Abstract
Myocardial infarction (MI) is a worldwide condition that affects millions of people. This is mainly caused by the adult human heart lacking the ability to regenerate upon injury, whereas zebrafish have the capacity through cardiomyocyte proliferation to fully regenerate the heart following injury such as apex resection (AR). But a systematic overview of the methods used to evidence heart regrowth and regeneration in the zebrafish is lacking. Herein, we conducted a systematical search in Embase and Pubmed for studies on heart regeneration in the zebrafish following injury and identified 47 AR studies meeting the inclusion criteria. Overall, three different methods were used to assess heart regeneration in zebrafish AR hearts. 45 out of 47 studies performed qualitative (37) and quantitative (8) histology, whereas immunohistochemistry for various cell cycle markers combined with cardiomyocyte specific proteins was used in 34 out of 47 studies to determine cardiomyocyte proliferation qualitatively (6 studies) or quantitatively (28 studies). For both methods, analysis was based on selected heart sections and not the whole heart, which may bias interpretations. Likewise, interstudy comparison of reported cardiomyocyte proliferation indexes seems complicated by distinct study designs and reporting manners. Finally, six studies performed functional analysis to determine heart function, a hallmark of human heart injury after MI. In conclusion, our data implies that future studies should consider more quantitative methods eventually taking the 3D of the zebrafish heart into consideration when evidencing myocardial regrowth after AR. Furthermore, standardized guidelines for reporting cardiomyocyte proliferation and sham surgery details may be considered to enable inter study comparisons and robustly determine the effect of given genes on the process of heart regeneration.
1. Introduction
Heart failure (HF) is a global condition that has been estimated to affect more than 38 million people [1]. The pathology of HF includes a loss of cardiomyocytes (CM) that causes the heart to pump an insufficient amount of blood to the body. The most common cause of HF is myocardial infarction (MI) [2]. MI is defined as an area that has reduced or absent oxygen perfusion, which can cause the initiation of necrosis, apoptosis and autophagy leading to death of myocardial cells [2,3,4]. It is generally accepted that the mammalian heart cannot regenerate after ischemia or injury [5]. Thus, the compensatory mechanism after ischemia or injury, are assumed to favor fibrotic healing and hypertrophy instead of regeneration and cardiomyocyte proliferation [2,5]. Unfortunately, fibrotic healing may result in loss of heart contractility, leading to various complications and symptoms such as arrhythmias that also contribute to an increase in morbidity and mortality [5]. Today, only heart transplantation and palliative medication is offered as treatment of MI. Thus, a substantial effort is ongoing worldwide to develop regenerative therapies that can reduce mortality and improve life quality for millions of people [6].
2. The Regenerating Zebrafish Heart
In search for new treatment options for MI, researchers have focused their attention on certain lower vertebrates such as amphibians (e.g. axolotl, salamander) and some fish species such as zebrafish, that have shown the capacity to fully regenerate their hearts following myocardial injury. Even though, the zebrafish anatomy differs from the human heart by only having one ventricle and one atrium (Figure 1), several similarities exist. The zebrafish heart itself consists of a pericardial sac as also seen in humans, the bulbous arteriosus that can be compared to the human aortic arch, and the sinus venosus that is analogous to the human vena cava [7]. However, a dissimilarity, beside a lack of pulmonary vasculature, is that the zebrafish have an absence of a coronary artery network that is found in the human heart. Instead, zebrafish have small coronary capillary like vessels localized to the compact muscle layer, which has to be taken into consideration when performing heart injury models [8]. In 2002, Poss et al. [9] pioneered the field of zebrafish heart regeneration when demonstrating that the zebrafish heart fully regenerates following removal of ~10–20% of the ventricular apex (AR) (Figure 1A–C). Poss and colleagues showed that the zebrafish cardiac muscle had gained its full contractile function and normal appearance after 60 days post injury (dpi) [9,10]. Since then, other adult injury models such as cryoinjury, genetic cell ablation and hypoxia models have also been established. Whereas regeneration time and scar formation differs between injury types (i.e Apex resection (AR), cryoinjury, genetic ablation), the defining process of regeneration in various zebrafish injury models remains the same, and is the requirement of fully differentiated cardiomyocytes to revert to a developmental like state and re-enter the cell cycle in order to replenish the lost myocardium [11,12]. Overall, the AR model consists of different stages [6]. At 3–12 h dpi, a fibrin clot is formed that eventually dissolves, and localized apoptosis and necrosis occur, along with activation of the immune system. In parallel, the endocardial and epicardial cells become activated. At 3 dpi, myofibroblasts occupy the scar and secrete extracellular material, while endo- and epicardial cells undergo proliferation to cover the interior and exterior of the heart within the injury site, with epicardial cells actively invading the injury site later in the regeneration process [13]. The scar is transient, and while being dissolved, cardiomyocytes start proliferating, peaking at day 7–14 dpi and then migrating into the injury site. Hereafter, the myocardial wall will undergo thickening until the AR heart has become fully recovered at 60 dpi. In contrast, cryo-injury includes more substantial scar formation and requires a longer recovery time of 80-130 days [6]. Thus, given the ability of the zebrafish heart to regenerate relatively quickly following AR, the mechanisms underlying cardiomyocyte proliferation in AR hearts are of high interest for researchers to shed new lights on how to treat MI in humans.
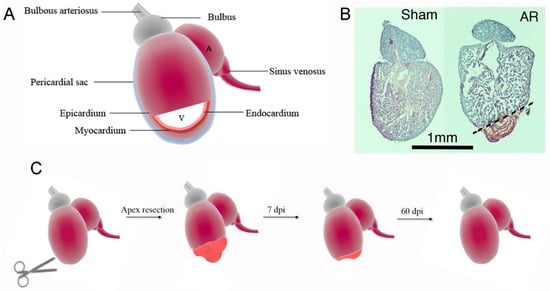
Figure 1.
Apex resected (AR) zebrafish hearts regenerates during a 60 days time course. (A) The zebrafish heart consists of an atrium (A) and a ventricle (V). The cardiac layers, starting from the inner, is called endocardium, myocardium and epicardium. The heart is furthermore covered by a pericardial sac. The blood comes from the sinus venosus into the atrium and goes to the ventricle and out through bulbous arteriosus. (B) Hematoxylin staining of zebrafish hearts following sham surgery and AR of 10–20% of the heart. (C) Schematic representation of the overall processes occurring following AR in the zebrafish heart. Initially, a fibrin clot is formed in the apex region, whereafter cardiomyocytes starts to proliferate, peaking at 7–14 dpi, and regenerates the heart until 60 dpi, when it is fully recovered.
Despite the rapid identification of proliferative signals since the introduction of the zebrafish model (For a detailed review refer to [6]), one current shortcoming in the field is that the techniques used for evaluation of heart regrowth and regeneration after AR are limited. This complicates a robust evaluation of genes and processes affecting zebrafish heart regeneration. To clarify this issue in detail and spur enthusiasm to develop new techniques for assessing zebrafish heart regeneration, we herein set out to perform a systematical literature analysis illuminating the quality and precision of the most commonly used methods for evidencing heart regrowth. We focused on studies exploiting the AR model since this injury type is the most prominent heart regeneration model studied in zebrafish.
