Targeting the NPL4 Adaptor of p97/VCP Segregase by Disulfiram as an Emerging Cancer Vulnerability Evokes Replication Stress and DNA Damage while Silencing the ATR Pathway
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
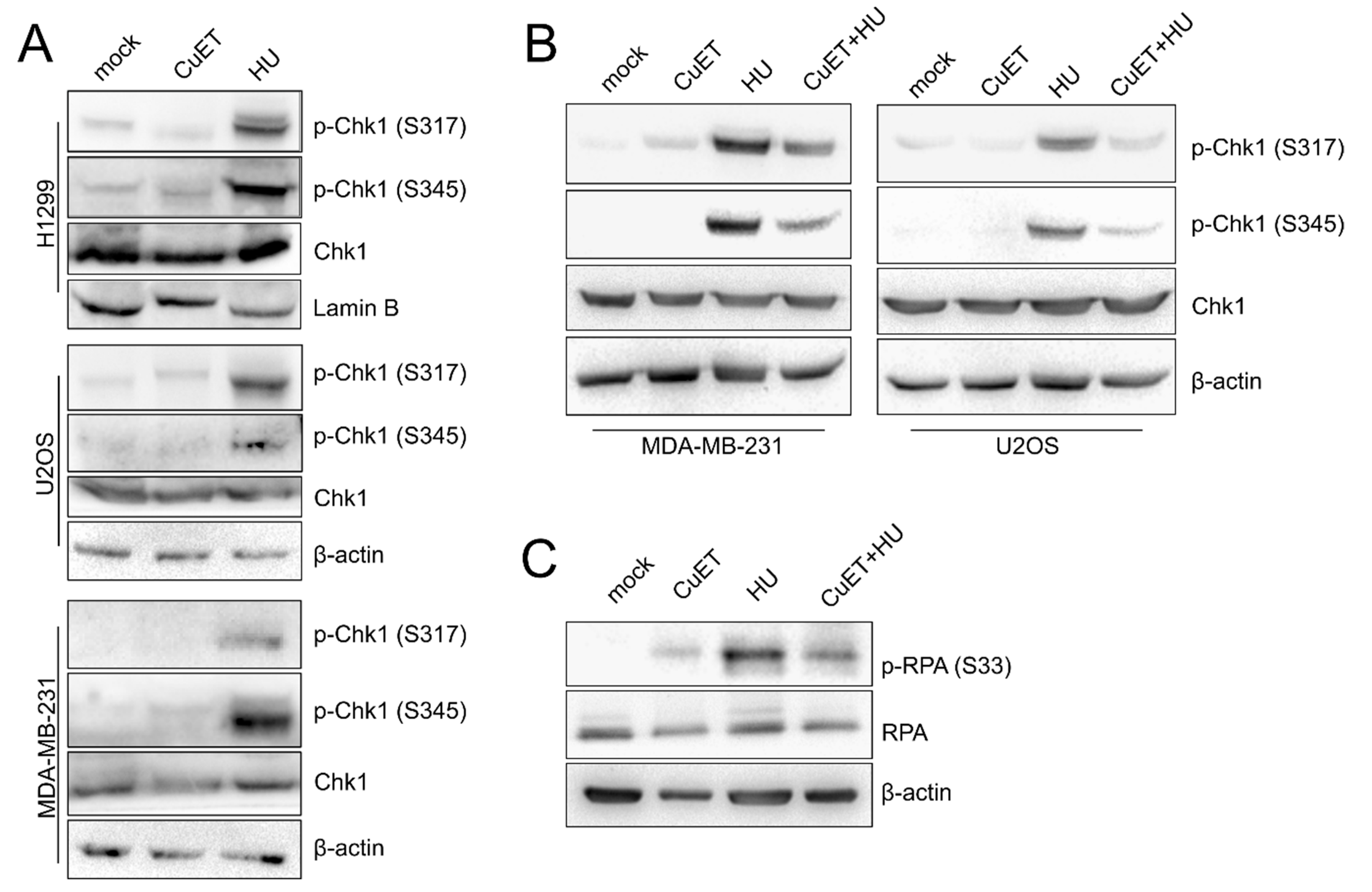
2.2. Immunoblotting
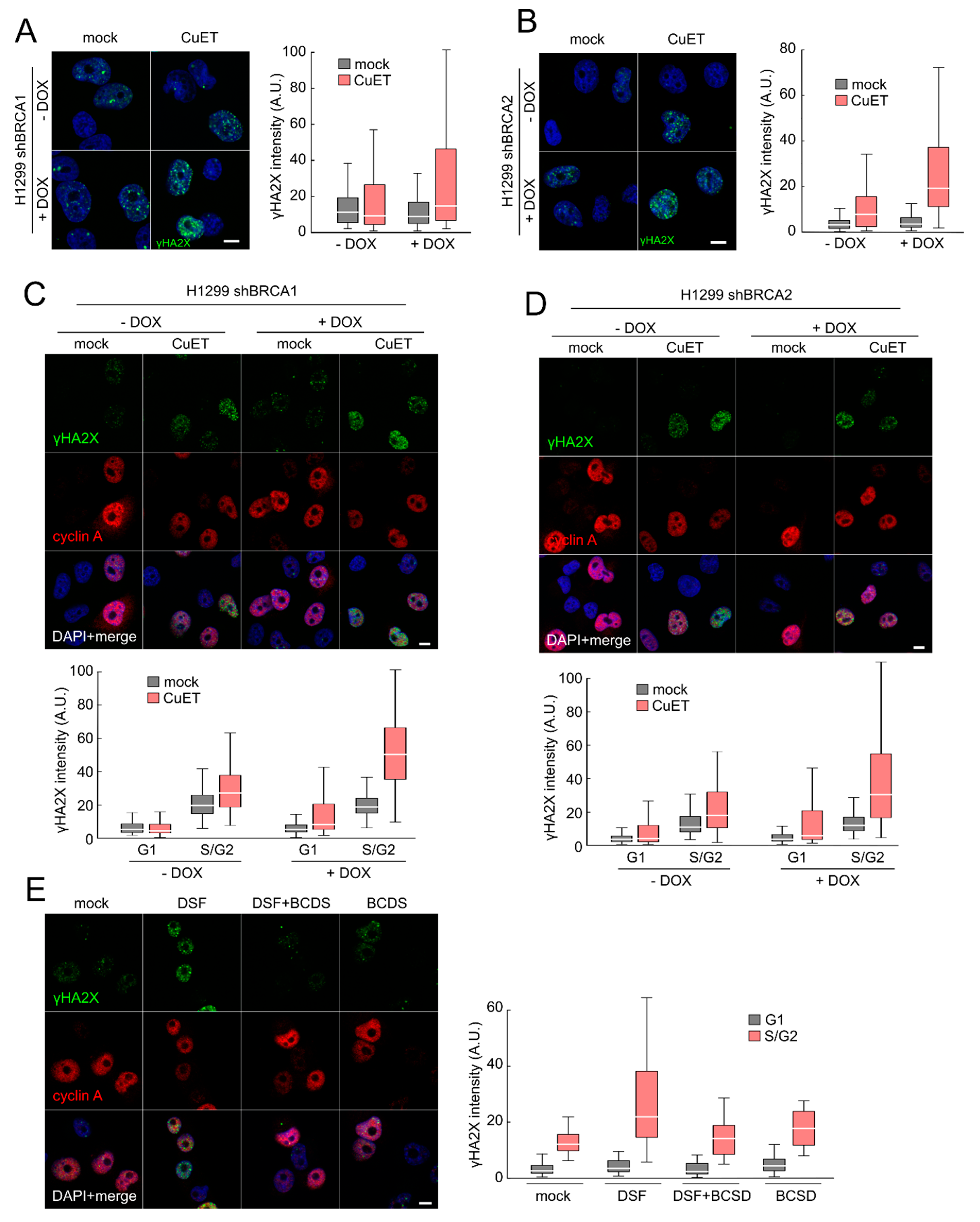
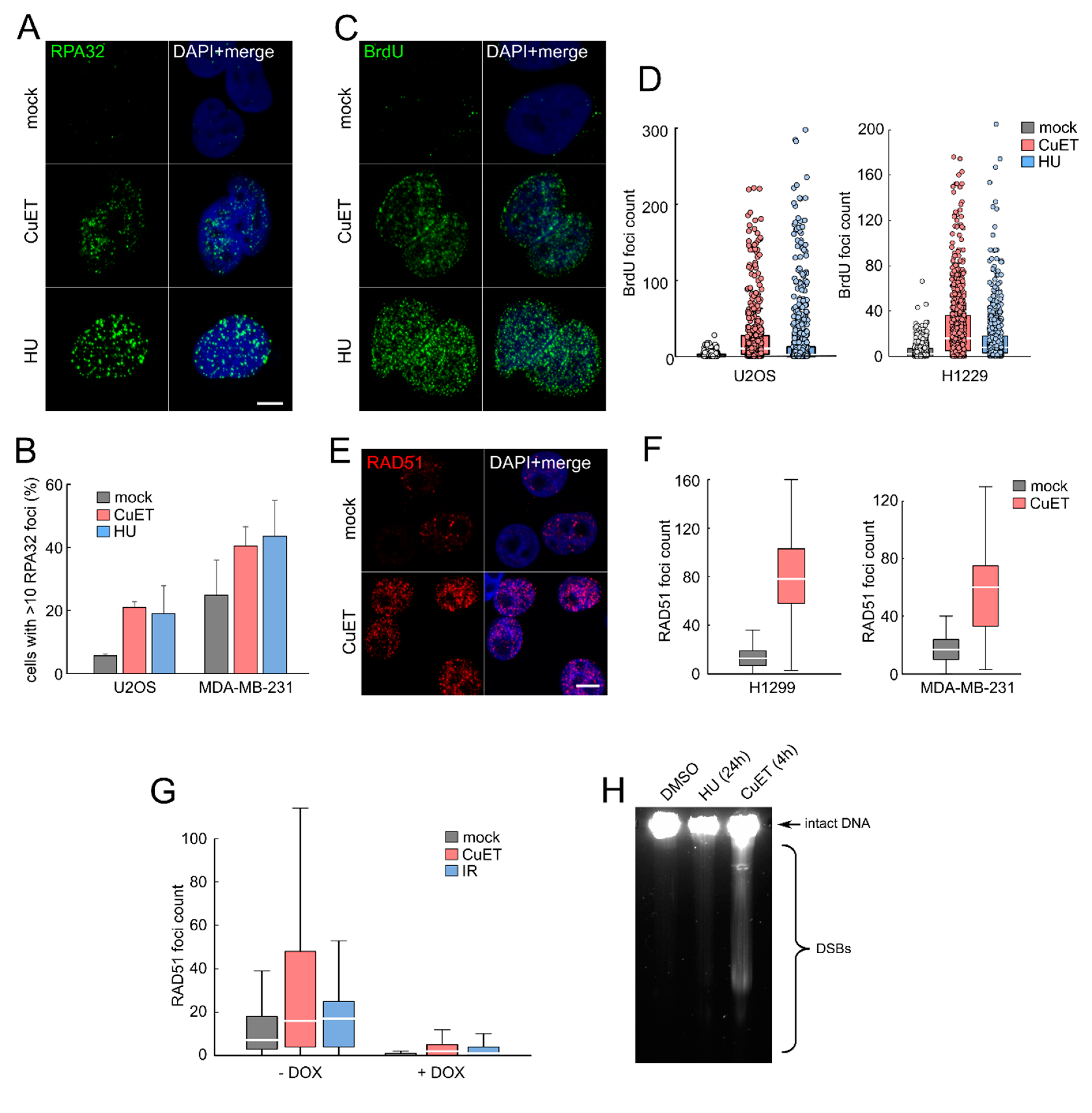
2.3. Immunofluorescence
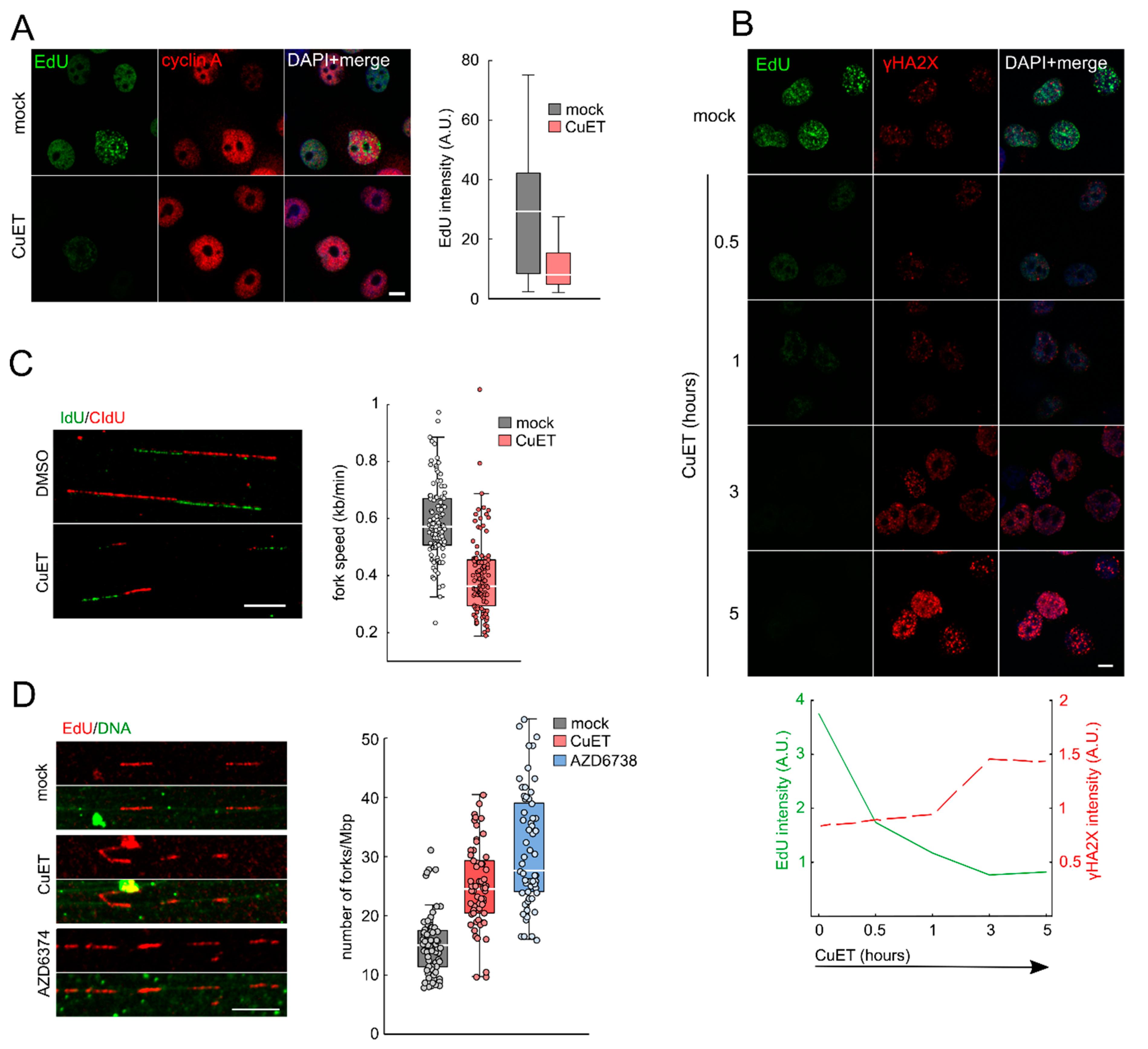
2.4. Ethynyldeoxyuridine (EdU) and Bromodeoxyuridine (BrdU) Incorporation and Detection
2.5. Image Quantification
2.6. DNA Combing
2.7. Estimation of DNA Replication Origin Density
2.8. Cell Fractionation for Triton X Insoluble Pellets
2.9. Laser Micro-Irradiation
2.10. Antibodies and Chemicals
2.11. Field Inversion Gel Electrophoresis (FIGE)
2.12. Alkaline Comet Assay
3. Results
3.1. CuET Causes DNA Damage Preferentially Detectable in S/G2-Phase Cells
3.2. CuET Treatment Decreases DNA Replication Fork Velocity and Increases the Number of Active Replication Origins
3.3. CuET-Induced Replication Stress Leads to DNA Damage that Triggers Homologous Recombination Repair Pathway
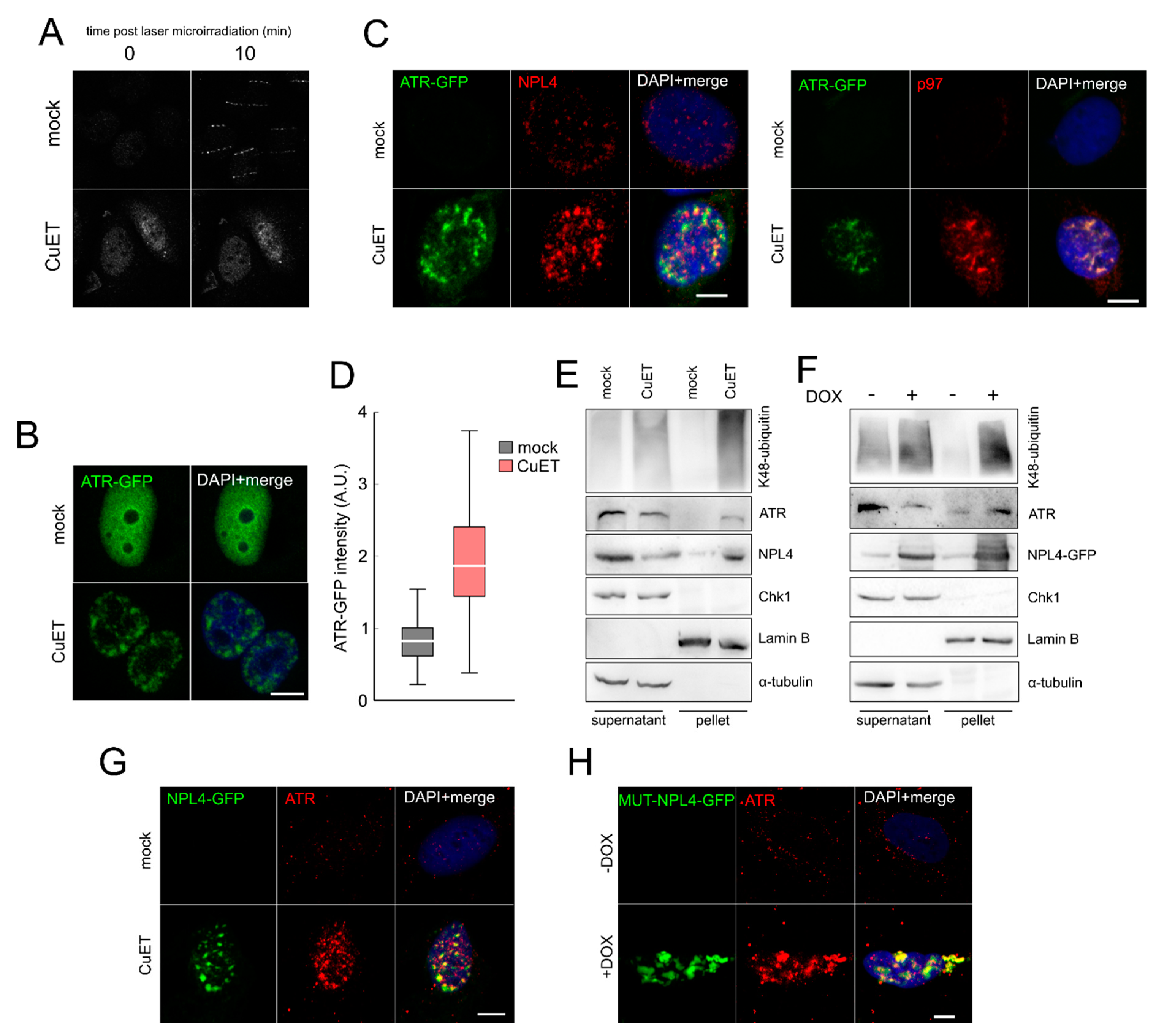
3.4. The ATR Signaling Pathway is Compromised in CuET-Treated Cells
3.5. The ATR Signaling Pathway is Compromised in CuET-Treated Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Collins, F.S. Mining for therapeutic gold. Nat. Rev. Drug Discov. 2011, 10, 397. [Google Scholar] [CrossRef] [PubMed]
- McMahon, A.; Chen, W.; Li, F. Old wine in new bottles: Advanced drug delivery systems for disulfiram-based cancer therapy. J. Control. Release 2020, 319, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Skrott, Z.; Mistrik, M.; Andersen, K.K.; Friis, S.; Majera, D.; Gursky, J.; Ozdian, T.; Bartkova, J.; Turi, Z.; Moudry, P.; et