, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Hannafon, B.; Ding, W.-Q. Intercellular communication by exosome-derived microRNAs in cancer. Int. J. Mol. Sci. 2013, 14, 14240–14269. [Google Scholar] [CrossRef] [PubMed]
- Vlassov, A.V.; Magdaleno, S.; Setterquist, R.; Conrad, R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta 2012, 1820, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Harshman, S.W.; Canella, A.; Ciarlariello, P.D.; Rocci, A.; Agarwal, K.; Smith, E.M.; Talabere, T.; Efebera, Y.A.; Hofmeister, C.C.; Benson, D.M., Jr. Characterization of multiple myeloma vesicles by label-free relative quantitation. Proteomics 2013, 13, 3013–3029. [Google Scholar] [CrossRef]
- Gonzalez-Begne, M.; Lu, B.; Liao, L.; Xu, T.; Bedi, G.; Melvin, J.E.; Yates, J.R., III. Characterization of the human submandibular/sublingual saliva glycoproteome using lectin affinity chromatography coupled to multidimensional protein identification technology. J. Proteome Res. 2011, 10, 5031–5046. [Google Scholar] [CrossRef]
- Gonzalez-Begne, M.; Lu, B.; Han, X.; Hagen, F.K.; Hand, A.R.; Melvin, J.E.; Yates, J.R., III. Proteomic analysis of human parotid gland exosomes by multidimensional protein identification technology (MudPIT). J. Proteome Res. 2009, 8, 1304–1314. [Google Scholar] [CrossRef]
- Abd Elmageed, Z.Y.; Yang, Y.; Thomas, R.; Ranjan, M.; Mondal, D.; Moroz, K.; Fang, Z.; Rezk, B.M.; Moparty, K.; Sikka, S.C. Neoplastic reprogramming of patient-derived adipose stem cells by prostate cancer cell-associated exosomes. Stem Cells 2014, 32, 983–997. [Google Scholar] [CrossRef]
- Webber, J.; Steadman, R.; Mason, M.D.; Tabi, Z.; Clayton, A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 2010, 70, 9621–9630. [Google Scholar] [CrossRef]
- Webber, J.P.; Spary, L.K.; Sanders, A.J.; Chowdhury, R.; Jiang, W.G.; Steadman, R.; Wymant, J.; Jones, A.T.; Kynaston, H.; Mason, M.D. Differentiation of tumour-promoting stromal myofibroblasts by cancer exosomes. Oncogene 2015, 34, 290. [Google Scholar] [CrossRef] [PubMed]
- Tomasek, J.J.; Gabbiani, G.; Hinz, B.; Chaponnier, C.; Brown, R.A. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat. Rev. Mol. Cell Biol. 2002, 3, 349. [Google Scholar] [CrossRef] [PubMed]
- Calvo, F.; Ege, N.; Grande-Garcia, A.; Hooper, S.; Jenkins, R.P.; Chaudhry, S.I.; Harrington, K.; Williamson, P.; Moeendarbary, E.; Charras, G. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat. Cell Biol. 2013, 15, 637. [Google Scholar] [CrossRef] [PubMed]
- Shimoda, M.; Principe, S.; Jackson, H.W.; Luga, V.; Fang, H.; Molyneux, S.D.; Shao, Y.W.; Aiken, A.; Waterhouse, P.D.; Karamboulas, C. Loss of the Timp gene family is sufficient for the acquisition of the CAF-like cell state. Nat. Cell Biol. 2014, 16, 889. [Google Scholar] [CrossRef]
- Chowdhury, R.; Webber, J.P.; Gurney, M.; Mason, M.D.; Tabi, Z.; Clayton, A. Cancer exosomes trigger mesenchymal stem cell differentiation into pro-angiogenic and pro-invasive myofibroblasts. Oncotarget 2015, 6, 715. [Google Scholar] [CrossRef]
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Mark, M.T.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329. [Google Scholar] [CrossRef]
- Tatarov, O.; Mitchell, T.J.; Seywright, M.; Leung, H.Y.; Brunton, V.G.; Edwards, J. SRC family kinase activity is up-regulated in hormone-refractory prostate cancer. Clin. Cancer Res. 2009, 15, 3540–3549. [Google Scholar] [CrossRef]
- Chang, C.Y.-M.; Kung, H.-J.; Evans, C.P. Nonreceptor Tyrosine Kinases in Prostate. Neoplasia 2007, 9, 90–100. [Google Scholar] [CrossRef][Green Version]
- Marx, M.; Warren, S.L.; Madri, J.A. pp60c-src modulates microvascular endothelial phenotype and in vitro angiogenesis. Exp. Mol. Pathol. 2001, 70, 201–213. [Google Scholar] [CrossRef]