3. Methods
3.1. Search Strategy
The systematical approach for literature search, study selection and data extraction follow the PRISMA guidelines. However, since our aim was to make a systematical assessment of existing literature regarding the methods used but not the reported results, we did not evaluate for risk of bias as otherwise recommended for meta-analysis etc. [14]. H.J.B. performed the systematical screening (Figure 2) in the Embase and Pubmed databases at 03/12/2018 and W.H. repeated the search at 04/11/2019. The search included three key MeSH and Emtree terms “Heart regeneration”, “Cardiac regeneration” and “Zebrafish”. The complete search strings in Pubmed (https://www.ncbi.nlm.nih.gov/pubmed/) and Embase (Ovid Interface; http://ovidsp.dc1.ovid.com.proxy1-bib.sdu.dk:2048/sp-3.33.0b/ovidweb.cgi) were as follows: Pubmed Search ((((((("journal article" [Publication Type]) AND zebrafish [MeSH Terms]) AND heart regeneration [MeSH Terms]) AND cardiac regeneration [MeSH Terms]) NOT "review" [Publication Type]) NOT "biography" [Publication Type]) NOT "editorial" [Publication Type]) NOT "comment" [Publication Type] and Embase search (Zebrafish and Heart regeneration and Cardiac regeneration).mp. not review.pt. Notably, MeSH and Emtree terms rely on annotation resulting in a two-three months delay from publication to annotation. Thus, we also checked literature using searches based on related free text terms, and hereby identified and included one article [15] that followed the criteria.
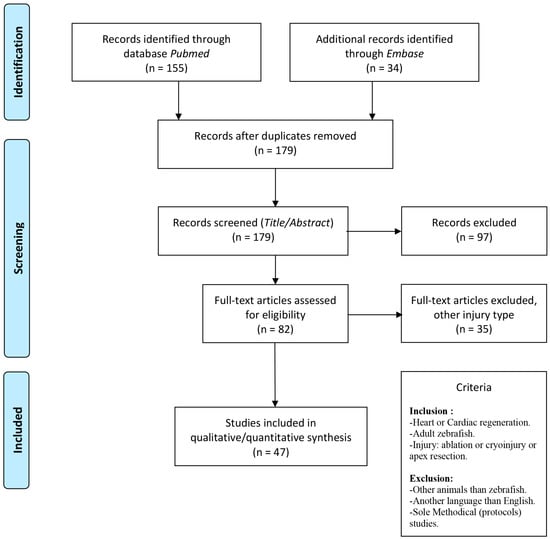
Figure 2.
Flowchart of the literature search including inclusion and exclusion criteria. See methods and materials for details. As modified from PRISMA guidelines.
3.2. Study Selection
Search identified studies were imported into the systematic data management program Covidence [16] where duplicates were initially removed by the program and then screened manually. Then Covidence was used to screen first on title and abstract, and then in a second round by full-text screening against inclusion and exclusion criteria (Figure 2, Supplementary Materials). Inclusion criteria accounted only for studies that had used adult zebrafish as a model for cardiac injury and evaluated cardiac regeneration. Only studies using injury types corresponding to ablation, cryoinjury or AR were extracted. Thus, studies using scratching, squeezing of the ventricle or hypoxia as heart injury were excluded from the search [17,18]. Methodical studies that primarily introduced a protocol for e.g. how to induce injury, were not included in the search [19,20]. Finally, we excluded studies if relevant information was lacking, if the full article was inaccessible online, or if a study was published in another language than English [21,22,23,24].
3.3. Extraction of Data
All data were extracted by H.J.B., and then re-checked independently by W.H. Any disagreements were solved by discussion between the disagreeing parties and by involving D.C.A. to solve the controversy. For each study, detailed data on injury type was first extracted and then in a second round of data extraction only AR studies were further evaluated for extraction on data concerning author research group and methods used for quantification of heart regeneration and regrowth. This included both methods related to reestablishment of the cardiomyocyte pool and the myocardium, as well as, measures of scarring and function. Moreover, we extracted count data and continuous data on days of injury, proliferation markers used, and measures as well as design relating to a cardiomyocyte proliferation index. By using the Pivot Table function in Microsoft excel, the identified data were categorized, sorted and counted. Data are either presented as count or continuous data.
4. Results
4.1. Included Studies on Zebrafish Heart Regeneration Following Apex Resection
A total of 34 studies from Embase and 155 from Pubmed were retrieved. Ten duplicates were identified, and the remaining 179 studies were screened against inclusion criteria based on title and abstract using Covidence (Figure 2). Further full text screening was then conducted for 82 articles, where approximately half (47 studies) used apex resection (AR), 27 exploited cryo-injury, and only 8 studies performed genetic ablation (Table 1).

Table 1.
Systematically identified zebrafish studies divided by heart injury type.
The 47 studies that used AR as an injury type, were performed by 24 different research groups, with only three groups headed by Poss (11 studies), Belmonte (5 studies), and Kawakami (3 studies) publishing more than two studies (Table 2). We identified that the majority of AR studies (45/47) used qualitative analysis of heart sections for evaluating viable myocardium, where 8 of these extended this evaluation to include quantitative measures such as scarring measures (Table 2). 34 out of the 47 studies assessed more directly heart regeneration by analyzing cardiomyocyte proliferation, whereas 6 out of 47 studies performed functional heart analysis (Table 2).

Table 2.
Methods used to evaluate heart regrowth and regeneration after apex resection (AR) in zebrafish.
4.2. Qualitative and Quantitative Analysis of Cardiac Outgrowth
A total number of 45 out of 47 articles used histology on sections for assessing heart regrowth and regeneration after AR (Table 2). The two articles [87,88] that did not use histology were focused on evaluating whether heart function as measured by echocardiography reflect heart regrowth. Thus, histology in general seems to be an accepted qualitative method for the evaluation of heart regeneration in zebrafish. Histology was performed either by Acid Fuchsin Orange G (AFOG; 23 studies), Masson’s Trichrome (MT; 5 studies), or Hematoxylin/Eosin (HE; 5 studies) or single reagents hereof [44]. Whereas HE [9,15,32,46,59] stains nucleic acids and proteins (nonspecifically e.g., cytoplasm and extracellular matrix) [97], AFOG and MT staining highlight the difference between muscle and collagen. The preference for AFOG staining may likely be explained by a higher sensitivity for Collagen in particular in zebrafish as suggested by Poss et al. [9]. Eight out of the 45 studies [9,29,36,66,73,89,90,94] evaluated the extent of heart regeneration by quantifying the amount of fibrosis on heart tissue sections in 2D where the area of collagen was compared to the total area of the ventricle.