al. Alcohol-abuse drug disulfiram targets cancer via p97 segregase adaptor NPL4. Nature 2017, 552, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Skrott, Z.; Majera, D.; Gursky, J.; Buchtova, T.; Hajduch, M.; Mistrik, M.; Bartek, J. Disulfiram’s anti-cancer activity reflects targeting NPL4, not inhibition of aldehyde dehydrogenase. Oncogene 2019, 38, 6711–6722. [Google Scholar] [CrossRef]
- Tacconi, E.M.; Lai, X.; Folio, C.; Porru, M.; Zonderland, G.; Badie, S.; Michl, J.; Sechi, I.; Rogier, M.; Matía García, V.; et al. BRCA1 and BRCA2 tumor suppressors protect against endogenous acetaldehyde toxicity. EMBO Mol. Med. 2017, 9, 1398–1414. [Google Scholar] [CrossRef]
- Lorenti Garcia, C.; Mechilli, M.; Proietti De Santis, L.; Schinoppi, A.; Katarzyna, K.; Palitti, F. Relationship between DNA lesions, DNA repair and chromosomal damage induced by acetaldehyde. Mutat. Res. Mol. Mech. Mutagen. 2009, 662, 3–9. [Google Scholar] [CrossRef]
- Bartkova, J.; Hořejší, Z.; Koed, K.; Krämer, A.; Tort, F.; Zieger, K.; Guldberg, P.; Sehested, M.; Nesland, J.M.; Lukas, C.; et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 2005, 434, 864–870. [Google Scholar] [CrossRef]
- Bartkova, J.; Rezaei, N.; Liontos, M.; Karakaidos, P.; Kletsas, D.; Issaeva, N.; Vassiliou, L.-V.F.; Kolettas, E.; Niforou, K.; Zoumpourlis, V.C.; et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 2006, 444, 633–637. [Google Scholar] [CrossRef]
- Gorgoulis, V.G.; Vassiliou, L.-V.F.; Karakaidos, P.; Zacharatos, P.; Kotsinas, A.; Liloglou, T.; Venere, M.; DiTullio, R.A.; Kastrinakis, N.G.; Levy, B.; et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 2005, 434, 907–913. [Google Scholar] [CrossRef]
