Clustered DNA Damage: Electronic Properties and Their Influence on Charge Transfer. 7,8-Dihydro-8-Oxo-2′-Deoxyguaosine Versus 5′,8-Cyclo-2′-Deoxyadenosines: A Theoretical Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Computation Methodology of QM/MM (Quantum Mechanics/Molecular Mechanics) Studies
2.2. Computation Methodology of DFT Study
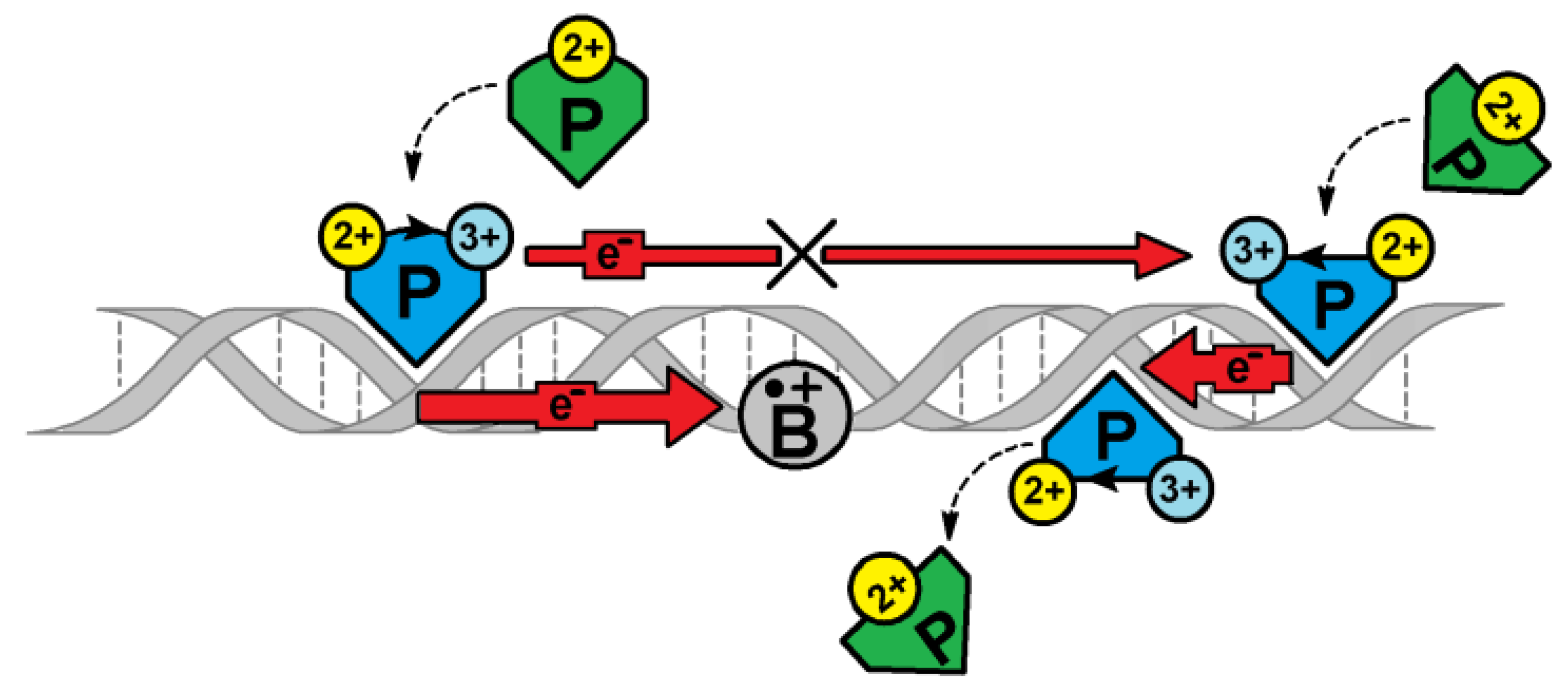
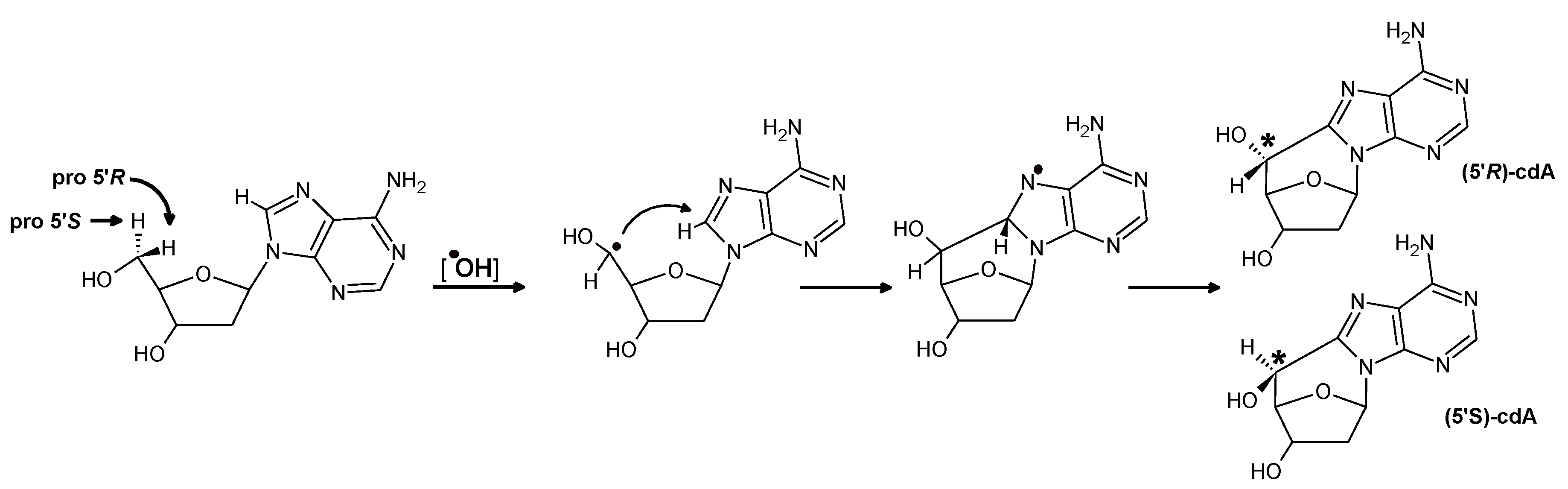
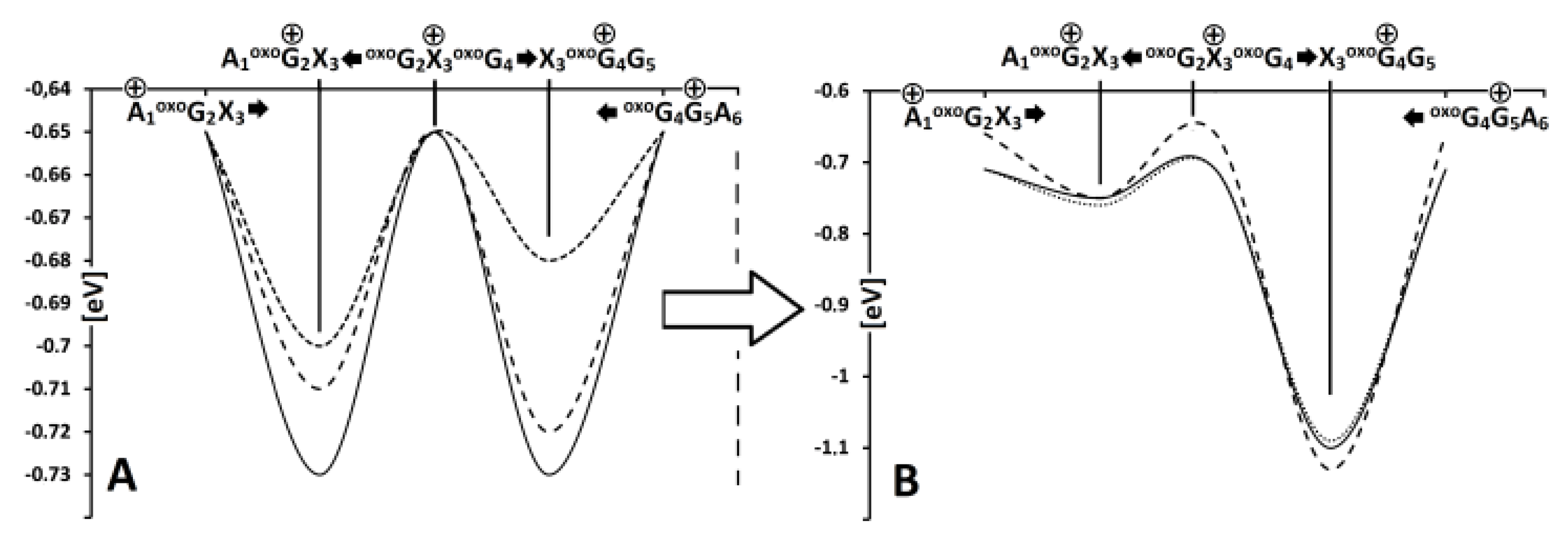
3. Results and Discussion
4. Conclusions
Supplementary Materials
Funding
Conflicts of Interest
References
- Hakem, R. DNA-damage repair; the good, the bad, and the ugly. EMBO J. 2008, 27, 589–605. [Google Scholar] [CrossRef] [PubMed]
- Eccles, L.J.; O’Neill, P.; Lomax, M.E. Delayed repair of radiation induced clustered DNA damage: Friend or foe? Mutat. Res. 2011, 711, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Albiser, G.; Lamiri, A.; Premilat, S. The A-B transition: Temperature and base composition effects on hydration of DNA. Int. J. Biol. Macromol. 2001, 28, 199–203. [Google Scholar] [CrossRef]
- Terato, H.; Ide, H. Clustered DNA damage induced by heavy ion particles. Biol. Sci. Space. 2004, 18, 206–215. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sy, D.; Savoye, C.; Begusova, M.; Michalik, V.; Charlier, M.; Spotheim-Maurizot, M. Sequence-dependent variations of DNA structure modulate radiation-induced strand breakage. Int. J. Radiat. Biol. 1997, 72, 147–155. [Google Scholar] [PubMed]
- Kumar, A.; Pottiboyina, V.; Sevilla, M.D. One-Electron Oxidation of Neutral Sugar Radicals of 2′-Deoxyguanosine and 2′-Deoxythymidine: A Density Functional Theory (DFT) Study. J. Phys. Chem. B. 2012, 116, 9409–9416. [Google Scholar] [CrossRef]
- Boussicault, F.; Kaloudis, P.; Caminal, C.; Mulazzani, Q.G.; Chatgilialoglu, C. The fate of C5’ radicals of purine nucleosides under oxidative conditions. J. Am. Chem. Soc. 2008, 130, 8377–8385. [Google Scholar] [CrossRef]
- Kim, Y.J.; Wilson III, D.M. Overview of base excision repair biochemistry. Curr. Mol. Pharmacol. 2012, 5, 3–13. [Google Scholar] [CrossRef]
- Murray, J.M.; Carr, A.M. Integrating DNA damage repair with the cell cycle. Curr. Opin. Cell Biol. 2018, 52, 120–125. [Google Scholar] [CrossRef]
- Sage, E.; Harrison, L. Clustered DNA lesion repair in eukaryotes: Relevance to mutagenesis and cell survival. Mutat Res. 2011, 711, 123–133. [Google Scholar] [CrossRef]
