Abstract
New players in plant signaling are described in detail in this review: extracellular ATP (eATP) and uncommon nucleotides such as dinucleoside polyphosphates (NpnN’s), adenosine 5′-phosphoramidate (NH2-pA), and extracellular NAD+ and NADP+ (eNAD(P)+). Recent molecular, physiological, and biochemical evidence implicating concurrently the signaling role of eATP, NpnN’s, and NH2-pA in plant biology and the mechanistic events in which they are involved are discussed. Numerous studies have shown that they are often universal signaling messengers, which trigger a signaling cascade in similar reactions and processes among different kingdoms. We also present here, not described elsewhere, a working model of the NpnN’ and NH2-pA signaling network in a plant cell where these nucleotides trigger induction of the phenylpropanoid and the isochorismic acid pathways yielding metabolites protecting the plant against various types of stresses. Through these signals, the plant responds to environmental stimuli by intensifying the production of various compounds, such as anthocyanins, lignin, stilbenes, and salicylic acid. Still, more research needs to be performed to identify signaling networks that involve uncommon nucleotides, followed by omic experiments to define network elements and processes that are controlled by these signals.
1. Introduction
Plant signaling is a set of phenomena that enables the transduction of external and internal signals into physiological responses such as modification of enzyme activity, cytoskeleton structure, and gene expression. It is known that in plants there exist mechanisms involved in the signal transduction pathways. Plants have evolved signaling networks providing reactions to environmental stimuli through signaling proteins such as plasma membrane receptors and ion transporters and by cascades of kinases and other enzymes up to effectors. For many years plant hormones were considered to be dominant molecules in plant signaling. Nowadays this term embraces many other compounds including second messengers, such as cytosolic Ca2+ [1], reactive oxygen (ROS) and nitrogen species (RNS) [2] or cyclic nucleotides such as adenosine 3′,5′-cyclic monophosphate (cAMP) and guanosine 3′,5′-cyclic monophosphate (cGMP) [3]. Nowadays there is more and more information about synthesis, degradation, and function of cAMP and cGMP in plant [3,4,5], and they are currently accepted as key signaling molecules in many processes in plants including growth and differentiation, photosynthesis, and biotic and abiotic defense [6]. However, recently it was shown that nucleotides, such as ATP and (di)nucleoside polyphosphates, also can play signaling roles in plant cells. In this review, we focus on the extracellular ATP (eATP) and uncommon nucleotides, such as mono- (pnNs) and dinucleoside polyphosphates (NpnN’s), and their new function as signaling molecules.
2. eATP as a Signaling Molecule
Adenosine 5′-triphosphate (ATP), as well as the other nucleoside triphosphates, are established as agents providing energy in various reactions inside cells, both in animal and plant organisms [7]. As ATP is omnipresent in all living cells, it is often called the essential energy currency molecule. In the 1970s it was first hypothesized that ATP might be released into the extracellular environment and act as a signaling compound for animal cells [8]. However, it is worth mentioning that the first effect of eATP on a cell was noted much earlier, i.e., in 1929, during research on heart muscle contraction [9]. In animals, three possible ways of ATP release from the cells into the extracellular matrix have been proposed; they involve multiple channels, transporters, and exocytosis. The interest in the eATP signaling function accelerated after the first purinoreceptor was cloned and characterized in rat brain tissue [10]. Currently, eATP, as well as some of the other nucleotides, are considered as signaling molecules mediating numerous animal cellular processes. For decades the role of nucleotides as signaling molecule functioning similarly in plants as it was demonstrated in animals was viewed with skepticism. A real breakthrough in this topic came with the discovery of the existence of a plant transmembrane receptor protein with serine/threonine kinase activity having a high affinity for extracellular nucleotides [11]. However, the evidence for the mechanism of ATP release from the cytosol into the extracellular matrix in plants appeared earlier [12,13,14,15]. In plants, there are several possible ways of ATP outlet. The ATP release triggered by environmental stimuli appears via the wounded cell membrane [16], exocytosis [12], the p-glycoprotein (PGP1) belonging to the ATP-binding cassette ABC transporters [17], and plasma membrane-localized nucleotide transporters (PM-ANT1) [18]. The receptor-eATP interaction begins a cascade reaction that leads to further downstream physiological changes protecting the plant against both biotic and abiotic stresses but also guarantees proper plant growth and development [19].