- Halazonetis, T.D.; Gorgoulis, V.G.; Bartek, J. An Oncogene-Induced DNA Damage Model for Cancer Development. Science 2008, 319, 1352–1355. [Google Scholar] [CrossRef]
- Bartek, J.; Bartkova, J.; Lukas, J. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene 2007, 26, 7773–7779. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Gaillard, H.; García-Muse, T.; Aguilera, A. Replication stress and cancer. Nat. Rev. Cancer 2015, 15, 276–289. [Google Scholar] [CrossRef]
- Bartek, J.; Mistrik, M.; Bartkova, J. Thresholds of replication stress signaling in cancer development and treatment. Nat. Struct. Mol. Biol. 2012, 19, 5–7. [Google Scholar] [CrossRef]
- Zeman, M.K.; Cimprich, K.A. Causes and consequences of replication stress. Nat. Cell Biol. 2014, 16, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Berti, M.; Vindigni, A. Replication stress: Getting back on track. Nat. Struct. Mol. Biol. 2016, 23, 103–109. [Google Scholar] [CrossRef]
- Eykelenboom, J.K.; Harte, E.C.; Canavan, L.; Pastor-Peidro, A.; Calvo-Asensio, I.; Llorens-Agost, M.; Lowndes, N.F. ATR Activates the S-M Checkpoint during Unperturbed Growth to Ensure Sufficient Replication Prior to Mitotic Onset. Cell Rep. 2013, 5, 1095–1107. [Google Scholar] [CrossRef]
- Toledo, L.I.; Altmeyer, M.; Rask, M.-B.; Lukas, C.; Larsen, D.H.; Povlsen, L.K.; Bekker-Jensen, S.; Mailand, N.; Bartek, J.; Lukas, J. ATR Prohibits Replication Catastrophe by Preventing Global Exhaustion of RPA. Cell 2014, 156, 374. [Google Scholar] [CrossRef]
- Bekker-Jensen, S.; Lukas, C.; Kitagawa, R.; Melander, F.; Kastan, M.B.; Bartek, J.; Lukas, J. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 2006, 173, 195–206. [Google Scholar] [CrossRef]