- DeRita, R.M.; Zerlanko, B.; Singh, A.; Lu, H.; Iozzo, R.V.; Benovic, J.L.; Languino, L.R. c-Src, insulin-like growth factor I receptor, G-protein-coupled receptor kinases and focal adhesion kinase are enriched into prostate cancer cell exosomes. J. Cell. Biochem. 2017, 118, 66–73. [Google Scholar] [CrossRef]
- Angelucci, A.; D’ascenzo, S.; Festuccia, C.; Gravina, G.L.; Bologna, M.; Dolo, V.; Pavan, A. Vesicle-associated urokinase plasminogen activator promotes invasion in prostate cancer cell lines. Clin. Exp. Metastasis 2000, 18, 163. [Google Scholar] [CrossRef] [PubMed]
- Drew, A.F.; Tucker, H.L.; Kombrinck, K.W.; Simon, D.I.; Bugge, T.H.; Degen, J.L. Plasminogen is a critical determinant of vascular remodeling in mice. Circ. Res. 2000, 87, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Lundholm, M.; Schröder, M.; Nagaeva, O.; Baranov, V.; Widmark, A.; Mincheva-Nilsson, L.; Wikström, P. Prostate tumor-derived exosomes down-regulate NKG2D expression on natural killer cells and CD8+ T cells: Mechanism of immune evasion. PLoS ONE 2014, 9, e108925. [Google Scholar] [CrossRef] [PubMed]
- Naito, Y.; Yoshioka, Y.; Yamamoto, Y.; Ochiya, T. How cancer cells dictate their microenvironment: Present roles of extracellular vesicles. Cell. Mol. Life Sci. 2017, 74, 697–713. [Google Scholar] [CrossRef]
- Steinbichler, T.B.; Dudas, J.; Riechelmann, H.; Skvortsova, I.-I. The role of exosomes in cancer metastasis. Semin. Cancer Biol. 2017, 44, 170–181. [Google Scholar] [CrossRef]
- Meehan, B.; Rak, J.; Di Vizio, D. Oncosomes–large and small: What are they, where they came from? J. Extracell. Vesicles 2016, 5. [Google Scholar] [CrossRef]
- Minciacchi, V.R.; Spinelli, C.; Reis-Sobreiro, M.; Cavallini, L.; You, S.; Zandian, M.; Li, X.; Mishra, R.; Chiarugi, P.; Adam, R.M. MYC mediates large oncosome-induced fibroblast reprogramming in prostate cancer. Cancer Res. 2017, 77, 2306–2317. [Google Scholar] [CrossRef]
- Silva, T.A.; Smuczek, B.; Valadao, I.C.; Dzik, L.M.; Iglesia, R.P.; Cruz, M.C.; Zelanis, A.; de Siqueira, A.S.; Serrano, S.M.; Goldberg, G.S.; et al. AHNAK enables mammary carcinoma cells to produce extracellular vesicles that increase neighboring fibroblast cell motility. Oncotarget 2016, 7, 49998–50016. [Google Scholar] [CrossRef]
- Mizutani, K.; Terazawa, R.; Kameyama, K.; Kato, T.; Horie, K.; Tsuchiya, T.; Seike, K.; Ehara, H.; Fujita, Y.; Kawakami, K. Isolation of prostate cancer-related exosomes. Anticancer Res. 2014, 34, 3419–3423. [Google Scholar]
- Lehmann, B.D.; Paine, M.S.; Brooks, A.M.; McCubrey, J.A.; Renegar, R.H.; Wang, R.; Terrian, D.M. Senescence-associated exosome release from human prostate cancer cells. Cancer Res. 2008, 68, 7864–7871. [Google Scholar] [CrossRef]
- Itoh, T.; Ito, Y.; Ohtsuki, Y.; Tsukamasa, Y.; Yamada, N.; Naoe, T.; Akao, Y. Microvesicles released from hormone-refractory prostate cancer cells facilitate mouse pre-osteoblast differentiation. J. Mol. Histol. 2012, 43, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Laderach, D.J.; Gentilini, L.D.; Giribaldi, L.; Delgado, V.C.; Nugnes, L.; Croci, D.O.; Al Nakouzi, N.; Sacca, P.; Casas, G.; Mazza, O. A unique galectin signature in human prostate cancer progression suggests galectin-1 as a key target for treatment of advanced disease. Cancer Res. 2013, 73, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, K.; Fuhita, Y.; Kato, T.; Mizutani, K.; Kameyama, K.; Tsumoto, H.; Miura, Y.; Deguchi, T.; Ito, M. Integrin beta4 and vinculin contained in exosomes are potential markers for progression of prostate cancer associated with taxane-resistance. Int. J. Oncol 2015, 47, 384–390. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Mizutani, K.; Kameyama, K.; Kawakami, K.; Fujita, Y.; Nakane, K.; Kanimoto, Y.; Ehara, H.; Ito, H.; Seishima, M. Serum exosomal P-glycoprotein is a potential marker to diagnose docetaxel resistance and select a taxoid for patients with prostate cancer. Urol. Oncol. 