In order to maintain proper cell growth and function, eATP concentration must be controlled. During regeneration after the stress factor has disappeared, eATP degradation is conducted by hydrolytic enzymes called apyrases [20]. The human apyrases are the best characterized and described apyrases among different kingdoms. Their cellular localization includes plasma membrane, Golgi apparatus, and endoplasmic reticulum. The human apyrases present in plasma membrane show ecto-apyrases activity mediating regulation of the eATP in the extracellular environment. The Arabidopsis thaliana apyrase family consists of seven enzymes among which two closely related ones, APY1 and APY2, are the most extensively characterized. These enzymes mediate the luminal glycosylation and can be a component of regulation of the eATP level. It was demonstrated that these two enzymes are an integral component of the Golgi apparatus membrane where they indirectly control the eATP level by modulating the luminal concentration of ATP in secretory vesicles [21,22,23] (Figure 1). Both APY1 and APY2 are also essential enzymes for proper plant growth and development. These processes are auxin-dependent. Among various factors, auxin transport also depends on the expression of the genes encoding APY1 and APY2. Suppression of the APY1 and APY2 expression causes dwarfism, impaired polar auxin transport and eATP over-accumulation in Arabidopsis thaliana [24,25]. However, it was suggested that some fractions of the APY1 and APY2 population with ecto-apyrases activity might by localized also in the plasma membrane [19]. Although there are many data regarding subcellular localization of Arabidopsis apyrases (APY1 and APY2), the apyrases from soybean (GS52), pea (PsAPY1), and potato (StAPY3) occur outside the cell (ecto-apyrases) [26,27,28]. A study conducted by Wu and co-workers indicated that the externally applied APY1 and APY2 inhibitors cause the increase of eATP and physiological changes typical for the plant reaction to stress [24]. Furthermore, there are some reports showing particular plant species secreting individual apyrase members out of the endomembrane system. Taking all these circumstances into account, the existence of a plasma membrane-localized Arabidopsis thaliana apyrase (Figure 1) cannot be excluded [29].
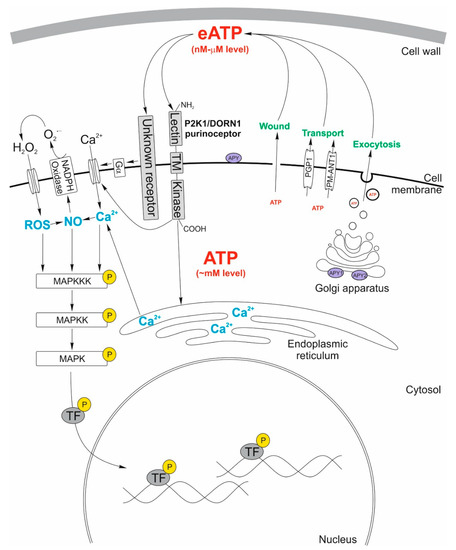
Figure 1.
Model of changes occurring in the plant cell triggered by the extracellular ATP (eATP). In this model, three possible ways of ATP release into the extracellular matrix are demonstrated. It considers the wounded cell membrane, exocytosis, and two transporters: the p-glycoprotein (PGP1) belonging to the ATP-binding cassette ABC transporters, and the plasma membrane-localized nucleotide transporters (PM-ANT1). Two apyrases, APY1, and APY2, localized in the Golgi apparatus membrane of Arabidopsis thaliana regulate the concentration of eATP. Additionally, the hypothesized apyrase (APY) located at the extracellular surface of the plasma membrane can decrease eATP concentration directly in the extracellular matrix. The released eATP acts as a signaling molecule triggering elevation of the cytosolic Ca2+ level by activation of the P2K1 receptor, which in turn activates the Ca2+ channel. The hypothetical non-P2K1 receptor, whose binding with eATP leads to activation of the Gα subunit of the heterotrimeric G-protein, activates the cell membrane Ca2+ channel. High cytosolic Ca2+ concentration causes an increase in production of nitric oxide (NO), reactive oxygen species (ROS), and mitogen-activated protein kinases (MAPKs), which finally leads to various physiological responses. The ROS boosted production is due to the activation of the RBOHD subunit of the plasma membrane-localized NADPH oxidase. The contribution of the transcription factors in the regulation of gene expression is of high importance.