- Jackson, D.A.; Pombo, A. Replicon Clusters Are Stable Units of Chromosome Structure: Evidence That Nuclear Organization Contributes to the Efficient Activation and Propagation of S Phase in Human Cells. J. Cell Biol. 1998, 140, 1285–1295. [Google Scholar] [CrossRef]
- Mistrik, M.; Vesela, E.; Furst, T.; Hanzlikova, H.; Frydrych, I.; Gursky, J.; Majera, D.; Bartek, J. Cells and Stripes: A novel quantitative photo-manipulation technique. Sci. Rep. 2016, 6, 19567. [Google Scholar] [CrossRef] [PubMed]
- Maya-Mendoza, A.; Moudry, P.; Merchut-Maya, J.M.; Lee, M.; Strauss, R.; Bartek, J. High speed of fork progression induces DNA replication stress and genomic instability. Nature 2018, 559, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Budzowska, M.; Kanaar, R. Mechanisms of Dealing with DNA Damage-Induced Replication Problems. Cell Biochem. Biophys. 2009, 53, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Bianco, J.N.; Poli, J.; Saksouk, J.; Bacal, J.; Silva, M.J.; Yoshida, K.; Lin, Y.-L.; Tourrière, H.; Lengronne, A.; Pasero, P. Analysis of DNA replication profiles in budding yeast and mammalian cells using DNA combing. Methods 2012, 57, 149–157. [Google Scholar] [CrossRef]
- Quinet, A.; Carvajal-Maldonado, D.; Lemacon, D.; Vindigni, A. DNA Fiber Analysis: Mind the Gap! Methods Enzymology 2017, 591, 55–82. [Google Scholar]
- Couch, F.B.; Bansbach, C.E.; Driscoll, R.; Luzwick, J.W.; Glick, G.G.; Betous, R.; Carroll, C.M.; Jung, S.Y.; Qin, J.; Cimprich, K.A.; et al. ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev. 2013, 27, 1610–1623. [Google Scholar] [CrossRef]
- Sogo, J.M. Fork Reversal and ssDNA Accumulation at Stalled Replication Forks Owing to Checkpoint Defects. Science 2002, 297, 599–602. [Google Scholar] [CrossRef]