- Lomax, M.E.; Cunniffe, S.; O’Neill, P. 8-OxoG retards the activity of the ligase III/XRCC1 complex during the repair of a single-strand break, when present within a clustered DNA damage site. DNA Repair. 2004, 3, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Cannan, W.J.; Pederson, D.S. Mechanisms and Consequences of Double-Strand DNA Break Formation in Chromatin. J. Cell Physiol. 2016, 231, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Houck, A.L.; Seddighi, S.; Driver, J.A. At the Crossroads Between Neurodegeneration and Cancer: A Review of Overlapping Biology and Its Implications. Curr. Aging Sci. 2018, 11, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Georgakilas, A.G.; O’Neill, P.; Stewart, R.D. Induction and Repair of Clustered DNA Lesions: What Do We Know So Far? Radiat. Res. 2013, 180, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.H.W.; Kuo, M.T. Improving radiotherapy in cancer treatment: Promises and challenges. Oncotarget 2017, 37, 62742–62758. [Google Scholar] [CrossRef] [PubMed]
- Pouget, J.-P.; Frelon, S.; Ravanat, J.-L.; Testard, I.; Odin, F.; Cadet, J. Formation of Modified DNA Bases in Cells Exposed either to Gamma Radiation or to High-LET Particles. Radiat Res. 2002, 157, 589–595. [Google Scholar] [CrossRef]
- Savolainen, S.; Kortesniemi, M.; Timonen, M.; Reijonen, V.; Kuusela, L.; Uusi-Simola, J.; Salli, E.; Koivunoro, H.; Seppala, T.; Lonnroth, N.; et al. Boron neutron capture therapy (BNCT) in Finland: Technological and physical prospects after 20 years of experiences. Phys. Medica. 2013, 29, 233–248. [Google Scholar] [CrossRef]
- Sauerwei, W.A.G. Principles and Roots of Neutron Capture Therapy: In Neutron Capture Therapy Principles and Applications., 1st ed.; Sauerwein, W., Wittig, A., Moss, R., Nakagawa, Y., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 1–16. [Google Scholar]
- Kankaanranta, L.; Seppälä, T.; Koivunoro, H.; Saarilahti, K.; Atula, T.; Collan, J.; Salli, E.; Kortesniemi, M.; Uusi-Simola, J.; Välimäki, P.; et al. Boron Neutron Capture Therapy in the Treatment of Locally Recurred Head-and-Neck Cancer: Final Analysis of a Phase I/II Trial. Int. J. Radiat Onc. Biol. Phys. 2012, 82, 67–75. [Google Scholar] [CrossRef]
- Minchom, A.; Aversa, C.; Lopez, J. Dancing with the DNA damage response: Next-generation anti-cancer therapeutic strategies. Ther. Adv. Med. Oncol. 2018, 10, 1–18. [Google Scholar] [CrossRef]