4.3. Quantification of Cardiomyocyte Proliferation
34 studies out of the 47 studies used CM proliferation either alone or in combination with the above mentioned method as a measurement for quantifying heart regeneration [9,11,12,13,15,29,30,31,32,35,36,44,46,51,59,60,64,68,69,73,74,79,80,81,83,84,85,86,89,92,93,94,95,96]. Accordingly, expression of proliferating cell nuclear antigen (PCNA) (20 out of 34 studies [15,29,31,35,44,59,60,68,69,74,79,80,81,83,84,85,89,94,95,96]), phosphohistone-H3 (PHH3) (7 out of 34 studies [9,11,29,30,31,64,68]) as well as incorporation of 5-Bromo-2′-deoxyuridine (BrdU) (17 out of 34 studies [9,11,12,13,29,30,31,32,36,44,46,51,64,73,86,92,93]) or 5-Ethynyl-2′-deoxyuridine (EdU)(2 out of 34 studies [59,85]) have been used to assess CM proliferation (Table 3, Table 4 and Table 5) in combination with a cardiomyocyte marker (myocyte enhancer factor 2 (Mef2c), Myosin heavy chain 1 (MYH1), α-Sarcomeric Actin, or a transgene CM reporter (such as cmlc2::DsRed2). One study used Phalloidin to define cardiac cells and one study based their evaluation on structural characteristics. The Assessment of CM proliferation was mainly reported as a CM proliferation index in percentage or as an absolute number, whereas only a few studies rely on qualitative evaluations (Table 5). Yet, besides the use of different proliferation- and CM markers, studies were distinct in study designs with respect to BrdU/EdU exposure time and the number of injections (Table 5).

Table 3.
Proliferation markers used to quantify cardiomyocyte proliferation after apex resection (AR) in zebrafish.

Table 4.
Timepoint (days past injury: dpi) used for assessment of cardiomyocyte proliferation after apex resection (AR) in zebrafish.

Table 5.
Overview of design and reporting manners for cardiomyocyte proliferation studies after Apex resection (AR) in zebrafish.
4.4. Analysis of Heart Function
The zebrafish heart, despite having only two chambers and a lack of pulmonary vasculature, shows a similar pattern of electrical activity and pump function as measured in humans when using electrocardiography (ECG) and echocardiography (ECHO), respectively. Yet, the parametric values are different between species, and data are thus mainly suited for relative measures between groups such as AR and sham zebrafish [88,98].
In our systematical analysis we identified a total number of six studies [7,12,49,87,88,89] that used a functional heart test as a measurement for cardiac recovery after AR. Three of these studies [7,49,89] used electrocardiography (ECG) while the other half [12,87,88] performed echocardiography.
The three studies that used ECG evaluated functional recovery after AR either by measuring variation in the R-R interval, (the time between each heartbeat) [89], or by measuring variation in the QT intervals [7,49].
5. Discussion
To the best of our knowledge, we here provide the first systematical analysis of methods used for evaluating heart regrowth and regeneration after apex resection in zebrafish. This is important since zebrafish offers an opportunity to identify mechanisms underlying heart regeneration, which may then be translated into the non-regenerative mammalian heart to improve heart repair after MI. 47 zebrafish studies performing AR were identified and showed that 95.7% of these studies used histology either alone or in combination with immunofluorescence detection of cardiomyocyte proliferation to assess zebrafish heart regeneration. 82.2% of the histology studies were merely qualitative, while 82.4% of the CM proliferation studies were quantitative. Only 12.8% of all studies included functional heart test, and no study used height or volume of the regenerating myocardium, which may provide even more robust evidence for heart outgrowth as discussed below.
Overall, histology was used in the studies to discriminate between viable myocardium and collagen rich scar tissue. The latter is a major outcome after MI in mammals, and as such it cannot compensate functionally for the CM loss. However, despite being informative on the presence or absence of fibrosis in the AR zebrafish hearts, such histology stains do not evidence heart outgrowth and regeneration capacity. This likely explains why only 8 out of 47 studies indeed perform quantitative histology (e.g. scarring index), whereas remaining studies only report qualitative evaluations. Moreover, one major caveat of this type of assay is that quantifications are based on a few sections, which then represent the whole heart. In this regard, it is important to note that consensus in the field exist, where the two-three largest sections of the ventricle are chosen to make an averaged and thus more robust analysis [87,95,96]. Yet, this could also bias heart outgrowth analysis, since one will select towards the size of the sections and not the injury site itself. With the numerous new imaging modalities 3D analysis or at least 2D serial analysis throughout the heart could be an alternative to minimize such bias and enable more robust quantifications as discussed below. Still the use of histology stains should be treated with cautions when claiming heart outgrowth. For instance, the degree of fibrosis and the amount of viable myocardium may depend on initial heart size and lesion performance, both parameters that may vary a lot in a heart that is only 1mm wide on average [6].
In contrast to fibrosis as a measure of heart regeneration, CM proliferation seems more appropriate as this biological process is considered the main underlying mechanism of heart regeneration [99]. As in general, zebrafish CMs go through the four phases of the cell cycle: G1, S, G2 and M, but in contrast to mammals the proportion of binucleated and polyploid CMs are <5% [36,99]. The latter may partly explain why zebrafish accomplish heart regeneration per se, but it is also important to consider when quantifying CM proliferation. In mammals, the majority of proliferation markers cannot distinguish whether a CM indeed undergoes division or simply binucleates or becomes polyploid (as reviewed elsewhere [100,101]), but in zebrafish it must be assumed that expression of these markers mainly represents the formation of two daughter CMs, although binucleation may occur at a low rate. However, the reported rates of CM proliferation may be difficult to compare across studies (Table 5), since the experimental design and reporting manner vary (Table 5). Also, the choice of cardiomyocyte- and proliferation markers as well as the day of analysis post injury are different between studies (Table 3, Table 4 and Table 5).
The inclusion of a cardiomyocyte marker is a prerequisite since also fibroblasts, hematopoietic cells as well as epicardial-, endocardial- and vascular cells proliferate following AR injury [6,8,102]. While Mef2c is a nuclear transcription factor, MYH1 and α-Sarcomeric Actin are cytoplasmic proteins present, and all can be used to label cardiomyocytes in the [103,104]. Visualization of these proteins mainly relies on antibody detection and thus depends on the quality of the reagents and protocols as in general. Since the used proliferation markers PCNA, BrdU/EdU, and PHH3 represent proteins residing in the nucleus, Mef2c co-localization may be more specific as compared to co-localizations with MYH1, α-Sarcomeric Actin, and the cardiac myosin light chain cmlc2 reporters that may require confocal imaging in 3D to validate single CM co-localization. The use of Phalloidin and CM structural characteristics [32,46] may be less robust in defining CMs. The used proliferation markers PCNA, BrdU/EdU, PHH3 are detected via immunofluorescence in the identified studies. Yet, it is important to note that they differ in their expression during the cell cycle which can have an impact upon analysis and interstudy comparison [100,101]. Pulse chase labelling using the thymidine analogues BrdU or EdU are golden standards for assessing cell proliferation through DNA labeling [105,106]. When comparing with PCNA, one must consider that PCNA marks cardiomyocytes from G1 phase until S phase, whereas thymidine analogues marks cardiomyocytes from S phase and onward. This will result in higher number of proliferating cardiomyocytes detected by PCNA per cell cycle [100,101,107]. In the identified studies, evaluation of CM proliferation was performed at different timepoints with 7- and 14 dpi being most commonly used (see Table 4 and Table 5) [9,60]. Since longer periods of BrdU/EdU incorporation and repetitive labelling pulses will increase the number of labelled cardiomyocytes, the quantifications depend on the specific BrdU/EdU setup [107] and should be taken into consideration upon interstudy comparisons.