- Byun, T.S.; Pacek, M.; Yee, M.C.; Walter, J.C.; Cimprich, K.A. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005, 19, 1040–1052. [Google Scholar] [CrossRef]
- Lee, Z.; Elledge, S.J. Sensing DNA Damage Through ATRIP Recognition of RPA-ssDNA Complexes. Science 2003, 300, 1542–1548. [Google Scholar] [CrossRef]
- Liao, H.; Ji, F.; Helleday, T.; Ying, S. Mechanisms for stalled replication fork stabilization: New targets for synthetic lethality strategies in cancer treatments. EMBO Rep. 2018, 19. [Google Scholar] [CrossRef]
- Petermann, E.; Orta, M.L.; Issaeva, N.; Schultz, N.; Helleday, T. Hydroxyurea-Stalled Replication Forks Become Progressively Inactivated and Require Two Different RAD51-Mediated Pathways for Restart and Repair. Mol. Cell 2010, 37, 492–502. [Google Scholar] [CrossRef] [PubMed]
- Whelan, D.R.; Lee, W.T.C.; Yin, Y.; Ofri, D.M.; Bermudez-Hernandez, K.; Keegan, S.; Fenyo, D.; Rothenberg, E. Spatiotemporal dynamics of homologous recombination repair at single collapsed replication forks. Nat. Commun. 2018, 9, 3882. [Google Scholar] [CrossRef] [PubMed]
- Davies, A.A.; Masson, J.Y.; McIlwraith, M.J.; Stasiak, A.Z.; Stasiak, A.; Venkitaraman, A.R.; West, S.C. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell 2001, 7, 273–282. [Google Scholar] [CrossRef]
- Majera, D.; Skrott, Z.; Bouchal, J.; Bartkova, J.; Simkova, D.; Gachechiladze, M.; Steigerova, J.; Kurfurstova, D.; Gursky, J.; Korinkova, G.; et al. Targeting genotoxic and proteotoxic stress-response pathways in human prostate cancer by clinically available PARP inhibitors, vorinostat and disulfiram. Prostate 2019, 79, 352–362. [Google Scholar] [CrossRef]