- Nikjoo, H.; O’Neill, P.; Wilson, W.E.; Goodhead, D.T. Computational approach for determining the spectrum of DNA damage induced by ionizing radiation. Radiat Res. 2001, 156, 577–583. [Google Scholar] [CrossRef]
- Sutherland, B.M.; Bennett, P.V.; Sidorkina, O.; Laval, J. Clustered Damages and Total Lesions Induced in DNA by Ionizing Radiation: Oxidized Bases and Strand Breaks. Biochemistry. 2000, 39, 8026–8031. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Yu, K.N. Ionizing Radiation, DNA Double Strand Break and Mutation. In Advances in Genetics Research; Urbano, K.V., Ed.; Nova Science Publishers, Inc.: New York, USA, 2010; Volume 4, pp. 1–13. [Google Scholar]
- Jeggo, P.A.; Löbrich, M. DNA double-strand breaks: Their cellular and clinical impact? Oncogene 2007, 26, 7717–7719. [Google Scholar] [CrossRef] [PubMed]
- Bignon, E.; Gattuso, H.; Morell, C.; Dehez, F.; Georgakilas, A.G.; Monari, A.; Dumont, E. Correlation of bistranded clustered abasic DNA lesion processing with structural and dynamic DNA helix distortion. Nucleic Acids Res. 2016, 44, 8588–8599. [Google Scholar] [CrossRef] [PubMed]
- MacKerell, A.D., Jr.; Nilsson, L. Theoretical Studies of Nucleic Acids and Nucleic Acid-Protein Complexes using Charmm. In Computational Studies of RNA and DNA; Sporen, J., Lankas, F., Eds.; Springer: Dordrecht, The Netherlands, 2006; p. 7394. [Google Scholar]
- Olson, M.V. The human genome project. Proc. Nat. Acad. Sci. USA 1993, 90, 4338–4344. [Google Scholar] [CrossRef] [PubMed]
- Blattner, F.R.; Plunkett 3rd, G.; Bloch, C.A.; Perna, N.T.; Burland, V.; Riley, M.; Collado-Vides, J.; Glasner, J.D.; Rode, C.K.; Mayhew, G.F.; et al. The complete genome sequence of Escherichia coli K-12. Science 1997, 277, 1453–1462. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, A.L.; Schär, P. DNA glycosylases: In DNA repair and beyond. Chromosoma 2012, 121, 1–20. [Google Scholar] [CrossRef]
- Fromme, J.C.; Banerjee, A.; Huang, S.J.; Verdine, G.L. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature 2004, 427, 652–656. [Google Scholar] [CrossRef]
- Merino, E.J.; Boal, A.K.; Barton, J.K. Biological contexts for DNA charge transport chemistry. Curr. Opin. Chem. Biol. 2008, 12, 229–237. [Google Scholar] [CrossRef][Green Version]