In humans, the heart pump function is the ultimate determinant of a patient’s well-being after MI with arrythmias after infarction being a dangerous complication, and in mouse heart regeneration studies functional analysis such as MRI, ECHO or PET often are included. The similar pattern of electrical activity between zebrafish and humans despite the obvious anatomical difference makes it also a candidate for similar functional studies in zebrafish. However, in zebrafish AR studies, only 6 out of the 47 identified studies used functional analysis of heart function likely due to the small size of the zebrafish heart and the sensitivity of available techniques. ECHO is a non-invasive method in zebrafish, that uses ultrasound for imaging of cardiac structures and visualizes systolic and diastolic function [108], representing ventricular emptying and ventricular filling, respectively. In two [87,88] out of the three ECHO studies identified, ECHO was the sole measure of heart recovery and regeneration. In this regards it is important to acknowledge that ECHO is highly subjective and depends on user training and equipment quality among other parameters [109]. ECG, another non-invasive method, on the other hand might be more reproducible, and favorable for studying arrythmias, as indicated by QT- and T-wave abnormalities that can appear after AR. Prolongation of QT interval would suggest alterations in the ventricular repolarization that should normalize over time when the heart is healed and can thus be used as a readout for complete functional regeneration. Interestingly study showed a type of arrythmia with a prolongated QT interval after AR, reflecting ventricular repolarization and which is also seen after cryoinjury [49,51]. This may still persist even after complete heart regeneration as evidenced by histology [49]. In contrast, another article reported a shortened QT interval, another type of arrythmia after AR [7]. With only six studies using a functional readout and varying results, there is room for an improvement in implementation of techniques such as ECHO and ECG analysis in zebrafish.
6. Conclusions and Perspectives
Finally, we cannot emphasize enough that the present analysis is restricted to the data collected for the present study and we also cannot exclude that a few eligible studies have been unnoticed by our search strategy, and we apologize to authors of those studies. Other factors than those analyzed and discussed above may as well have an impact on the evaluation of zebrafish heart regrowth after AR. Particular we observed that only 3 out of 47 articles [46,49,74] explained the procedure of sham operations in detail and described clearly whether the sham was matched with a particular dpi specimen. Since, CM proliferation can be induced by injury or activation of tissues adjacent to the myocardium such as the epicardium [60,102], sham animals at a matched day seem obligate to consider when evaluating heart regeneration after AR.
Furthermore, our data suggest that some of the more advanced new imaging modalities could improve the field and accuracy of determining heart regrowth after AR. For instance, one may consider measuring the myocardial volume or height after AR and compare with a day matched sham. In this regard, the volume or height of myocardial tissue could ideally be quantified by two-photon imaging of a persistent cardiomyocyte reporter following tissue clearance. Very recently, Mercader and co-workers used a similar approach for analyzing Sox10+ CMs following cell specific ablation within the zebrafish heart [110].
Thus, although some consensus exists in the field, more standard guidelines for testing and reporting as well as the use of new less biased approaches could improve the field and provide more close evidence of heart regrowth after AR in zebrafish.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4409/9/3/548/s1, Prisma 2009 Checklist.
Author Contributions
H.J.B. and W.H.: Collection of data, Data analysis and interpretation, Manuscript writing and Final approval of manuscript. D.C.A.: Conception and design, Data interpretation, Manuscript writing, Final approval of manuscript, and Financial support. All authors have read and agreed to the published version of the manuscript.
Funding
The work was supported by research grants from The Novo Nordisk Foundation Section for Basic Stem Cell Biology, National Collaboration; The Novo Nordisk Foundation (#NNF17OC0028764); The Danish National Research Council (Sapere Aude; # 8045-00019B); and financial support from the Dep. Of Clinical Biochemistry and Pharmacology/Odense University Hospital and Clinical Institute, University of Southern Denmark.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cahill, T.J.; Choudhury, R.P.; Riley, P.R. Heart regeneration and repair after myocardial infarction: Translational opportunities for novel therapeutics. Nat. Rev. Drug Discov. 2017, 16, 699–717. [Google Scholar] [CrossRef] [PubMed]
- Burke, A.P.; Virmani, R. Pathophysiology of acute myocardial infarction. Med. Clin. N. Am. 2007, 91, 553–572; ix. [Google Scholar] [CrossRef] [PubMed]
- Talman, V.; Ruskoaho, H. Cardiac fibrosis in myocardial infarction-from repair and remodeling to regeneration. Cell Tissue Res. 2016, 365, 563–581. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.F.; Cui, M.X.; Yang, S.W.; Zhou, Y.J.; Hu, D.Y. Cell death, dysglycemia and myocardial infarction. Biomed. Rep. 2013, 1, 341–346. [Google Scholar] [CrossRef]
- Van Amerongen, M.J.; Engel, F.B. Features of cardiomyocyte proliferation and its potential for cardiac regeneration. J. Cell. Mol. Med. 2008, 12, 2233–2244. [Google Scholar] [CrossRef]
- Gonzalez-Rosa, J.M.; Burns, C.E.; Burns, C.G. Zebrafish heart regeneration: 15 years of discoveries. Regeneration 2017, 4, 105–123. [Google Scholar] [CrossRef]
- Sun, P.; Zhang, Y.; Yu, F.; Parks, E.; Lyman, A.; Wu, Q.; Ai, L.; Hu, C.H.; Zhou, Q.; Shung, K.; et al. Micro-electrocardiograms to study post-ventricular amputation of zebrafish heart. Ann. Biomed. Eng. 2009, 37, 890–901. [Google Scholar] [CrossRef]
- Tahara, N.; Brush, M.; Kawakami, Y. Cell migration during heart regeneration in zebrafish. Dev. Dyn. 2016, 245, 774–787. [Google Scholar] [CrossRef]
- Poss, K.D.; Wilson, L.G.; Keating, M.T. Heart regeneration in zebrafish. Science 2002, 298, 2188–2190. [Google Scholar] [CrossRef]
- Uygur, A.; Lee, R.T. Mechanisms of Cardiac Regeneration. Dev. Cell 2016, 36, 362–374. [Google Scholar] [CrossRef]
- Jopling, C.; Sleep, E.; Raya, M.; Marti, M.; Raya, A.; Izpisua Belmonte, J.C. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 2010, 464, 606–609. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, K.; Holdway, J.E.; Werdich, A.A.; Anderson, R.M.; Fang, Y.; Egnaczyk, G.F.; Evans, T.; Macrae, C.A.; Stainier, D.Y.; Poss, K.D. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 2010, 464, 601–605. [Google Scholar] [CrossRef] [PubMed]
- Lepilina, A.; Coon, A.N.; Kikuchi, K.; Holdway, J.E.; Roberts, R.W.; Burns, C.G.; Poss, K.D. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell 2006, 127, 607–619. [Google Scholar] [CrossRef]
- Viswanathan, M.; Ansari, M.T.; Berkman, N.D.; Chang, S.; Hartling, L.; McPheeters, M.; Santaguida, P.L.; Shamliyan, T.; Singh, K.; Tsertsvadze, A.; et al. Assessing the risk of bias of individual studies in systematic reviews of health care interventions. In Methods Guide for Comparative Effectiveness Reviews; Agency for Healthcare Research and Quality: Rockville, MD, USA, 2008. [Google Scholar]
- Itou, J.; Kawakami, H.; Burgoyne, T.; Kawakami, Y. Life-long preservation of the regenerative capacity in the fin and heart in zebrafish. Biol. Open 2012, 1, 739–746. [Google Scholar] [CrossRef] [PubMed]
- Covidence Systematic Review Software. Veritas Health Innovation, Melbourne, Australia. Available online: www.covidence.org (accessed on 15 January 2020).