- Ramadan, K. p97/VCP- and Lys48-linked polyubiquitination form a new signaling pathway in DNA damage response. Cell Cycle 2012, 11, 1062–1069. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ramadan, K.; Halder, S.; Wiseman, K.; Vaz, B. Strategic role of the ubiquitin-dependent segregase p97 (VCP or Cdc48) in DNA replication. Chromosoma 2017, 126, 17–32. [Google Scholar] [CrossRef]
- Meerang, M.; Ritz, D.; Paliwal, S.; Garajova, Z.; Bosshard, M.; Mailand, N.; Janscak, P.; Hübscher, U.; Meyer, H.; Ramadan, K. The ubiquitin-selective segregase VCP/p97 orchestrates the response to DNA double-strand breaks. Nat. Cell Biol. 2011, 13, 1376. [Google Scholar] [CrossRef]
- Bergink, S.; Ammon, T.; Kern, M.; Schermelleh, L.; Leonhardt, H.; Jentsch, S. Role of Cdc48/p97 as a SUMO-targeted segregase curbing Rad51–Rad52 interaction. Nat. Cell Biol. 2013, 15, 526–532. [Google Scholar] [CrossRef]
- Singh, A.N.; Oehler, J.; Torrecilla, I.; Kilgas, S.; Li, S.; Vaz, B.; Guérillon, C.; Fielden, J.; Hernandez-Carralero, E.; Cabrera, E.; et al. The p97-Ataxin 3 complex regulates homeostasis of the DNA damage response E3 ubiquitin ligase RNF8. EMBO J. 2019, 38, e102361. [Google Scholar] [CrossRef]
- Davis, E.J.; Lachaud, C.; Appleton, P.; Macartney, T.J.; Näthke, I.; Rouse, J. DVC1 (C1orf124) recruits the p97 protein segregase to sites of DNA damage. Nat. Struct. Mol. Biol. 2012, 19, 1093–1100. [Google Scholar] [CrossRef]
- Kumar, A.; Mazzanti, M.; Mistrik, M.; Kosar, M.; Beznoussenko, G.V.; Mironov, A.A.; Garrè, M.; Parazzoli, D.; Shivashankar, G.V.; Scita, G.; et al. ATR Mediates a Checkpoint at the Nuclear Envelope in Response to Mechanical Stress. Cell 2014, 158, 633–646. [Google Scholar] [CrossRef] [PubMed]