- Henderson, P.T.; Jones, D.; Hampikian, G.; Kan, Y.Z.; Schuster, G.B. Long-distance charge transport in duplex DNA: The phonon-assisted polaron-like hopping mechanism. Proc. Natl. Acad. Sci. USA 1999, 96, 8353–8358. [Google Scholar] [CrossRef]
- Sontz, P.A.; Mui, T.P.; Fuss, J.O.; Tainer, J.A.; Barton, J.K. DNA charge transport as a first step in coordinating the detection of lesions by repair proteins. Proc. Nat. Acad. Sci. USA 2012, 109, 1856–1861. [Google Scholar] [CrossRef]
- Blancafort, L.; Voityuk, A.A. Thermally induced hopping model for long-range triplet excitation energy transfer in DNA. Phys. Chem. Chem. Phys. 2018, 20, 4997–5000. [Google Scholar] [CrossRef] [PubMed]
- Curutchet, C.; Voityuk, A.A. Triplet–triplet energy transfer in DNA: A process that occurs on the nanosecond timescale. Angew. Chem. Int. Ed. 2011, 50, 1820–1822. [Google Scholar] [CrossRef] [PubMed]
- Crespo-Hernández, C.E.; Cohen, B.; Hare, P.M.; Kohler, B. Ultrafast Excited-State Dynamics in Nucleic Acids. Chem. Rev. 2004, 104, 1977–2020. [Google Scholar] [CrossRef]
- Gut, I.G.; Wood, P.D.; Redmond, R.W. Interaction of Triplet Photosensitizers with Nucleotides and DNA in Aqueous Solution at Room Temperature. J. Am. Chem. Soc. 1996, 118, 2366–2373. [Google Scholar] [CrossRef]
- Eisinger, J.; Lamola, A.A. The excited states of nucleic acids. In Excited States of Proteins and Nucleic Acids.; Steiner, R.F., Weinryb, I., Eds.; Springer: New York, NY, USA, 1971; pp. 107–198. [Google Scholar]
- Palivec, V.; Pluharova, E.; Unger, I.; Winter, B.; Jungwirth, P. DNA Lesion Can Facilitate Base Ionization: Vertical Ionization Energies of Aqueous 8-Oxoguanine and its Nucleoside and Nucleotide. J. Phys. Chem. B 2014, 118, 13833–13837. [Google Scholar] [CrossRef]
- Guerrero, C.R.; Wang, J.; Wang, Y. Induction of 8,5’-cyclo-2’-deoxyadenosine and 8,5’-cyclo-2’-deoxyguanosine in isolated DNA by Fenton-type reagents. Chem. Res. Toxicol. 2013, 26, 1361–1366. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chatgilialoglu, C.H.; Ferreri, C.; Geacintov, N.E.; Krokidis, M.G.; Liu, Y.; Masi, A.; Shafirovich, V.; Terzidis, M.A.; Tsegay, P.S. 5’,8-Cyclopurine lesions in DNA damage: Chemical, analytical, biological, and diagnostic significance. Cells 2019, 8, 1–34. [Google Scholar] [CrossRef]
- Cadet, J.; Di Mascio, P.; Wagner, J.R. (5’R)-and (5’S)-purine 5’,8-cyclo-2’-deoxyribonucleosides: Reality or artifactual measurements? A reply to Chatgilialoglu’s comments (this issue). Free. Radic. Res. 2019, 53, 1014–1018. [Google Scholar] [CrossRef]