2015, 33, 385. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wilson, M.J.; Slaton, J.W.; Sinha, A.A.; Ewing, S.L.; Pei, D. Increased Aggressiveness of Human Prostate PC-3 Tumor Cells Expressing Cell Surface Localized Membrane Type-1 Matrix Metalloproteinase (MT1-MMP). J. Androl. 2009, 30, 259–274. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Huang, A.C.; Zhang, W.; Zhang, G.; Wu, M.; Xu, W.; Yu, Z.; Yang, J.; Wang, B.; Sun, H.; et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 2018, 560, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Oh, W.K.; Vargas, R.; Jacobus, S.; Leitzel, K.; Regan, M.M.; Hamer, P.; Pierce, K.; Brown-Shimer, S.; Carney, W.; Ali, S.M. Elevated plasma tissue inhibitor of metalloproteinase-1 levels predict decreased survival in castration-resistant prostate cancer patients. Cancer 2011, 117, 517–525. [Google Scholar] [CrossRef]
- Trerotola, M.; Ganguly, K.K.; Fazli, L.; Fedele, C.; Lu, H.; Dutta, A.; Liu, Q.; De Angelis, T.; Riddell, L.W.; Riobo, N.A. Trop-2 is up-regulated in invasive prostate cancer and displaces FAK from focal contacts. Oncotarget 2015, 6, 14318. [Google Scholar] [CrossRef]
- El-Sayed, I.Y.; Daher, A.; Destouches, D.; Firlej, V.; Kostallari, E.; Maillé, P.; Huet, E.; Haidar-Ahmad, N.; Jenster, G.; de la Taille, A. Extracellular vesicles released by mesenchymal-like prostate carcinoma cells modulate EMT state of recipient epithelial-like carcinoma cells through regulation of AR signaling. Cancer Lett. 2017, 410, 100–111. [Google Scholar] [CrossRef]
- Charrin, S.; Manié, S.; Thiele, C.; Billard, M.; Gerlier, D.; Boucheix, C.; Rubinstein, E. A physical and functional link between cholesterol and tetraspanins. Eur. J. Immunol. 2003, 33, 2479–2489. [Google Scholar] [CrossRef]
- Record, M.; Carayon, K.; Poirot, M.; Silvente-Poirot, S. Exosomes as new vesicular lipid transporters involved in cell–cell communication and various pathophysiologies. Biochim. Biophys. Acta 2014, 1841, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Zakharova, L.; Svetlova, M.; Fomina, A.F. T cell exosomes induce cholesterol accumulation in human monocytes via phosphatidylserine receptor. J. Cell. Physiol. 2007, 212, 174–181. [Google Scholar] [CrossRef]
- Wang, D.; DuBois, R.N. Eicosanoids and cancer. Nat. Rev. Cancer 2010, 10, 181. [Google Scholar] [CrossRef] [PubMed]
- Beloribi, S.; Ristorcelli, E.; Breuzard, G.; Silvy, F.; Bertrand-Michel, J.; Beraud, E.; Verine, A.; Lombardo, D. Exosomal lipids impact notch signaling and induce death of human pancreatic tumoral SOJ-6 cells. PLoS ONE 2012, 7, e47480. [Google Scholar] [CrossRef]
- Beloribi-Djefaflia, S.; Siret, C.; Lombardo, D. Exosomal lipids induce human pancreatic tumoral MiaPaCa-2 cells resistance through the CXCR4-SDF-1α signaling axis. Oncoscience 2015, 2, 15. [Google Scholar] [PubMed]
- Llorente, A.; Skotland, T.; Sylvänne, T.; Kauhanen, D.; Róg, T.; Orłowski, A.; Vattulainen, I.; Ekroos, K.; Sandvig, K. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim. Biophys. Acta 2013, 1831, 1302–1309. [Google Scholar] [CrossRef]
- Prantl, L.; Muehlberg, F.; Navone, N.M.; Song, Y.H.; Vykoukal, J.; Logothetis, C.J.; Alt, E.U. Adipose tissue-derived stem cells promote prostate tumor growth. Prostate 2010, 70, 1709–1715. [Google Scholar] [CrossRef]
- Gajos-Michniewicz, A.; Duechler, M.; Czyz, M. MiRNA in melanoma-derived exosomes. Cancer Lett. 2014, 347, 29–37. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef]
- Song, J.J.; Smith, S.K.; Hannon, G.J.; Joshua-Tor, L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 2004, 305, 1434–1437. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Hu, S.; Liu, Q.; Qian, C.; Liu, Z.; Luo, D. Exosomal microRNA remodels the tumor microenvironment. Peer J. 2017, 5, e4196. [Google Scholar] [CrossRef] [PubMed]