- Borchardt, T.; Looso, M.; Bruckskotten, M.; Weis, P.; Kruse, J.; Braun, T. Analysis of newly established EST databases reveals similarities between heart regeneration in newt and fish. BMC Genom. 2010, 11, 4. [Google Scholar] [CrossRef] [PubMed]
- Parente, V.; Balasso, S.; Pompilio, G.; Verduci, L.; Colombo, G.I.; Milano, G.; Guerrini, U.; Squadroni, L.; Cotelli, F.; Pozzoli, O.; et al. Hypoxia/reoxygenation cardiac injury and regeneration in zebrafish adult heart. PLoS ONE 2013, 8, e53748. [Google Scholar] [CrossRef]
- Liu, X.X.; Zhang, Y.T.; Zhang, B. A surgery protocol to construct zebrafish heart damage and regeneration model. Yi Chuan Hered. 2013, 35, 529–532. [Google Scholar] [CrossRef]
- Wang, J.; Poss, K.D. Methodologies for Inducing Cardiac Injury and Assaying Regeneration in Adult Zebrafish. Methods Mol. Biol. 2016, 1451, 225–235. [Google Scholar] [CrossRef]
- Zuppo, D.A.; Missinato, M.A.; DeMoya, R.A.; SaydMohammed, M.; Tsang, M. Cardiac transcriptome profiling during regeneration in zebrafish. In Molecular Biology of the Cell (volume 27); ASCB: Betesda, MD, USA, 2017. [Google Scholar]
- Sleep, E.; Boue, S.; Jopling, C.; Raya, M.; Raya, A.; Izpisua Belmonte, J.C. Transcriptomics approach to investigate zebrafish heart regeneration. J. Cardiovasc. Med. 2010, 11, 369–380. [Google Scholar] [CrossRef]
- Schoffstall, B.; DeVerteuil, P.; Jean, M.; Lopez, N.; Tapia, J. The regenerative response of zebrafish hearts to long-term induced exercise stress. In Molecular Biology of the Cell (volume 22); ASCB: Betesda, MD, USA, 2011. [Google Scholar]
- Mias, C.; Genet, G.; Pathak, A.; Senard, J.M.; Gales, C. Adult resident cardiomyocytes wake up: New axis for cardiac tissue regeneration. Med. Sci. 2012, 28, 1103–1109. [Google Scholar]
- Grajevskaja, V.; Camerota, D.; Bellipanni, G.; Balciuniene, J.; Balciunas, D. Analysis of a conditional gene trap reveals that tbx5a is required for heart regeneration in zebrafish. PLoS ONE 2018, 13, e0197293. [Google Scholar] [CrossRef] [PubMed]
- Rodius, S.; Nazarov, P.V.; Nepomuceno-Chamorro, I.A.; Jeanty, C.; Gonzalez-Rosa, J.M.; Ibberson, M.; da Costa, R.M.; Xenarios, I.; Mercader, N.; Azuaje, F. Transcriptional response to cardiac injury in the zebrafish: Systematic identification of genes with highly concordant activity across in vivo models. BMC Genom. 2014, 15, 852. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rodius, S.; Androsova, G.; Gotz, L.; Liechti, R.; Crespo, I.; Merz, S.; Nazarov, P.V.; de Klein, N.; Jeanty, C.; Gonzalez-Rosa, J.M.; et al. Analysis of the dynamic co-expression network of heart regeneration in the zebrafish. Sci. Rep. 2016, 6, 26822. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Navis, A.; Cox, B.D.; Dickson, A.L.; Gemberling, M.; Karra, R.; Bagnat, M.; Poss, K.D. Single epicardial cell transcriptome sequencing identifies Caveolin 1 as an essential factor in zebrafish heart regeneration. Development 2016, 143, 232–243. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, A.; Montserrat, N.; Zacchigna, S.; Nivet, E.; Hishida, T.; Krause, M.N.; Kurian, L.; Ocampo, A.; Vazquez-Ferrer, E.; Rodriguez-Esteban, C.; et al. In vivo activation of a conserved microRNA program induces mammalian heart regeneration. Cell Stem Cell 2014, 15, 589–604. [Google Scholar] [CrossRef]
- Jopling, C.; Sune, G.; Faucherre, A.; Fabregat, C.; Izpisua Belmonte, J.C. Hypoxia induces myocardial regeneration in zebrafish. Circulation 2012, 126, 3017–3027. [Google Scholar] [CrossRef] [PubMed]
- Jopling, C.; Sune, G.; Morera, C.; Izpisua Belmonte, J.C. p38alpha MAPK regulates myocardial regeneration in zebrafish. Cell Cycle 2012, 11, 1195–1201. [Google Scholar] [CrossRef]
- Raya, A.; Koth, C.M.; Buscher, D.; Kawakami, Y.; Itoh, T.; Raya, R.M.; Sternik, G.; Tsai, H.J.; Rodriguez-Esteban, C.; Izpisua-Belmonte, J.C. Activation of Notch signaling pathway precedes heart regeneration in zebrafish. Proc. Natl. Acad. Sci. USA 2003, 100, 11889–11895. [Google Scholar] [CrossRef]
- Wu, C.C.; Kruse, F.; Vasudevarao, M.D.; Junker, J.P.; Zebrowski, D.C.; Fischer, K.; Noel, E.S.; Grun, D.; Berezikov, E.; Engel, F.B.; et al. Spatially Resolved Genome-wide Transcriptional Profiling Identifies BMP Signaling as Essential Regulator of Zebrafish Cardiomyocyte Regeneration. Dev. Cell 2016, 36, 36–49. [Google Scholar] [CrossRef]
- Sanchez-Iranzo, H.; Galardi-Castilla, M.; Minguillon, C.; Sanz-Morejon, A.; Gonzalez-Rosa, J.M.; Felker, A.; Ernst, A.; Guzman-Martinez, G.; Mosimann, C.; Mercader, N. Tbx5a lineage tracing shows cardiomyocyte plasticity during zebrafish heart regeneration. Nat. Commun 2018, 9, 428. [Google Scholar] [CrossRef]
- Zhao, L.; Borikova, A.L.; Ben-Yair, R.; Guner-Ataman, B.; MacRae, C.A.; Lee, R.T.; Burns, C.G.; Burns, C.E. Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration. Proc. Natl. Acad. Sci. USA 2014, 111, 1403–1408. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Rosa, J.M.; Sharpe, M.; Field, D.; Soonpaa, M.H.; Field, L.J.; Burns, C.E.; Burns, C.G. Myocardial Polyploidization Creates a Barrier to Heart Regeneration in Zebrafish. Dev. Cell 2018, 44, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.K.; Sarathchandra, P.; Chester, A.; Yacoub, M.; Brand, T.; Butcher, J.T. Cardiac regeneration following cryoinjury in the adult zebrafish targets a maturation-specific biomechanical remodeling program. Sci. Rep. 2018, 8, 15661. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Panakova, D.; Kikuchi, K.; Holdway, J.E.; Gemberling, M.; Burris, J.S.; Singh, S.P.; Dickson, A.L.; Lin, Y.F.; Sabeh, M.K.; et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development 2011, 138, 3421–3430. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Poss, K.D. Explant culture of adult zebrafish hearts for epicardial regeneration studies. Nat. Protoc. 2016, 11, 872–881. [Google Scholar] [CrossRef]