- Karwowski, B.T. Formation of 5′,8-cyclo-2′-deoxyadenosine in single strand DNA. Theoretical quantum mechanics study. Org. Biomol. Chem. 2010, 7, 1603–1609. [Google Scholar] [CrossRef]
- Imoto, S.; Bransfield, L.A.; Croteau, D.L.; Van Houten, B.; Greenberg, M.M. DNA tandem lesion repair by strand displacement synthesis and nucleotide excision repair. Biochemistry 2008, 47, 4306–4316. [Google Scholar] [CrossRef][Green Version]
- Paul, C.R.; Budzinski, E.E.; Maccubbin, A.; Wallace, J.C.; Box, H.C. Characterization of radiation- induced damage in d(TpApCpG). Int. J. Radiat. Biol. 1990, 58, 759–768. [Google Scholar] [CrossRef] [PubMed]
- Bourdat, A.-G.; Douki, T.; Frelon, S.; Gasparutto, D.; Cadet, J. Tandem base lesions are generated by hydroxyl radical within isolated DNA in aerated aqueous solution. J. Am. Chem. Soc. 2000, 122, 4549–4556. [Google Scholar] [CrossRef]
- Karwowski, B.T.; Grand, A.; Cadet, J. 5’,8-Cyclo-2’-deoxyadenosine (cdA) formation by γ-radiation. Theoretical quantum mechanics study. Acta Biochim. Pol. 2009, 56, 655–662. [Google Scholar] [CrossRef]
- Dickerson, M.-L.; Blakely, F.W.; Holwitt, E.; Dizdaroglu, M. Effect of DNA Conformation on the hydroxyl radical-induced formation of 8,5′-cyclopurine 2′-deoxyribonucleoside residues in DNA. Int. J. Radiat. Biol. 1988, 54, 195–204. [Google Scholar] [CrossRef]
- Kuraoka, I.; Bender, C.; Romieu, A.; Cadet, J.; Wood, R.D.; Lindahl, T. Removal of oxygen free-radical-induced 5’,8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc Natl. Acad. Sci. USA 2000, 97, 3832–3837. [Google Scholar] [CrossRef] [PubMed]
- Von Sonntag, C. Nucleobases, Nucleosides and Nucleotides. In Free-Radical-Induced DNA Damage and Its Repair A Chemical Perspective; Springer: Berlin/Heidelberg, Germany, 2006; pp. 211–334. [Google Scholar]
- Sugiyama, H.; Saito, I. Theoretical studies of GG-specific photocleavage of DNA via electron transfer: Significant lowering of ionization potential and 5′-localization of HOMO of stacked GG bases in B-form DNA. J. Am. Chem. Soc. 1996, 118, 7063–7068. [Google Scholar] [CrossRef]
- Senthilkumar, K.; Grozema, F.C.; Fonseca Guerra, C.; Bickelhaupt, F.M.; Siebbeles, L.D.A. Mapping the Sites for Selective Oxidation of Guanines in DNA. J. Am. Chem. Soc. 2003, 125, 13658–13659. [Google Scholar] [CrossRef]