- Pang, W.; Su, J.; Wang, Y.; Feng, H.; Dai, X.; Yuan, Y.; Chen, X.; Yao, W. Pancreatic cancer-secreted miR-155 implicates in the conversion from normal fibroblasts to cancer-associated fibroblasts. Cancer Sci. 2015, 106, 1362–1369. [Google Scholar] [CrossRef] [PubMed]
- Renzulli, J.F., II; Del Tatto, M.; Dooner, G.; Aliotta, J.; Goldstein, L.; Dooner, M.; Colvin, G.; Chatterjee, D.; Quesenberry, P. Microvesicle induction of prostate specific gene expression in normal human bone marrow cells. J. Urol. 2010, 184, 2165–2171. [Google Scholar] [CrossRef]
- Sánchez, C.A.; Andahur, E.I.; Valenzuela, R.; Castellon, E.A.; Fulla, J.A.; Ramos, C.G.; Trivino, J.C. Exosomes from bulk and stem cells from human prostate cancer have a differential microRNA content that contributes cooperatively over local and pre-metastatic niche. Oncotarget 2016, 7, 3993. [Google Scholar] [CrossRef] [PubMed]
- Shiao, S.L.; Chu, G.C.-Y.; Chung, L.W. Regulation of prostate cancer progression by the tumor microenvironment. Cancer Lett. 2016, 380, 340–348. [Google Scholar] [CrossRef]
- Chu, G.C.-Y.; Zhau, H.E.; Wang, R.; Rogatko, A.; Feng, X.; Zayzafoon, M.; Liu, Y.; Farach-Carson, M.C.; You, S.; Kim, J. RANK-and c-Met-mediated signal network promotes prostate cancer metastatic colonization. Endocr. Relat. Cancer 2014, 21, 311. [Google Scholar] [CrossRef]
- Li, Q.; Li, Q.; Nuccio, J.; Liu, C.; Duan, P.; Wang, R.; Jones, L.W.; Chung, L.W.; Zhau, H.E. Metastasis initiating cells in primary prostate cancer tissues from transurethral resection of the prostate (TURP) predicts castration-resistant progression and survival of prostate cancer patients. Prostate 2015, 75, 1312–1321. [Google Scholar] [CrossRef][Green Version]
- Sugatani, T.; Vacher, J.; Hruska, K.A. A microRNA expression signature of osteoclastogenesis. Blood 2011, 117, 3648–3657. [Google Scholar] [CrossRef]
- Zhang, H.-L.; Qin, X.-J.; Cao, D.-L.; Zhu, Y.; Yao, X.-D.; Zhang, S.-L.; Dai, B.; Ye, D.-W. An elevated serum miR-141 level in patients with bone-metastatic prostate cancer is correlated with more bone lesions. Asian J. Androl. 2013, 15, 231. [Google Scholar] [CrossRef]
- Morhayim, J.; van de Peppel, J.; Demmers, J.A.; Kocer, G.; Nigg, A.L.; van Driel, M.; Chiba, H.; van Leeuwen, J.P. Proteomic signatures of extracellular vesicles secreted by nonmineralizing and mineralizing human osteoblasts and stimulation of tumor cell growth. Faseb J. 2014, 29, 274–285. [Google Scholar] [CrossRef] [PubMed]
- Dragomir, M.; Chen, B.; Calin, G.A. Exosomal lncRNAs as new players in cell-to-cell communication. Transl. Cancer Res. 2018, 7, S243. [Google Scholar] [CrossRef] [PubMed]
- Ahadi, A.; Brennan, S.; Kennedy, P.J.; Hutvagner, G.; Tran, N. Long non-coding RNAs harboring miRNA seed regions are enriched in prostate cancer exosomes. Sci. Rep. 2016, 6, 24922. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, X.; Li, R.; Wang, L.; Gu, Y.; Zhao, Y.; Huang, K.H.; Cheng, T.; Yuan, Y.; Gao, S. Long non-coding RNA MYU promotes prostate cancer proliferation by mediating the miR-184/c-Myc axis. Oncol. Rep. 2018, 40, 2814–2825. [Google Scholar] [CrossRef] [PubMed]
- Işın, M.; Uysaler, E.; Özgür, E.; Köseoğlu, H.; Şanlı, Ö.; Yücel, Ö.B.; Gezer, U.; Dalay, N. Exosomal lncRNA-p21 levels may help to distinguish prostate cancer from benign disease. Front. Genet. 2015, 6, 168. [Google Scholar]
- Kim, J.; Morley, S.; Le, M.; Bedoret, D.; Umetsu, D.T.; Di Vizio, D.; Freeman, M.R. Enhanced shedding of extracellular vesicles from amoeboid prostate cancer cells: Potential effects on the tumor microenvironment. Cancer Biol. Ther. 2014, 15, 409–418. [Google Scholar] [CrossRef]