- Wang, J.; Cao, J.; Dickson, A.L.; Poss, K.D. Epicardial regeneration is guided by cardiac outflow tract and Hedgehog signalling. Nature 2015, 522, 226–230. [Google Scholar] [CrossRef]
- Wang, J.; Karra, R.; Dickson, A.L.; Poss, K.D. Fibronectin is deposited by injury-activated epicardial cells and is necessary for zebrafish heart regeneration. Dev. Biol. 2013, 382, 427–435. [Google Scholar] [CrossRef]
- Chen, C.H.; Durand, E.; Wang, J.; Zon, L.I.; Poss, K.D. zebraflash transgenic lines for in vivo bioluminescence imaging of stem cells and regeneration in adult zebrafish. Development 2013, 140, 4988–4997. [Google Scholar] [CrossRef]
- Cao, J.; Wang, J.; Jackman, C.P.; Cox, A.H.; Trembley, M.A.; Balowski, J.J.; Cox, B.D.; De Simone, A.; Dickson, A.L.; Di Talia, S.; et al. Tension Creates an Endoreplication Wavefront that Leads Regeneration of Epicardial Tissue. Dev. Cell 2017, 42, 600–615. [Google Scholar] [CrossRef]
- Huang, W.C.; Yang, C.C.; Chen, I.H.; Liu, Y.M.; Chang, S.J.; Chuang, Y.J. Treatment of Glucocorticoids Inhibited Early Immune Responses and Impaired Cardiac Repair in Adult Zebrafish. PLoS ONE 2013, 8, e66613. [Google Scholar] [CrossRef]
- Bednarek, D.; Gonzalez-Rosa, J.M.; Guzman-Martinez, G.; Gutierrez-Gutierrez, O.; Aguado, T.; Sanchez-Ferrer, C.; Marques, I.J.; Galardi-Castilla, M.; de Diego, I.; Gomez, M.J.; et al. Telomerase Is Essential for Zebrafish Heart Regeneration. Cell Rep. 2015, 12, 1691–1703. [Google Scholar] [CrossRef] [PubMed]
- Hofsteen, P.; Mehta, V.; Kim, M.S.; Peterson, R.E.; Heideman, W. TCDD inhibits heart regeneration in adult zebrafish. Toxicol. Sci. 2013, 132, 211–221. [Google Scholar] [CrossRef]
- Hein, S.J.; Lehmann, L.H.; Kossack, M.; Juergensen, L.; Fuchs, D.; Katus, H.A.; Hassel, D. Advanced echocardiography in adult zebrafish reveals delayed recovery of heart function after myocardial cryoinjury. PLoS ONE 2015, 10, e0122665. [Google Scholar] [CrossRef] [PubMed]
- Hein, S.; Lehmann, L.; Katus, H.A.; Hassel, D. High frequency echocardiography and speckle tracking based strain analysis revealed delayed functional recovery after myocardial cryoinjury in adult zebrafish. Circ. Conf. 2014, 130. [Google Scholar]
- Yu, F.; Li, R.; Parks, E.; Takabe, W.; Hsiai, T.K. Electrocardiogram signals to assess zebrafish heart regeneration: Implication of long QT intervals. Ann. Biomed. Eng. 2010, 38, 2346–2357. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Cao, H.; Kang, B.J.; Jen, N.; Yu, F.; Lee, C.A.; Fei, P.; Park, J.; Bohlool, S.; Lash-Rosenberg, L.; et al. emodynamics and ventricular function in a zebrafish model of injury and repair. Zebrafish 2014, 11, 447–454. [Google Scholar] [CrossRef]
- Chablais, F.; Jazwinska, A. The regenerative capacity of the zebrafish heart is dependent on TGFbeta signaling. Development 2012, 139, 1921–1930. [Google Scholar] [CrossRef]
- Pfefferli, C.; Jazwinska, A. The careg element reveals a common regulation of regeneration in the zebrafish myocardium and fin. Nat. Commun. 2017, 8, 15151. [Google Scholar] [CrossRef]
- Chablais, F.; Veit, J.; Rainer, G.; Jazwinska, A. The zebrafish heart regenerates after cryoinjury-induced myocardial infarction. BMC Dev. Biol 2011, 11, 21. [Google Scholar] [CrossRef]
- De Preux Charles, A.S.; Bise, T.; Baier, F.; Marro, J.; Jazwinska, A. Distinct effects of inflammation on preconditioning and regeneration of the adult zebrafish heart. Open Biol. 2016, 6. [Google Scholar] [CrossRef]
- De Preux Charles, A.S.; Bise, T.; Baier, F.; Sallin, P.; Jazwinska, A. Preconditioning boosts regenerative programmes in the adult zebrafish heart. Open Biol. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Marro, J.; Pfefferli, C.; de Preux Charles, A.S.; Bise, T.; Jazwinska, A. Collagen XII Contributes to Epicardial and Connective Tissues in the Zebrafish Heart during Ontogenesis and Regeneration. PLoS ONE 2016, 11, e0165497. [Google Scholar] [CrossRef] [PubMed]
- Sallin, P.; de Preux Charles, A.S.; Duruz, V.; Pfefferli, C.; Jazwinska, A. A dual epimorphic and compensatory mode of heart regeneration in zebrafish. Dev. Biol. 2015, 399, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Sallin, P.; Jazwinska, A. Acute stress is detrimental to heart regeneration in zebrafish. Open Biol. 2016, 6, 160012. [Google Scholar] [CrossRef] [PubMed]
- Itou, J.; Oishi, I.; Kawakami, H.; Glass, T.J.; Richter, J.; Johnson, A.; Lund, T.C.; Kawakami, Y. Migration of cardiomyocytes is essential for heart regeneration in zebrafish. Development 2012, 139, 4133–4142. [Google Scholar] [CrossRef] [PubMed]
- Itou, J.; Akiyama, R.; Pehoski, S.; Yu, X.; Kawakami, H.; Kawakami, Y. Regenerative responses after mild heart injuries for cardiomyocyte proliferation in zebrafish. Dev. Dyn. 2014, 243, 1477–1486. [Google Scholar] [CrossRef]
- Gonzalez-Rosa, J.M.; Martin, V.; Peralta, M.; Torres, M.; Mercader, N. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development 2011, 138, 1663–1674. [Google Scholar] [CrossRef]
- Gonzalez-Rosa, J.M.; Guzman-Martinez, G.; Marques, I.J.; Sanchez-Iranzo, H.; Jimenez-Borreguero, L.J.; Mercader, N. Use of echocardiography reveals reestablishment of ventricular pumping efficiency and partial ventricular wall motion recovery upon ventricular cryoinjury in the Zebrafish. PLoS ONE 2014, 9, e115604. [Google Scholar] [CrossRef]