- Lin, H.; Truhlar, D.G. QM/MM: What have we learned, where are we, and where do we go from here? Theor. Chem. Acc. 2007, 117, 185–199. [Google Scholar] [CrossRef]
- Mayhall, N.J.; Raghavachari, K. Charge transfer across ONIOMQM: QM boundaries: The impact of model system preparation. J. Chem. TheoryComput. 2010, 6, 3131–3136. [Google Scholar]
- Chung, W.A.; Sameera, W.M.C.; Ramozzi, R.; Page, A.J.; Hatanaka, M.; Petrova, G.P.; Harris, T.V.; Li, X.; Ke, Z.; Liu, F.; et al. The ONIOM Method and Its Applications. Chem. Rev. 2015, 115, 5678–5796. [Google Scholar] [CrossRef]
- Lin, H.; Truhlar, D.G. Redistributed charge and dipole schemes for combined quantum mechanical and molecular mechanical calculations. J. Phys. Chem. A. 2005, 109, 3991–4004. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Xie, Y.; Schaefer, H.F., 3rd. Electron attachment to nucleotides in aqueous solution. Chem. Phys. Chem. 2006, 7, 1885–1887. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Xie, Y.; Schaefer, H.F., 3rd. Electron attachment to DNA single strands: Gas phase and aqueous solution. Nucleic Acids Res. 2007, 35, 5165–5172. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gu, J.; Wang, L.J. Electron attachment-induced DNA single-strand breaks at the pyrimidine sites. Nucleic Acids Res. 2010, 38, 5280–5290. [Google Scholar] [CrossRef]
- Zhao, Y.; Pu, J.; Lynch, B.J.; Truhlar, D.G. Tests of second-generationand third-generation density functionals for thermochemical kinetics. Phys. Chem. Chem. Phys. 2004, 6, 673–676. [Google Scholar] [CrossRef]
- Davidson, E.R.; Feller, D. Basis set selection for molecular calculations. Chem. Rev. 1986, 86, 681–696. [Google Scholar] [CrossRef]
- Runge, E.; Gross, E.K. Density-Functional Theory for Time-Dependent Systems. Phys. Rev. Lett. 1984, 2, 997–1000. [Google Scholar] [CrossRef]
- Miertus, S.; Tomasi, J. Approximate evaluations of the electrostatic freeenergy and internal energy changes in solution processes. Chem. Phys. 1982, 65, 239–245. [Google Scholar] [CrossRef]
- Marenich, A.V.; Jerome, S.V.; Cramer, C.J.; Truhlar, D.G. Charge Model5: An extension of Hirshfeld population analysis for the accurate description of molecular interactions in gaseous and condensed phases. J. Chem. Theory Comput. 2012, 8, 527–541. [Google Scholar] [CrossRef]