2.1. Plant eATP Receptors
The eATP animal receptors were discovered in 1976 [30]. Initially, they were called ‘purinergic receptors’. The name was changed to ‘nucleotide receptors’, as both purine and pyrimidine nucleotides trigger their activation [31]. These receptors belong to two groups, P1 and P2, but only P2 receptors are activated by ATP [32]. P2 receptors are divided into two classes: ligand-gated ion channels (P2X) and G protein-coupled (P2Y) receptors [33]. It is important to emphasize that P2X and P2Y receptors do not exist in plant organisms [34]. For many years it was hard to prove the existence of a similar receptor in plants although there were papers indicating changes in plants’ development, growth and response to stresses under exogenously applied ATP [11]. Research on dorn1 (DOes not Respond to Nucleotides) mutants of Arabidopsis thaliana revealed the first plant receptor with a high affinity to bind eATP. Mutants showed a lower cytosolic Ca2+ level, lack of mitogen-activated protein kinases (MAPKs) activation and as a consequence decline of the defense-related gene expression level. Initially, the newly discovered receptor was named DORN1, but later the name was changed to P2K1 because the research showed that the DORN1 gene encodes the L-type lectin receptor-like kinase I.9 (LecRK-I.9). This receptor has three domains: an extracellular ATP-binding lectin domain, a single transmembrane domain, and an intracellular kinase domain (Figure 1). In the case of dorn1 mutants defective in the kinase domain, eATP is not able to connect with the receptor and as a result is not able to trigger downstream responses. Decreased expression of the LecRK-I.9 gene caused weaker eATP responses, while intensified LecRK-I.9 gene expression enhanced eATP responses [35]. DORN1 is a plant purine receptor that belongs to the lectin-receptor kinase family and is denoted as P2K1 to distinguish it from the animal P2 receptors: P2X and P2Y [11,36,37]. Interestingly, P2K1 gene expression level is relatively high during the major stages of plant development [37], suggesting the essential role of the eATP in processes such as seedling growth, stomata movement, pollen tube development, root hair growth, gravitropism, and biotic and abiotic stress responses [38].
Recently, the existence of another, unidentified, non-P2K1 receptor with affinity to eATP was suggested [39]. It was reported that Arabidopsis thaliana dorn1 null mutants demonstrated an increased level of cytosolic Ca2+ under the exogenously applied ATP. It was a result of the heterotrimeric G-protein Gα subunit activation followed by the opening of a plasma-membrane Ca2+ channel. This finding suggests new, P2K1-independent responses to eATP, involved among others in the root-bending mechanism [39]. Moreover DORN1 could underpin several calcium-related responses but it may not be the only receptor for eATP in Arabidopsis thaliana [40].
2.2. Plant eATP Signal Transduction Pathway
In plants, eATP takes part in cell signaling as a messenger, which triggers a signaling cascade, when binding to the P2K1 receptor (Figure 1). Cytosolic nitric oxide (NO), Ca2+, and ROS form a secondary messenger trio, which occurs in various signaling pathways leading to the transient phosphorylation of MAPK, especially MPK3 and MPK6, and expression of defense-related genes [41]. The multibranched reaction starts with the elevation of the cytosolic Ca2+ level triggered by eATP, leading to the accumulation of MAPK and NO, as well as to the phosphorylation and activation of the RBOHD (respiratory burst oxidase homolog protein D) subunit of the plasma membrane-localized NADPH oxidase [42]. This enzyme catalyzes the synthesis of extracellular ROS like superoxide (O2−), which is then converted into hydrogen peroxide (H2O2) in the extracellular milieu [16,33,43]. Extracellular H2O2 crosses the double-layered plasma membrane via various channels among which aquaporins are distinguishable [44]. Subsequently, ROS trigger changes in the expression of nuclear genes, responsible for defense responses [16], root hair growth [12] or regulation of Na+, H+, and K+ levels. Consequently, a disturbance in the ion homeostasis may lead to the mitochondria-independent type of programmed cell death by activation of the caspase-like proteases [45].