- Sanchez-Iranzo, H.; Galardi-Castilla, M.; Sanz-Morejon, A.; Gonzalez-Rosa, J.M.; Costa, R.; Ernst, A.; Sainz de Aja, J.; Langa, X.; Mercader, N. Transient fibrosis resolves via fibroblast inactivation in the regenerating zebrafish heart. Proc. Natl. Acad. Sci. USA 2018, 115, 4188–4193. [Google Scholar] [CrossRef]
- Lien, C.L.; Schebesta, M.; Makino, S.; Weber, G.J.; Keating, M.T. Gene expression analysis of zebrafish heart regeneration. Plos Biol. 2006, 4, 1386–1396. [Google Scholar] [CrossRef]
- Rovira, M.; Borras, D.M.; Marques, I.J.; Puig, C.; Planas, J.V. Physiological responses to swimming-induced exercise in the adult zebrafish regenerating heart. Front. Physiol. 2018, 9, 1362. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Morioka, M.; Kimura, S.; Tasaki, M.; Inohaya, K.; Kudo, A. Differential reparative phenotypes between zebrafish and medaka after cardiac injury. Dev. Dyn. 2014, 243, 1106–1115. [Google Scholar] [CrossRef] [PubMed]
- Munch, J.; Grivas, D.; Gonzalez-Rajal, A.; Torregrosa-Carrion, R.; de la Pompa, J.L. Notch signalling restricts inflammation and serpine1 expression in the dynamic endocardium of the regenerating zebrafish heart. Development 2017, 144, 1425–1440. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, A.I.; O’Meara, C.C.; Gemberling, M.; Zhao, L.; Bryant, D.M.; Zheng, R.; Gannon, J.B.; Cai, L.; Choi, W.Y.; Egnaczyk, G.F.; et al. Nerves Regulate Cardiomyocyte Proliferation and Heart Regeneration. Dev. Cell 2015, 34, 387–399. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, N.; Abbas, Y.; Bryant, D.M.; Gonzalez-Rosa, J.M.; Sharpe, M.; Uygur, A.; Cocco-Delgado, L.H.; Ho, N.N.; Gerard, N.P.; Gerard, C.J.; et al. Complement Receptor C5aR1 Plays an Evolutionarily Conserved Role in Successful Cardiac Regeneration. Circulation 2018, 137, 2152–2165. [Google Scholar] [CrossRef]
- Marin-Juez, R.; Marass, M.; Gauvrit, S.; Rossi, A.; Lai, S.L.; Materna, S.C.; Black, B.L.; Stainier, D.Y. Fast revascularization of the injured area is essential to support zebrafish heart regeneration. Proc. Natl. Acad. Sci. USA 2016, 113, 11237–11242. [Google Scholar] [CrossRef]
- Dogra, D.; Ahuja, S.; Kim, H.T.; Rasouli, S.J.; Stainier, D.Y.R.; Reischauer, S. Opposite effects of Activin type 2 receptor ligands on cardiomyocyte proliferation during development and repair. Nat. Commun. 2017, 8, 1–15. [Google Scholar] [CrossRef]
- Lai, S.L.; Marin-Juez, R.; Moura, P.L.; Kuenne, C.; Lai, J.K.H.; Tsedeke, A.T.; Guenther, S.; Looso, M.; Stainier, D.Y. Reciprocal analyses in zebrafish and medaka reveal that harnessing the immune response promotes cardiac regeneration. eLife 2017, 6. [Google Scholar] [CrossRef]
- Huang, Y.; Harrison, M.R.; Osorio, A.; Kim, J.; Baugh, A.; Duan, C.; Sucov, H.M.; Lien, C.L. Igf Signaling is Required for Cardiomyocyte Proliferation during Zebrafish Heart Development and Regeneration. PLoS ONE 2013, 8, e67266. [Google Scholar] [CrossRef]
- Schnabel, K.; Wu, C.C.; Kurth, T.; Weidinger, G. Regeneration of cryoinjury induced necrotic heart lesions in zebrafish is associated with epicardial activation and cardiomyocyte proliferation. PLoS ONE 2011, 6, e18503. [Google Scholar] [CrossRef]
- Ma, D.; Tu, C.; Sheng, Q.; Yang, Y.; Kan, Z.; Guo, Y.; Shyr, Y.; Scott, I.C.; Lou, X. Dynamics of Zebrafish Heart Regeneration Using an HPLC-ESI-MS/MS Approach. J. Proteome Res. 2018, 17, 1300–1308. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Liu, C.; Xie, F.; Tian, L.; Manno, S.H.; Manno, F.A.M., 3rd; Fallah, S.; Pelster, B.; Tse, G.; Cheng, S.H. Excessive inflammation impairs heart regeneration in zebrafish breakdance mutant after cryoinjury. Fish. Shellfish Immunol. 2019, 89, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Webb, S.E.; Lau, T.C.K.; Cheng, S.H. Matrix metalloproteinases (MMPs) mediate leukocyte recruitment during the inflammatory phase of zebrafish heart regeneration. Sci. Rep. 2018, 8, 7199. [Google Scholar] [CrossRef] [PubMed]
- Crippa, S.; Nemir, M.; Ounzain, S.; Ibberson, M.; Berthonneche, C.; Sarre, A.; Boisset, G.; Maison, D.; Harshman, K.; Xenarios, I.; et al. Comparative transcriptome profiling of the injured zebrafish and mouse hearts identifies miRNA-dependent repair pathways. Cardiovasc Res. 2016, 110, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Gupta, V.; Karra, R.; Holdway, J.E.; Kikuchi, K.; Poss, K.D. Translational profiling of cardiomyocytes identifies an early Jak1/Stat3 injury response required for zebrafish heart regeneration. Proc. Natl. Acad. Sci. USA 2013, 110, 13416–13421. [Google Scholar] [CrossRef] [PubMed]
- Gemberling, M.; Karra, R.; Dickson, A.L.; Poss, K.D. Nrg1 is an injury-induced cardiomyocyte mitogen for the endogenous heart regeneration program in zebrafish. eLife 2015, 4. [Google Scholar] [CrossRef]
- Karra, R.; Knecht, A.K.; Kikuchi, K.; Poss, K.D. Myocardial NF-kappaB activation is essential for zebrafish heart regeneration. Proc. Natl. Acad. Sci. USA 2015, 112, 13255–13260. [Google Scholar] [CrossRef]
- Kikuchi, K.; Gupta, V.; Wang, J.; Holdway, J.E.; Wills, A.A.; Fang, Y.; Poss, K.D. Tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development 2011, 138, 2895–2902. [Google Scholar] [CrossRef]
- Kikuchi, K.; Holdway, J.E.; Major, R.J.; Blum, N.; Dahn, R.D.; Begemann, G.; Poss, K.D. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev. Cell 2011, 20, 397–404. [Google Scholar] [CrossRef]
- Yin, V.P.; Lepilina, A.; Smith, A.; Poss, K.D. Regulation of zebrafish heart regeneration by miR-133. Dev. Biol. 2012, 365, 319–327. [Google Scholar] [CrossRef]
- Han, Y.; Chen, A.; Umansky, K.B.; Oonk, K.A.; Choi, W.Y.; Dickson, A.L.; Ou, J.; Cigliola, V.; Yifa, O.; Cao, J.; et al. Vitamin D Stimulates Cardiomyocyte Proliferation and Controls Organ Size and Regeneration in Zebrafish. Dev. Cell 2019, 48, 853–863. [Google Scholar] [CrossRef] [PubMed]