- Liu, C.M.; Hsieh, C.L.; Shen, C.N.; Lin, C.C.; Shigemura, K.; Sung, S.Y. Exosomes from the tumor microenvironment as reciprocal regulators that enhance prostate cancer progression. Int. J. Urol. 2016, 23, 734–744. [Google Scholar] [CrossRef]
- Li, S.L.; An, N.; Liu, B.; Wang, S.Y.; Wang, J.J.; Ye, Y. Exosomes from LNCaP cells promote osteoblast activity through miR-375 transfer. Oncol. Lett. 2019, 17, 4463–4473. [Google Scholar] [CrossRef]
- Lucotti, S.; Rainaldi, G.; Evangelista, M.; Rizzo, M. Fludarabine treatment favors the retention of miR-485-3p by prostate cancer cells: Implications for survival. Mol. Cancer 2013, 12, 52. [Google Scholar] [CrossRef]
- Olumi, A.F.; Grossfeld, G.D.; Hayward, S.W.; Carroll, P.R.; Tlsty, T.D.; Cunha, G.R. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 1999, 59, 5002–5011. [Google Scholar]
- Harper, J.; Sainson, R.C. Regulation of the anti-tumour immune response by cancer-associated fibroblasts. Semin. Cancer Biol. 2014, 25, 69–77. [Google Scholar] [CrossRef]
- Bhowmick, N.A.; Chytil, A.; Plieth, D.; Gorska, A.E.; Dumont, N.; Shappell, S.; Washington, M.K.; Neilson, E.G.; Moses, H.L. TGF-beta signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science 2004, 303, 848–851. [Google Scholar] [CrossRef]
- Bruzzese, F.; Hägglöf, C.; Leone, A.; Sjöberg, E.; Roca, M.S.; Kiflemariam, S.; Sjöblom, T.; Hammarsten, P.; Egevad, L.; Bergh, A.; et al. Local and systemic protumorigenic effects of cancer-associated fibroblast-derived GDF15. Cancer Res. 2014, 74, 3408–3417. [Google Scholar] [CrossRef] [PubMed]
- Hayward, S.W.; Wang, Y.; Cao, M.; Hom, Y.K.; Zhang, B.; Grossfeld, G.D.; Sudilovsky, D.; Cunha, G.R. Malignant transformation in a nontumorigenic human prostatic epithelial cell line. Cancer Res. 2001, 61, 8135–8142. [Google Scholar] [PubMed]
- Josson, S.; Gururajan, M.; Sung, S.; Hu, P.; Shao, C.; Zhau, H.; Liu, C.; Lichterman, J.; Duan, P.; Li, Q. Stromal fibroblast-derived miR-409 promotes epithelial-to-mesenchymal transition and prostate tumorigenesis. Oncogene 2015, 34, 2690. [Google Scholar] [CrossRef]
- Josson, S.; Gururajan, M.; Hu, P.; Shao, C.; Chu, G.C.-Y.; Zhau, H.E.; Liu, C.; Lao, K.; Lu, C.-L.; Lu, Y.-T. miR-409-3p/-5p promotes tumorigenesis, epithelial-to-mesenchymal transition, and bone metastasis of human prostate cancer. Clin. Cancer Res. 2014, 20, 4636–4646. [Google Scholar] [CrossRef]
- Yeung, C.L.A.; Co, N.-N.; Tsuruga, T.; Yeung, T.-L.; Kwan, S.-Y.; Leung, C.S.; Li, Y.; Lu, E.S.; Kwan, K.; Wong, K.-K. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat. Commun. 2016, 7, 11150. [Google Scholar] [CrossRef]
- Alcayaga-Miranda, F.; González, P.L.; Lopez-Verrilli, A.; Varas-Godoy, M.; Aguila-Díaz, C.; Contreras, L.; Khoury, M. Prostate tumor-induced angiogenesis is blocked by exosomes derived from menstrual stem cells through the inhibition of reactive oxygen species. Oncotarget 2016, 7, 44462. [Google Scholar] [CrossRef]
- De Visser, K.E.; Eichten, A.; Coussens, L.M. Paradoxical roles of the immune system during cancer development. Nat. Rev. Cancer 2006, 6, 24–37. [Google Scholar] [CrossRef]
- Eichmüller, S.B.; Osen, W.; Mandelboim, O.; Seliger, B. Immune modulatory microRNAs involved in tumor attack and tumor immune escape. J. Natl. Cancer Inst. 2017, 109. [Google Scholar]
- Fridlender, Z.G.; Sun, J.; Kim, S.; Kapoor, V.; Cheng, G.; Ling, L.; Worthen, G.S.; Albelda, S.M. Polarization of tumor-associated neutrophil phenotype by TGF-β: “N1” versus “N2” TAN. Cancer Cell 2009, 16, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, inflammation, and cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef] [PubMed]
- Psaila, B.; Lyden, D. The metastatic niche: Adapting the foreign soil. Nat. Rev. Cancer 2009, 9, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Gabrilovich, D.I.; Nagaraj, S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009, 9, 162–174. [Google Scholar] [CrossRef]