2.3. eATP Involvement in Plant Responses to Biotic and Abiotic Stresses
Aside from different eATP-induced factors and signaling molecules contributing to the plant resistance against pathogens and various abiotic stresses, classical defense hormones such as jasmonate, ethylene, and salicylic acid are of high importance [38]. Based on the pathogens’ lifestyles two groups of these organisms are distinguishable: biotrophs and necrotrophs. Interestingly, the efficiency of the used phytohormone against these two groups of pathogeneses differs. Salicylic acid induces plant defense against biotrophic pathogens, whereas jasmonate and ethylene are indispensable when necrotrophic pathogens and herbivorous insects attack the plant. Furthermore, salicylic acid regulates pathogen-induced systemic acquired resistance (SAR), whereas jasmonate and ethylene play a crucial role in rhizobacteria-mediated induced systemic resistance (ISR) [46]. Transcriptomic research of Arabidopsis thaliana mutants defective in the jasmonate, ethylene, and salicylic acid signaling pathway, which were treated with ATP, revealed crosstalk in the signaling of the typical plant hormones and eATP. These results showed that from among all of the defense-related genes, up to 50% were induced by eATP in cooperation with the typical plant defense hormones. This finding suggests a complex network of the plant defense mechanism that needs to be explored more deeply at different levels [38]. The contribution of transcription factors to ATP-responsive transcription is also considered. It was demonstrated that a calmodulin-binding transcription activator (CAMTA3) and MYC transcription factors are required for proper defense-related gene transcription, whose expression is induced by eATP [38].
In order to examine the importance of eATP in the plant reaction to pathogens two approaches might be considered: either alteration of the P2K1 receptor or manipulation at the eATP level. Overexpression of the P2K1 gene resulted in increased plant resistance to the bacterial pathogen Pseudomonas syringae and the oomycete pathogen Phytophthora brassicae [47,48]. Plants treated with ATP were found to be protected against various organisms such as the fungal pathogen Botrytis cinerea [7] and the bacterial pathogen Pseudomonas syringae [48]. Although the complete mechanisms of eATP signal transduction through the P2K1 receptor remain unclear, a plenitude of evidence shows the involvement of eATP in plant resistance to biotic stresses.
Studies show that eATP level, as well as P2K1 activity, are also of high importance in the plant-fungus symbiosis. Interactions of the filamentous root endophyte Serendipita indica with various experimental plant hosts, including Arabidopsis thaliana and Hordeum vulgare, have been examined. Serendipita indica colonization was found to be beneficial for plants, reflected in plant growth enhancement, assimilation of nitrate and phosphate improvement and better tolerance to both abiotic and biotic stresses [49]. Although Serendipita indica penetrates the host’s root cells, massive plant cell death does not occur. It is because of an enzymatically active ecto-5′-nucleotidase (E5′NT) enzyme secreting by Serendipita indica which is capable of hydrolyzing nucleotides in the apoplast. By the hydrolyzation of ATP, ADP, and AMP, Serendipita indica E5′NT modifies the eATP level, leading to switching off the plant defense reactions which promote proper fungal accommodation [50]. Moreover, it was observed that the rhizobial nodule factor stimulates the release of ATP outside the cell by the root hair tips of Medicago truncatula [12]. Treatment of plants with eATP caused also the change their susceptibility to pathogen infection [16]. It is hypothesized that ecto-apyrases may decrease the eleveted level of eATP upon symbiont infection and thereby prevents the activation of plant defense pathways that could limit symbiont invasion [51].