- Tekeli, I.; Garcia-Puig, A.; Notari, M.; Garcia-Pastor, C.; Aujard, I.; Jullien, L.; Raya, A. Fate predetermination of cardiac myocytes during zebrafish heart regeneration. Open Biol. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.C.; Su, T.H.; Shih, C.C. High-resolution tissue Doppler imaging of the zebrafish heart during its regeneration. Zebrafish 2015, 12, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.J.; Park, J.; Kim, J.; Kim, H.H.; Lee, C.; Hwang, J.Y.; Lien, C.L.; Shung, K.K. High-frequency dual mode pulsed wave Doppler imaging for monitoring the functional regeneration of adult zebrafish hearts. J. R. Soc. Interface 2015, 12. [Google Scholar] [CrossRef] [PubMed]
- Missinato, M.A.; Saydmohammed, M.; Zuppo, D.A.; Rao, K.S.; Opie, G.W.; Kuhn, B.; Tsang, M. Dusp6 attenuates Ras/MAPK signaling to limit zebrafish heart regeneration. Development 2018, 145. [Google Scholar] [CrossRef]
- Missinato, M.A.; Tobita, K.; Romano, N.; Carroll, J.A.; Tsang, M. Extracellular component hyaluronic acid and its receptor Hmmr are required for epicardial EMT during heart regeneration. Cardiovasc. Res. 2015, 107, 487–498. [Google Scholar] [CrossRef]
- Chen, W.C.; Wang, Z.; Missinato, M.A.; Park, D.W.; Long, D.W.; Liu, H.J.; Zeng, X.; Yates, N.A.; Kim, K.; Wang, Y. Decellularized zebrafish cardiac extracellular matrix induces mammalian heart regeneration. Sci. Adv. 2016, 2, e1600844. [Google Scholar] [CrossRef]
- Han, P.; Zhou, X.H.; Chang, N.; Xiao, C.L.; Yan, S.; Ren, H.; Yang, X.Z.; Zhang, M.L.; Wu, Q.; Tang, B.; et al. Hydrogen peroxide primes heart regeneration with a derepression mechanism. Cell Res. 2014, 24, 1091–1107. [Google Scholar] [CrossRef]
- Xiao, C.; Gao, L.; Hou, Y.; Xu, C.; Chang, N.; Wang, F.; Hu, K.; He, A.; Luo, Y.; Wang, J.; et al. Chromatin-remodelling factor Brg1 regulates myocardial proliferation and regeneration in zebrafish. Nat. Commun 2016, 7, 13787. [Google Scholar] [CrossRef]
- Beauchemin, M.; Smith, A.; Yin, V.P. Dynamic microRNA-101a and Fosab expression controls zebrafish heart regeneration. Development 2015, 142, 4026–4037. [Google Scholar] [CrossRef]
- Liu, P.; Zhong, T.P. MAPK/ERK signalling is required for zebrafish cardiac regeneration. Biotechnol. Lett. 2017, 39, 1069–1077. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; He, Q.; Li, G.; Ma, J.; Zhong, T.P. Rac1-PAK2 pathway is essential for zebrafish heart regeneration. Biochem. Biophys. Res. Commun. 2016, 472, 637–642. [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.H.; Jacobson, K.A.; Rose, J.; Zeller, R. Paraffin embedding tissue samples for sectioning. CSH Protoc. 2008, 2008. [Google Scholar] [CrossRef] [PubMed]
- Alders, M.; Bikker, H.; Christiaans, I. Long QT Syndrome. In GeneReviews; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Stephens, K., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Leone, M.; Engel, F.B. Advances in heart regeneration based on cardiomyocyte proliferation and regenerative potential of binucleated cardiomyocytes and polyploidization. Clin. Sci. 2019, 133, 1229–1253. [Google Scholar] [CrossRef]
- Leone, M.; Magadum, A.; Engel, F.B. Cardiomyocyte proliferation in cardiac development and regeneration: A guide to methodologies and interpretations. Am. J. Physiol. 2015, 309, 1237–1250. [Google Scholar] [CrossRef]
- Zebrowski, D.C.; Becker, R.; Engel, F.B. Towards regenerating the mammalian heart: Challenges in evaluating experimentally induced adult mammalian cardiomyocyte proliferation. Am. J. Physiol. 2016, 310, 1045–1054. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Poss, K.D. The epicardium as a hub for heart regeneration. Nat. Rev. Cardiol 2018, 15, 631–647. [Google Scholar] [CrossRef]
- Sofronescu, A.G.; Jin, Y.; Cattini, P.A. A myocyte enhancer factor 2 (MEF2) site located in a hypersensitive region of the FGF16 gene locus is required for preferential promoter activity in neonatal cardiac myocytes. DNA Cell Biol. 2008, 27, 173–182. [Google Scholar] [CrossRef]
- Yao, J.; Wang, X.; Ren, H.; Liu, G.; Lu, P. Ultrastructure of medial rectus muscles in patients with intermittent exotropia. Eye 2016, 30, 146–151. [Google Scholar] [CrossRef]
- Muskhelishvili, L.; Latendresse, J.R.; Kodell, R.L.; Henderson, E.B. Evaluation of cell proliferation in rat tissues with BrdU, PCNA, Ki-67(MIB-5) immunohistochemistry and in situ hybridization for histone mRNA. J. Histochem. Cytochem. 2003, 51, 1681–1688. [Google Scholar] [CrossRef] [PubMed]
- Gratzner, H.G. Monoclonal antibody to 5-bromo- and 5-iododeoxyuridine: A new reagent for detection of DNA replication. Science 1982, 218, 474–475. [Google Scholar] [CrossRef] [PubMed]
- Wildemann, B.; Schmidmaier, G.; Ordel, S.; Stange, R.; Haas, N.P.; Raschke, M. Cell proliferation and differentiation during fracture healing are influenced by locally applied IGF-I and TGF-β1: Comparison of two proliferation markers, PCNA and BrdU. J. Biomed. Mater. Res. 2003, 65, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Steeds, R.P. Echocardiography: Frontier imaging in cardiology. Br. J. Radiol. 2011, 84, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Marwick, T.H. The future of echocardiography. Eur. J. Echocardiogr. 2009, 10, 594–601. [Google Scholar] [CrossRef] [PubMed]
- Sande-Melon, M.; Marques, I.J.; Galardi-Castilla, M.; Langa, X.; Perez-Lopez, M.; Botos, M.A.; Sanchez-Iranzo, H.; Guzman-Martinez, G.; Ferreira Francisco, D.M.; Pavlinic, D.; et al. Adult sox10(+) Cardiomyocytes Contribute to Myocardial Regeneration in the Zebrafish. Cell Rep. 2019, 29, 1041–1054 e1045. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).