- Alderson, M.R.; Tough, T.W.; Davis-Smith, T.; Braddy, S.; Falk, B.; Schooley, K.A.; Goodwin, R.G.; Smith, C.; Ramsdell, F.; Lynch, D.H. Fas ligand mediates activation-induced cell death in human T lymphocytes. J. Exp. Med. 1995, 181, 71–77. [Google Scholar] [CrossRef]
- Van Parijs, L.; Abbas, A.K. Role of Fas-mediated cell death in the regulation of immune responses. Curr. Opin. Immunol. 1996, 8, 355–361. [Google Scholar] [CrossRef]
- Boyiadzis, M.; Whiteside, T. The emerging roles of tumor-derived exosomes in hematological malignancies. Leukemia 2017, 31, 1259–1268. [Google Scholar] [CrossRef]
- Filipazzi, P.; Bürdek, M.; Villa, A.; Rivoltini, L.; Huber, V. Recent advances on the role of tumor exosomes in immunosuppression and disease progression. Semin. Cancer Biol. 2012, 22, 342–349. [Google Scholar] [CrossRef]
- Whiteside, T.L. Exosomes and tumor-mediated immune suppression. J. Clin. Investig. 2016, 126, 1216–1223. [Google Scholar] [CrossRef]
- Chow, A.; Zhou, W.; Liu, L.; Fong, M.Y.; Champer, J.; Van Haute, D.; Chin, A.R.; Ren, X.; Gugiu, B.G.; Meng, Z. Macrophage immunomodulation by breast cancer-derived exosomes requires Toll-like receptor 2-mediated activation of NF-κB. Sci. Rep. 2014, 4, 1–11. [Google Scholar] [CrossRef]
- Marton, A.; Vizler, C.; Kusz, E.; Temesfoi, V.; Szathmary, Z.; Nagy, K.; Szegletes, Z.; Varo, G.; Siklos, L.; Katona, R.L. Melanoma cell-derived exosomes alter macrophage and dendritic cell functions in vitro. Immunol. Lett. 2012, 148, 34–38. [Google Scholar] [CrossRef]
- Sonda, N.; Simonato, F.; Peranzoni, E.; Calì, B.; Bortoluzzi, S.; Bisognin, A.; Wang, E.; Marincola, F.M.; Naldini, L.; Gentner, B. miR-142-3p prevents macrophage differentiation during cancer-induced myelopoiesis. Immunity 2013, 38, 1236–1249. [Google Scholar] [CrossRef]
- Bell, E.; Taylor, M.A. Functional roles for exosomal microRNAs in the tumour microenvironment. Comput. Struct. Biotechnol. J. 2017, 15, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.B.; Li, Z.L.; Luo, D.H.; Huang, B.J.; Chen, Y.S.; Zhang, X.S.; Cui, J.; Zeng, Y.X.; Li, J. Tumor-derived exosomes promote tumor progression and T-cell dysfunction through the regulation of enriched exosomal microRNAs in human nasopharyngeal carcinoma. Oncotarget 2014, 5, 5439. [Google Scholar] [CrossRef] [PubMed]
- Ding, G.; Zhou, L.; Qian, Y.; Fu, M.; Chen, J.; Chen, J.; Xiang, J.; Wu, Z.; Jiang, G.; Cao, L. Pancreatic cancer-derived exosomes transfer miRNAs to dendritic cells and inhibit RFXAP expression via miR-212-3p. Oncotarget 2015, 6, 29877. [Google Scholar] [CrossRef]
- Zhou, M.; Chen, J.; Zhou, L.; Chen, W.; Ding, G.; Cao, L. Pancreatic cancer derived exosomes regulate the expression of TLR4 in dendritic cells via miR-203. Cell. Immunol. 2014, 292, 65–69. [Google Scholar] [CrossRef]
- Rivoltini, L.; Chiodoni, C.; Squarcina, P.; Tortoreto, M.; Villa, A.; Vergani, B.; Burdek, M.; Botti, L.; Arioli, I.; Cova, A.; et al. TNF-Related Apoptosis-Inducing Ligand (TRAIL)-Armed Exosomes Deliver Proapoptotic Signals to Tumor Site. Clin. Cancer Res. 2016, 22, 3499–3512. [Google Scholar] [CrossRef] [PubMed]
- Catalano, V.; Turdo, A.; Di Franco, S.; Dieli, F.; Todaro, M.; Stassi, G. Tumor and its Microenvironment: A Synergistic Interplay. Semin. Cancer Biol. 2013, 23, 522–532. [Google Scholar]
- Roma-Rodrigues, C.; Fernandes, A.R.; Baptista, P.V. Exosome in tumour microenvironment: Overview of the crosstalk between normal and cancer cells. Biomed. Res. Int. 2014, 2014. [Google Scholar] [CrossRef]
- Ruivo, C.F.; Adem, B.; Silva, M.; Melo, S.A. The biology of cancer exosomes: Insights and new perspectives. Cancer Res. 2017, 77, 6480–6488. [Google Scholar] [CrossRef]