Several studies indicate the role of eATP in the response to different types of abiotic stresses. In addition to mechanical stimuli caused by wounding or touch, ATP is also released in response to treatment with molecules such as abscisic acid and L-glutamate [15,52]. A similar reaction was observed during both osmotic and salt stress [12,15,53] as well as under cadmium treatment [54]. Consequently, eATP accumulation triggers plant physiological changes leading to enhancement of resistance. It involves rapid closure of leaf stomata [55], probable seedling viability enhancement [56] and modification of root growth direction when encountering an obstacle [34]. In addition, hypertonic salt stress interferes with the photosynthesis machinery by decreasing the levels of maximal efficiency of photosystem II and depleting the photochemical quenching [54]. Moreover, abiotic stress caused by cadmium triggers a rapid increase in lipid peroxidation but also higher antioxidant and lipoxygenase activities in Arabidopsis thaliana cells [54]. These reactions are associated with boosted synthesis of jasmonic acid, which is one of the most important molecules involved in the different stress responses [57]. Nowadays, one of the major anthropogenic pollutants with the highest level of threat to human health is cadmium [58]. It is also considered as one of the most phytotoxic elements among heavy metals, causing a reduction in crop biomass by disrupting photosynthesis and respiration [59]. In addition, cadmium can induce other physiological changes including induction of oxidative stress by boosting the production of ROS [58], modifications in gene expression [60,61], as well as changes in enzyme activity [62], hence activating plant defense.
3. Extracellular Pyridine Nucleotides
The pyridine nucleotides nicotinamide adenine dinucleotide (NAD+) and NAD+ phosphate (NADP+) are commonly occurring electron carriers that are involved in metabolic reactions as well as intracellular signaling [63,64]. It is known that in plants NAD(P)+ can be released outside the cell [65]. Extracellular NAD(P)+ (eNAD(P)+) induces the expression of pathogen-related genes and the resistance to Pseudomonas syringae in Arabidopsis thaliana through pathways involving calcium- and salicylic acid-mediated defense signaling. It was also indicated that eNAD(P)+ induces transcriptional and metabolic changes in Arabidopsis thaliana similar to those caused by pathogen infection [66]. Moreover, the expression of the human NAD(P)+-hydrolyzing ecto-enzyme CD38 in Arabidopsis thaliana partially compromises systemic acquired resistance (SAR) and this suggests that eNAD(P)+ can be a SAR signal molecule [67]. Recent studies on the function of eNAD(P)+ focus on understanding their role in plants and on identifying their receptor(s). Analysis of transcriptome changes in Arabidopsis thaliana evoked by eNAD+ identified a lectin receptor kinase (LecRK) LecRK-I.8 as a potential eNAD+ receptor. LecRK-I.8 is located in the plasma membrane, has kinase activity, and specifically binds only NAD+, but not NADP+, ATP, ADP or AMP. Moreover, the expression of LecRK-I.8 is induced by the eNAD+ [68]. Another LecRK that can be a potential receptor for eNAD(P)+ is LecRK-VI.2, which was identified in Arabidopsis thaliana. LecRK-VI.2 is constitutively associated with Brassinosteroid Insensitive1-Associated Kinase1 (BAK1) and it was shown that complex LecRK-VI.2/BAK1 is involved in SAR [69].
4. Uncommon Nucleotides as Signaling Molecules
4.1. Mononucleoside Polyphosphates
4.1.1. Structure and Occurrence of Mononucleoside Polyphosphates
Despite mono- and dinucleoside polyphosphates having been discovered in the middle of the twentieth century, our knowledge about their biological function is still poor, especially in plants. Mononucleoside polyphosphates (pnNs) contain a nucleoside and oligophosphate chain. Examples of these nucleotides are adenosine 5′-tetraphosphate (p4A, ppppA, Figure 2) and adenosine 5′-pentaphosphate (p5A, pppppA). Both of them were discovered in commercial preparations of ATP obtained from bovine cells [70,71], horse muscle [72] and yeast [73]. Additionally, other purine and pyrimidine p4Ns were found as contamination of various nucleoside triphosphates (NTP) preparations: p4G [74,75], p4U [76], and p4C [77]. The existing of p4A and p5A was confirmed in biological materials such as rat liver [78,79], rabbit and horse muscle [80], bovine adrenal medulla [80,81,82,83], rabbit thrombocytes [84], and Saccharomyces cerevisiae [85]. However, the concentration of p4A in the above-mentioned animal samples was about 2 μM, but in chromaffin granules from the adrenal medulla it was about 800 μM, and it was even up to 4 orders of magnitude and 300-fold lower than ATP concentration, respectively [83]. Until now there is no information about the content of pnNs in the plant tissues.