- Woerner, A.C.; Frottin, F.; Hornburg, D.; Feng, L.R.; Meissner, F.; Patra, M.; Tatzelt, J.; Mann, M.; Winklhofer, K.F.; Hartl, F.U.; et al. Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science 2016, 351, 173–176. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, S.; Tsuchiya, H.; Kaiho, A.; Guo, Q.; Ikeuchi, K.; Endo, A.; Arai, N.; Ohtake, F.; Murata, S.; Inada, T.; et al. Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature 2020, 578, 296–300. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, W.; Patel, S.S.; Cong, J.; Zhang, N.; Sabbatino, F.; Liu, X.; Qi, Y.; Huang, P.; Lee, H.; et al. Blocking the formation of radiation induced breast cancer stem cells. Oncotarget 2014, 5, 3743–3755. [Google Scholar] [CrossRef]
- Lun, X.; Wells, J.C.; Grinshtein, N.; King, J.C.; Hao, X.; Dang, N.-H.; Wang, X.; Aman, A.; Uehling, D.; Datti, A.; et al. Disulfiram when Combined with Copper Enhances the Therapeutic Effects of Temozolomide for the Treatment of Glioblastoma. Clin. Cancer Res. 2016, 22, 3860–3875. [Google Scholar] [CrossRef]
- Huang, J.; Campian, J.L.; Gujar, A.D.; Tsien, C.; Ansstas, G.; Tran, D.D.; DeWees, T.A.; Lockhart, A.C.; Kim, A.H. Final results of a phase I dose-escalation, dose-expansion study of adding disulfiram with or without copper to adjuvant temozolomide for newly diagnosed glioblastoma. J. Neurooncol. 2018, 138, 105–111. [Google Scholar] [CrossRef]
- Huang, J.; Campian, J.L.; Gujar, A.D.; Tran, D.D.; Lockhart, A.C.; DeWees, T.A.; Tsien, C.I.; Kim, A.H. A phase I study to repurpose disulfiram in combination with temozolomide to treat newly diagnosed glioblastoma after chemoradiotherapy. J. Neurooncol. 2016, 128, 259–266. [Google Scholar] [CrossRef]
- Jakola, A.S.; Werlenius, K.; Mudaisi, M.; Hylin, S.; Kinhult, S.; Bartek, J.J.; Salvesen, O.; Carlsen, S.M.; Strandeus, M.; Lindskog, M.; et al. Disulfiram repurposing combined with nutritional copper supplement as add-on to chemotherapy in recurrent glioblastoma (DIRECT): Study protocol for a randomized controlled trial. F1000Research 2018, 7, 1797. [Google Scholar] [CrossRef]