- Busby, J.; McMenamin, Ú.; Spence, A.; Johnston, B.; Hughes, C.; Cardwell, C. Angiotensin receptor blocker use and gastro-oesophageal cancer survival: A population-based cohort study. Aliment. Pharmacol. Ther. 2018, 47, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Coulson, R.; Liew, S.H.; Connelly, A.A.; Yee, N.S.; Deb, S.; Kumar, B.; Vargas, A.C.; O’Toole, S.A.; Parslow, A.C.; Poh, A. The angiotensin receptor blocker, Losartan, inhibits mammary tumor development and progression to invasive carcinoma. Oncotarget 2017, 8, 18640. [Google Scholar] [CrossRef] [PubMed]
- Diop-Frimpong, B.; Chauhan, V.P.; Krane, S.; Boucher, Y.; Jain, R.K. Losartan inhibits collagen I synthesis and improves the distribution and efficacy of nanotherapeutics in tumors. Proc. Natl. Acad. Sci. USA 2011, 108, 2909–2914. [Google Scholar] [CrossRef] [PubMed]
- Cassinelli, G.; Lanzi, C.; Tortoreto, M.; Cominetti, D.; Petrangolini, G.; Favini, E.; Zaffaroni, N.; Pisano, C.; Penco, S.; Vlodavsky, I. Antitumor efficacy of the heparanase inhibitor SST0001 alone and in combination with antiangiogenic agents in the treatment of human pediatric sarcoma models. Biochem. Pharmacol. 2013, 85, 1424–1432. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, J.P.; Ramani, V.C.; Ren, Y.; Naggi, A.; Torri, G.; Casu, B.; Penco, S.; Pisano, C.; Carminati, P.; Tortoreto, M. SST0001, a chemically modified heparin, inhibits myeloma growth and angiogenesis via disruption of the heparanase/syndecan-1 axis. Clin. Cancer Res. 2011, 17, 1382–1393. [Google Scholar] [CrossRef]
- Paolicchi, E.; Gemignani, F.; Krstic-Demonacos, M.; Dedhar, S.; Mutti, L.; Landi, S. Targeting hypoxic response for cancer therapy. Oncotarget 2016, 7, 13464. [Google Scholar] [CrossRef]
- Yu, T.; Tang, B.; Sun, X. Development of inhibitors targeting hypoxia-inducible factor 1 and 2 for cancer therapy. Yonsei Med. J. 2017, 58, 489–496. [Google Scholar] [CrossRef]
- Duffy, A.; Melillo, G.; Turkbey, B.; Allen, D.; Choyke, P.; Chen, C.; Raffeld, M.; Doroshow, J.; Murgo, A.; Kummar, S. A pilot trial of oral topotecan (TPT) in patients with refractory advanced solid neoplasms expressing HIF-1α. J. Clin. Oncol. 2010, 28, e13518. [Google Scholar] [CrossRef]
- Sounni, N.E.; Noel, A. Targeting the tumor microenvironment for cancer therapy. Clin. Chem. 2013, 59, 85–93. [Google Scholar] [CrossRef]
- Fukumura, D.; Jain, R.K. Tumor microenvironment abnormalities: Causes, consequences, and strategies to normalize. J. Cell. Biochem. 2007, 101, 937–949. [Google Scholar] [CrossRef]
- Netea-Maier, R.T.; Smit, J.W.; Netea, M.G. Metabolic changes in tumor cells and tumor-associated macrophages: A mutual relationship. Cancer Lett. 2018, 413, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Waldmann, T.A. Cytokines in cancer immunotherapy. Cold Spring Harb. Perspect. Biol. 2018, 10, a028472. [Google Scholar] [CrossRef] [PubMed]
- Bastos, N.; Ruivo, C.F.; da Silva, S.; Melo, S.A. Exosomes in cancer: Use them or target them? Semin. Cell Dev. Biol. 2018, 78, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Luberto, C.; Hassler, D.F.; Signorelli, P.; Okamoto, Y.; Sawai, H.; Boros, E.; Hazen-Martin, D.J.; Obeid, L.M.; Hannun, Y.A.; Smith, G.K. Inhibition of tumor necrosis factor-induced cell death in MCF7 by a novel inhibitor of neutral sphingomyelinase. J. Biol. Chem. 2002, 277, 41128–41139. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Thery, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Datta, A.; Kim, H.; Lal, M.; McGee, L.; Johnson, A.; Moustafa, A.A.; Jones, J.C.; Mondal, D.; Ferrer, M.; Abdel-Mageed, A.B. Manumycin A suppresses exosome biogenesis and secretion via targeted inhibition of Ras/Raf/ERK1/2 signaling and hnRNP H1 in castration-resistant prostate cancer cells. Cancer Lett. 2017, 408, 73–81. [Google Scholar] [CrossRef]