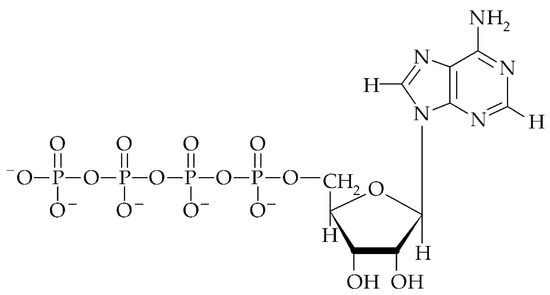
Figure 2.
Structure of adenosine 5′-tetraphosphate (p4A).
4.1.2. Synthesis and Degradation of Mononucleoside Polyphosphates
The level of pnNs in a cell depends on its biosynthesis and degradation, but also pnNs may be a product of the degradation of some dinucleoside polyphosphates (NpnN’s). Enzymes that can synthesize pnNs in vitro can be grouped into two categories: aminoacyl-tRNA synthetases (AARS) and non-aminoacyl-tRNA synthetases (non-AARS) [86]. Among enzymes synthesizing pnNs only one belongs to the AARS and it is lysyl-tRNA synthetase (LysRS) from Escherichia coli, which can synthesize p4A [87,88]. The majority of the enzymes belong to the non-AARS. Table 1 presents non-plant enzymes synthesizing pnNs.

Table 1.
Non-plant enzymes synthesizing mononucleoside polyphosphates (pnNs).
Among enzymes that can degrade pnNs some of them exhibit low substrate specificity, for example, alkaline (EC 3.1.3.1) and acid phosphatases (EC 3.1.3.2), that release phosphate residues up to adenosine [113]. Apyrase (EC 3.6.1.5) also can cut off phosphate residues, but only to AMP [113]. Phosphodiesterase I (EC 3.1.4.1) degrades p4A to triphosphate and AMP [100]. Additionally, adenosine-phosphate deaminase from Aspergillus oryzae and Helix pomatia is involved in p4A metabolism, converting it into inosine 5′-tetraphosphate (p4I) [114].
As mentioned above, pnNs can accumulate in the cell as a result of the degradation of NpnN’s. The way in which p4A can accumulate in a cell is the degradation of diadenosine 5′,5′′′-pentaphosphate (Ap5A) and diadenosine 5′,5′′′-hexaphosphate (Ap6A). The degrading enzymes include phosphodiesterase I (EC 3.1.4.1) occurring in prokaryotes and eukaryotes [115,116], symmetrical dinucleoside tetraphosphatase (EC 3.6.1.41) from bacteria [117], dinucleoside tetraphosphate phosphorylase (EC 2.7.7.53) from Saccharomyces cerevisiae and Euglena gracilis [118,119], and dinucleoside triphosphatase (EC 3.6.1.29) among others from Lupinus luteus [120].
Despite the lack of knowledge about the occurrence and the concentration of mononucleoside polyphosphates in higher plants, there are described a few enzymes which can synthesize pnNs. All of them are listed in Table 2. The first described plant enzyme that synthesizes pnNs is 4-coumarate:CoA ligase (4CL2) from Arabidopsis thaliana that catalyzes the reaction of the synthesis of both p4A and p5A [121]. This enzyme is a branch point in the phenylpropanoid pathway that leads to the biosynthesis of flavonoids, lignin, and stilbenes. It is known that the phenylpropanoid pathway is involved in plant responses to numerous environmental stimuli, especially under biotic and abiotic stresses [122]. Another enzyme synthesizing p4A is jasmonate:amino acid synthetase from Arabidopsis thaliana (JAR1) [123]. JAR1 is involved in the function of jasmonic acid (JA) as a plant hormone and it catalyzes the synthesis of several JA-amido conjugates, the most important of which appears to be jasmonic acid-isoleucine. Both of the above-mentioned plant enzymes belong to the acyl~adenylate-forming firefly luciferase superfamily [124] which catalyzes a two-step reaction. During the first step, acyl and ATP form an acyl-adenylate intermediate with the simultaneous release of pyrophosphate (PPi). In the second step, 4CL in the absence of CoA catalyzes the formation of uncommon mononucleoside polyphosphates such as p4A and p5A [121].