- Cave, R.J.; Newton, M.D. Generalization of the Mulliken-Hush treatmentfor the calculation of electron transfer matrix elements. Chem. Phys. Lett. 1996, 249, 15–19. [Google Scholar] [CrossRef]
- Karwowski, B.T. The influence of phosphorothioate on charge migration in single and double stranded DNA: A theoretical approach. Phys. Chem. Chem. Phys. 2015, 17, 21507–21516. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision, A.02; Gaussian Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Zheng, G.; Lu, X.J.; Olson, W.K. Web 3DNA–a web server for the analysis, reconstruction, and visualization of three-dimensional nucleic-acid structures. Nucleic Acids Res. 2009, 37, W240–W246. [Google Scholar] [CrossRef] [PubMed]
- Karwowski, B.T. The Influence of (5’R)- and (5’S)-5’,8-Cyclo-2’-Deoxyadenosine on UDG and hAPE1 Activity. Tandem Lesions are the Base Excision Repair System’s Nightmare. Cells 2019, 8, 1303. [Google Scholar] [CrossRef] [PubMed]
- Dapprich, S.; Komáromi, I.; Byun, K.S.; Morokuma, K.; Frisch, M.J. A new ONIOM implementation inGaussian98. Part, I. The calculation of energies, gradients, vibrational frequencies and electric field derivatives. J. Mol. Struct. 1999, 462, 1–21. [Google Scholar] [CrossRef]
- Zhao, Y.; Truhlar, M. Design of density functionals that are broadly accurate for thermochemistry, thermochemical kinetics, and nonbonded interactions. J. Phys. Chem. A 2005, 109, 5656–5667. [Google Scholar] [CrossRef] [PubMed]
- Diamantis, P.; Tavernelli, I.; Rothlisberger, U. Vertical Ionization Energies and Electron Affinities of Native and Damaged DNA Bases, Nucleotides, and Pairs from Density Functional Theory Calculations: Model Assessment and Implications for DNA Damage Recognition and Repair. J. Chem. Theory Comput. 2019, 15, 2042–2052. [Google Scholar] [CrossRef] [PubMed]
- Kawai, K.; Majima, T. Hole Transfer Kinetics of DNA. Acc. Chem. Res. 2013, 46, 2616–2625. [Google Scholar] [CrossRef]
- Karwowski, B.T. The AT Interstrand Cross-Link: Structure, Electronic Properties, and Influence on Charge Transfer in ds-DNA. Mol. Therapy Nucleic Acids 2018, 13, 665–685. [Google Scholar] [CrossRef]
- Marcus, R.A. Electron transfer reactions in chemistry theory and experiment. J. Electroanal. Chem. 1997, 438, 251–259. [Google Scholar] [CrossRef]