- Escrevente, C.; Keller, S.; Altevogt, P.; Costa, J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer 2011, 11, 108. [Google Scholar] [CrossRef]
| Protein(s) | Biological Function | Reference(s) |
|---|---|---|
| AHNAK | Cancer-associated exosomes increase motility of fibroblasts | [149] |
| ANXA2, CLSTN1, FASN, FLNC, FOLH1, and GDF15 | Correlated with PCa malignancy | [67] |
| CD9 | Increase the proliferation and chemo-resistance of PCa cells | [87,150] |
| B7-H3 | Immune checkpoint regulator | [151] |
| CD63, CD81, HSP90, HSP70, TNF1α, IL-6, MMP2, MMP9, Annexin II, TSG101, Akt, ILK1, and β-catenin | Increase stemness, metastasis and CAFs formation | [91] |
| c-Src, IGF-1R and FAK | PCa development and angiogenesis | [139,140,141] |
| Ets-1 | Induce osteoclast differentiation | [152] |
| FasL, TGF-β, galectin-9 and HSP72 | Evade immune responses | [145,146] |
| Galectin-1 | Promote angiogenesis | [153] |
| Integrin αvβ3 | Pro-inflammatory effect on stromal cells | [93,137] |
| Integrin β4, vinculin and P-gp | Associated with taxane and docetaxel resistance | [154,155] |
| MMP14 | Promote PCa cell growth | [156] |
| PD-L1 | Immune checkpoint regulator | [157] |
| Rab1a, Rab1b and Rab11a | Neoplastic transformation of pASCs | [130] |
| TGF-β | Differentiation of fibroblast to CAFs | [131,132,133,134,135] |
| TIMP-1 | Associated with PCa aggressiveness | [158] |
| Trop-2, vimentin, N-cadherin and Integrin αvβ3 | Induce PCa cell invasion | [92,136,159,160] |
| uPA | Vascularization remodeling of PCa microenvironment | [142,143] |
| VEGFA, HGF and MMP | Angiogenesis, EMT and tumor growth | [136] |
| Nucleic Acid(s) | Biological Function | Reference(s) |
|---|---|---|
| miR-125b, miR130b, miR-155 | Neoplastic transformation of PCa patients’ adipose stem cells | [130] |
| miR-125b | Downregulate AKT1 expression and induce PCa proliferation | [187] |
| miR-100, miR-21 and miR-139 | PCa growth and metastasis | [176] |
| miR-141, miR-21, miRNA-375 | Affect osteoclastogenesis and osteoblastogenesis and help PCa cells to overcome androgen deprivation in long distant metastasis | [180,181,188,189] |
| miR-409, miR-379 and miR-154 | Support PCa carcinogenesis and metastasis | [177] |
| miR-485-3p | Associated with fludarabine resistance in PCa cells | [190] |
| H-ras, K-ras (mRNA) | Neoplastic transformation of PC patients’ adipose stem cells. | [130] |
| LincRNA-p21 (lncRNA) | Associated with PC malignancy | [186] |
| lncRNA MY (lncRNA) | Promote adjacent cell proliferation and migration | [185] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saber, S.H.; Ali, H.E.A.; Gaballa, R.; Gaballah, M.; Ali, H.I.; Zerfaoui, M.; Abd Elmageed, Z.Y. Exosomes are the Driving Force in Preparing the Soil for the Metastatic Seeds: Lessons from the Prostate Cancer. Cells 2020, 9, 564. https://doi.org/10.3390/cells9030564
Saber SH, Ali HEA, Gaballa R, Gaballah M, Ali HI, Zerfaoui M, Abd Elmageed ZY. Exosomes are the Driving Force in Preparing the Soil for the Metastatic Seeds: Lessons from the Prostate Cancer. Cells. 2020; 9(3):564. https://doi.org/10.3390/cells9030564
Chicago/Turabian StyleSaber, Saber H., Hamdy E. A. Ali, Rofaida Gaballa, Mohamed Gaballah, Hamed I. Ali, Mourad Zerfaoui, and Zakaria Y. Abd Elmageed. 2020. "Exosomes are the Driving Force in Preparing the Soil for the Metastatic Seeds: Lessons from the Prostate Cancer" Cells 9, no. 3: 564. https://doi.org/10.3390/cells9030564
APA StyleSaber, S. H., Ali, H. E. A., Gaballa, R., Gaballah, M., Ali, H. I., Zerfaoui, M., & Abd Elmageed, Z. Y. (2020). Exosomes are the Driving Force in Preparing the Soil for the Metastatic Seeds: Lessons from the Prostate Cancer. Cells, 9(3), 564. https://doi.org/10.3390/cells9030564