Table 2.
Plant enzymes synthesizing mononucleoside polyphosphates (pnNs).
In plants there also exists an enzyme degrading p4A. It is nucleoside tetraphosphate hydrolase (EC 3.6.1.14) occurring in Lupinus luteus seeds. This enzyme hydrolyzes both p4A and p4G with the same rate while p5A is degraded up to 200-fold more slowly than p4A and p4G [120].
4.2. Adenosine 5′-Phosphoramidate
Structure, Occurrence, and Metabolism of Adenosine 5′-Phosphoramidate
Among naturally occurring uncommon mononucleotides is adenosine 5′-phosphoramidate (NH2-pA, Figure 3). This compound is believed to occur in all organisms; however, so far NH2-pA has only been detected among cellular nucleotides purified from the green alga Chlorella pyrenoidosa [125]. Similarly to pnNs, the level of NH2-pA in a cell is enzymatically controlled. It is known that NH2-pA can be synthesized by adenylyl sulfate:ammonia adenylyltransferase (EC 2.7.7.51) in the algae Chlorella pyrenoidosa, Euglena gracilis, amoeba Dictyostelium discoideum, bacteria Escherichia coli, and in higher plants Hordeum vulgare, Spinacia oleracea [126], and Lupinus luteus [127]. This transferase catalyzes the following reaction: SO4-pA + NH4+ → NH2-pA + SO42− + 2H+.
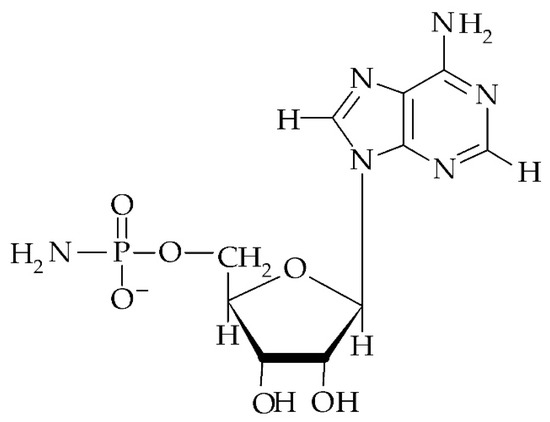
Figure 3.
Structure of adenosine 5′-phosphoramidate (NH2-pA).
The supposition that NH2-pA is a ubiquitous compound and that its concentration is enzymatically controlled may be supported by the existence of various enzymes that catalyze the cleavage of NH2-pA to ammonia and AMP (pA) by hydrolysis [128,129,130,131,132,133], or to ammonia and ADP (ppA) by phosphorolysis [130]. Both the synthesis [127] and the degradation [131] of NH2-pA can be controlled by HIT (histidine triad) proteins. One of the proteins belonging to the HIT family proteins is Fhit (fragile histidine triad). Based on the mechanism of the action of Fhit protein, which is able to hydrolyze the P-N bond in adenosine phosphoimidazolide [134], it is hypothesized that other uncommon nucleotides can be substrates for human and Arabidopsis thaliana Fhit. Other proteins belonging to HIT family proteins are Hint protein (having the activity of NH2-pA hydrolase) and GalT proteins (having the specific activity of nucleoside monophosphate transferase) [135]. The protein Fhit from human and Arabidopsis thaliana exhibited the activity attributed to adenylyl sulfate sulfohydrolase and nucleoside phosphoamidases releasing AMP from adenosine 5′-phosphosulfate (SO4-pA) and NH2-pA, respectively. Fhit protein also catalyzes the hydrolysis of the P-F bond in the synthetic nucleotide adenosine 5′-monophosphofluoride (F-pA) releasing AMP [136]. Recently it has been shown that NH2-pA can also be synthesized by Fhit proteins from Lupinus luteus seeds and Arabidopsis thaliana. It catalyzes the ammonolysis of SO4-pA, leading to the formation of NH2-pA [127].