- Rust, M.; Lappe, J.; Cave, R.J. Multistate effects in calculations of the electronic coupling element for electron transfer using the generalized Mulliken–Hush method. J. Phys. Chem. A. 2002, 106, 3930–3940. [Google Scholar] [CrossRef]
- Dreuw, A.; Head-Gordon, M. Single-reference ab initio methods for the calculation of excited states of large molecules. Chem. Rev. 2005, 105, 4009–4037. [Google Scholar] [CrossRef] [PubMed]
- Cauet, E.; Valiev, M.; Weare, J.H. Vertical ionization potentials of nucleobases in a fully solvated DNA environment. J. Phys. Chem. B 2010, 114, 5886–5894. [Google Scholar] [CrossRef] [PubMed]
- Georgakilas, A.G. Processing of DNA damage clusters in human cells: Current status of knowledge. Mol. BioSyst. 2008, 4, 30–35. [Google Scholar] [CrossRef] [PubMed]
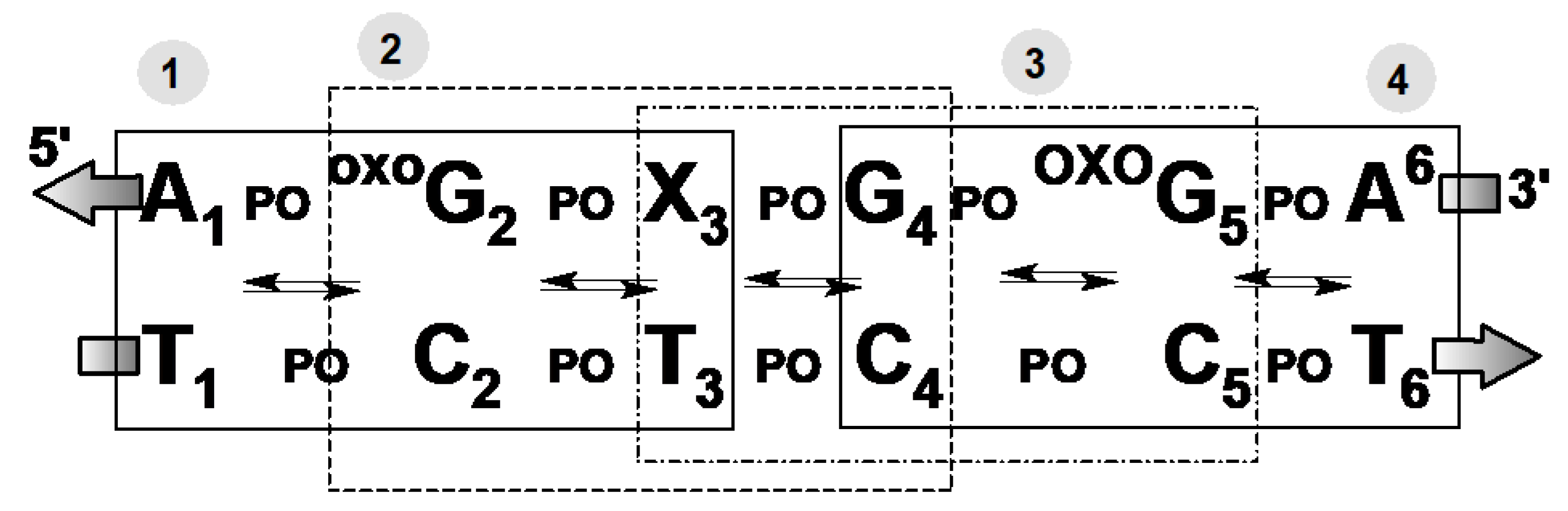
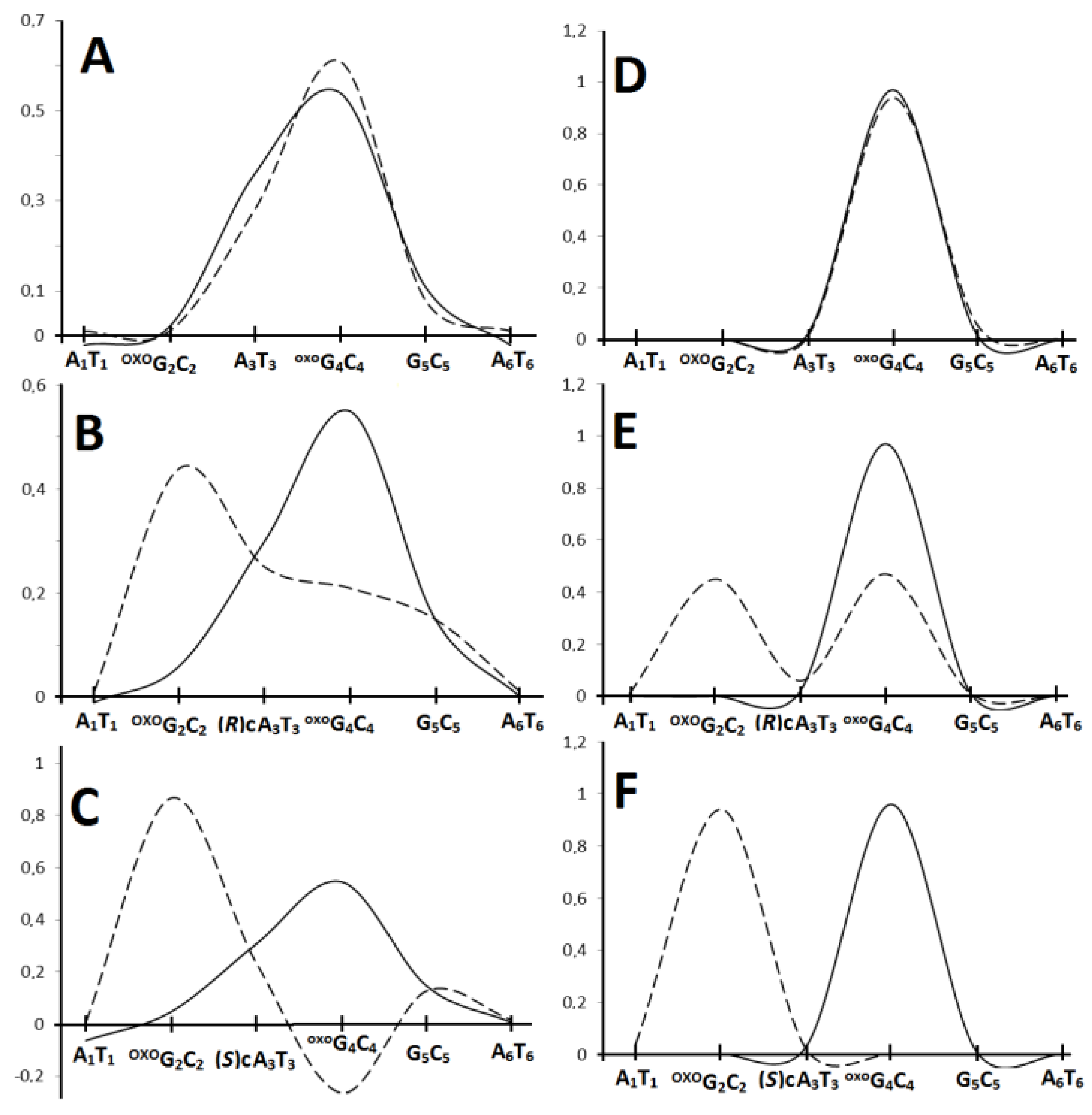
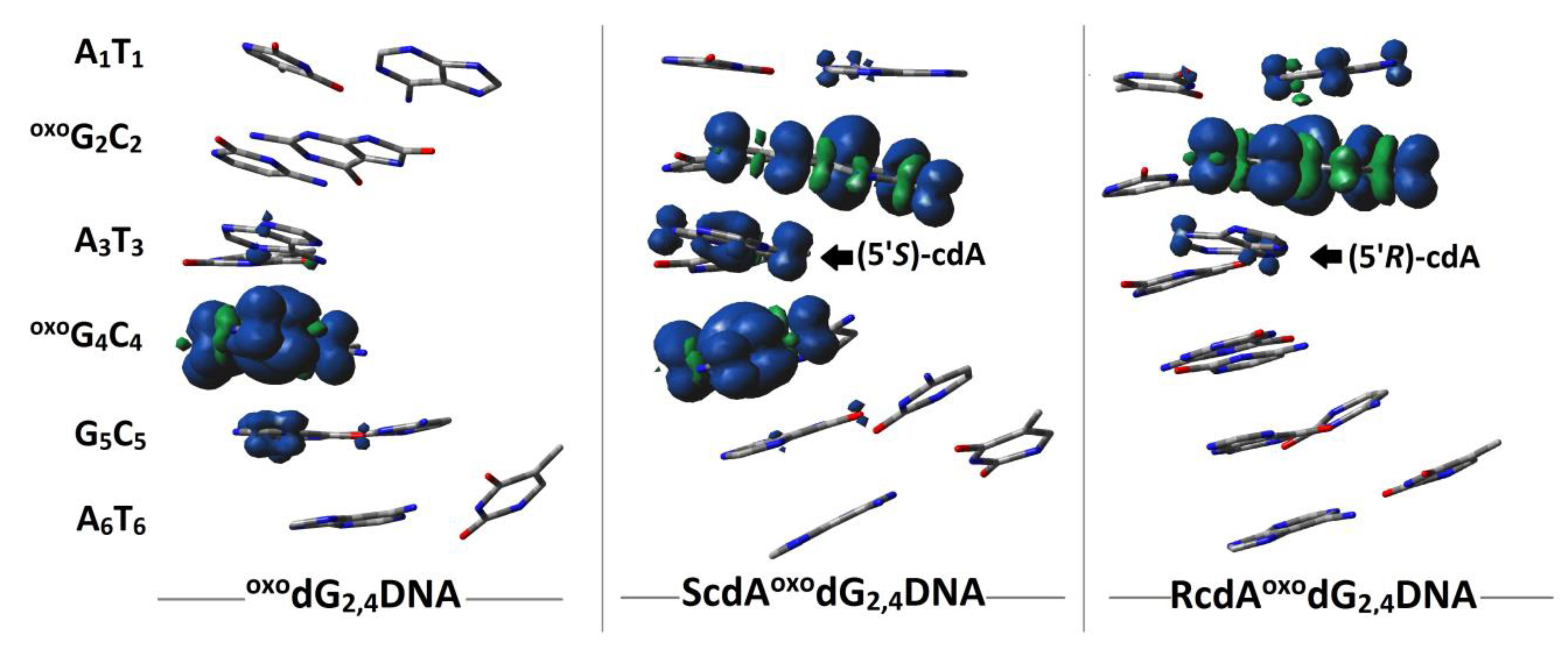
| HT System | oxodG2.4DNA | ScdAoxodG2.4DNA | RcdAoxodG2.4DNA | |||
|---|---|---|---|---|---|---|
| X = dA | X = (5′S)cdA | X = (5′R)cdA | ||||
| λ | KHT | λ | KHT | λ | KHT | |
| A1→oxoG2 | 0.04 | 0.00 | 0.02 | 0.00 | 0.04 | 0.00 |
| oxoG2←X3 | 0.03 | 0.00 | 0.01 | 0.00 | 0.05 | 0.00 |
| X3→oxoG4 | 0.41 | 9.8 × 109 | 0.38 | 5.65 × 109 | 0.41 | 2.53 × 1010 |
| oxoG4←G5 | 0.38 | 2.1 × 1014 | 0.37 | 6.95 × 1013 | 0.34 | 3.16 × 1013 |
| G5←A6 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karwowski, B.T. Clustered DNA Damage: Electronic Properties and Their Influence on Charge Transfer. 7,8-Dihydro-8-Oxo-2′-Deoxyguaosine Versus 5′,8-Cyclo-2′-Deoxyadenosines: A Theoretical Approach. Cells 2020, 9, 424. https://doi.org/10.3390/cells9020424
Karwowski BT. Clustered DNA Damage: Electronic Properties and Their Influence on Charge Transfer. 7,8-Dihydro-8-Oxo-2′-Deoxyguaosine Versus 5′,8-Cyclo-2′-Deoxyadenosines: A Theoretical Approach. Cells. 2020; 9(2):424. https://doi.org/10.3390/cells9020424
Chicago/Turabian StyleKarwowski, Boleslaw T. 2020. "Clustered DNA Damage: Electronic Properties and Their Influence on Charge Transfer. 7,8-Dihydro-8-Oxo-2′-Deoxyguaosine Versus 5′,8-Cyclo-2′-Deoxyadenosines: A Theoretical Approach" Cells 9, no. 2: 424. https://doi.org/10.3390/cells9020424
APA StyleKarwowski, B. T. (2020). Clustered DNA Damage: Electronic Properties and Their Influence on Charge Transfer. 7,8-Dihydro-8-Oxo-2′-Deoxyguaosine Versus 5′,8-Cyclo-2′-Deoxyadenosines: A Theoretical Approach. Cells, 9(2), 424. https://doi.org/10.3390/cells9020424