- Nechushtan, H.; Hamamreh, Y.; Nidal, S.; Gotfried, M.; Baron, A.; Shalev, Y.I.; Nisman, B.; Peretz, T.; Peylan-Ramu, N. A Phase IIb Trial Assessing the Addition of Disulfiram to Chemotherapy for the Treatment of Metastatic Non-Small Cell Lung Cancer. Oncologist 2015, 20, 366–367. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Majera, D.; Skrott, Z.; Chroma, K.; Merchut-Maya, J.M.; Mistrik, M.; Bartek, J. Targeting the NPL4 Adaptor of p97/VCP Segregase by Disulfiram as an Emerging Cancer Vulnerability Evokes Replication Stress and DNA Damage while Silencing the ATR Pathway. Cells 2020, 9, 469. https://doi.org/10.3390/cells9020469
Majera D, Skrott Z, Chroma K, Merchut-Maya JM, Mistrik M, Bartek J. Targeting the NPL4 Adaptor of p97/VCP Segregase by Disulfiram as an Emerging Cancer Vulnerability Evokes Replication Stress and DNA Damage while Silencing the ATR Pathway. Cells. 2020; 9(2):469. https://doi.org/10.3390/cells9020469
Chicago/Turabian StyleMajera, Dusana, Zdenek Skrott, Katarina Chroma, Joanna Maria Merchut-Maya, Martin Mistrik, and Jiri Bartek. 2020. "Targeting the NPL4 Adaptor of p97/VCP Segregase by Disulfiram as an Emerging Cancer Vulnerability Evokes Replication Stress and DNA Damage while Silencing the ATR Pathway" Cells 9, no. 2: 469. https://doi.org/10.3390/cells9020469
APA StyleMajera, D., Skrott, Z., Chroma, K., Merchut-Maya, J. M., Mistrik, M., & Bartek, J. (2020). Targeting the NPL4 Adaptor of p97/VCP Segregase by Disulfiram as an Emerging Cancer Vulnerability Evokes Replication Stress and DNA Damage while Silencing the ATR Pathway. Cells, 9(2), 469. https://doi.org/10.3390/cells9020469