4.3. Dinucleoside Polyphosphates
4.3.1. Structure and Occurrence of Dinucleoside Polyphosphates
Dinucleoside polyphosphates (NpnN’s) consist of an oligophosphate chain that links the two 5′-esterified nucleosides. Among NpnN’s that are most frequently tested are adenine nucleotides: diadenosine triphosphate (Ap3A) and diadenosine tetraphosphate (Ap4A, Figure 4). The first reports about the existence of NpnN’s come from the 1950s. It was discovered that during the chemical synthesis of adenosine-uridine monophosphate (ApU) there were synthesized byproducts such as diadenosine diphosphate (Ap2A) and diuridine diphosphate (Up2U) [137]. Nevertheless, the first NpnN discovered in the biological material was diguanosine tetraphosphate (Gp4G). The presence of Gp4G and Gp3G was observed in biological preparations in encysted gastrulae of the brine shrimp and Artemia salina in millimolar concentrations [138,139]. The most common NpnN is Ap4A, which for the first time was detected in rat liver, and its concentration was estimated at 30 nM [97]. Subsequently Ap4A and other NpnN’s have been observed in submicromolar concentrations in many investigated cells and tissues [140,141]. NpnNs were identified in bacteria [142,143], yeast [144,145], and animal and human cells [140,146,147,148,149,150]. So far, NpnN’s have not been identified in plants.
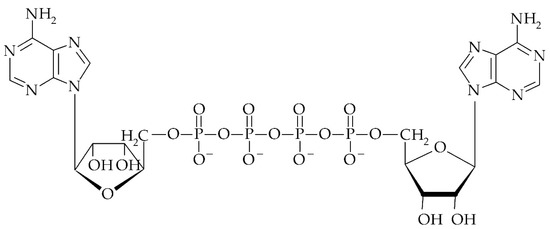
Figure 4.
Structure of diadenosine 5′, 5′′′-tetraphosphate (Ap4A).
4.3.2. Synthesis and Degradation of Dinucleoside Polyphosphates
The level of NpnN’s in cells may be a result of the synthesis and degradation of these compounds. Table 3 lists non-plant enzymes synthesizing various NpnN’s. Enzymes synthesizing NpnN’s are some ligases [89,92,106,121,151,152], luciferase from Photinus pyralis [153], and some transferases [100,119,154]. Among aminoacyl-tRNA synthetases, the most effective in ApnN synthesis are lysyl- (EC 6.1.1.6), phenylalanyl- (EC 6.1.1.20), alanyl- (EC 6.1.1.7), and prolyl- (EC 6.1.1.15) t-RNA synthetases [86]. Moreover, the RNA-dependent RNA polymerase elongation complex from hepatitis C virus (HCV) can use in vitro nucleoside triphosphates (NTPs) to excise the terminal nucleotide in nascent RNA and mismatched ATP, UTP, or CTP could mediate excision of 3′-terminal CMP to generate the dinucleoside tetraphosphate products Ap4C, Up4C, and Cp4C, respectively [155].

Table 3.
Non-plant enzymes synthesizing dinucleoside polyphosphates (NpnN’s).
So far, only three enzymes synthesizing NpnN’s have been described in plants (Table 4). Among aminoacyl-tRNA synthetases only phenylalanyl- (EC 6.1.1.20) and seryl-tRNA (EC 6.1.1.11) synthetases from Lupinus luteus [152] synthesize ApnN. The mechanism of the synthesis of ApnNs catalyzed by plant aminoacyl-tRNA synthetases is based on the formation of aminoacyl~pA and the transfer of adenylate to pppN [152]. The third enzyme which is involved in the synthesis of ApnN in plant cells is 4:coumarate-CoA ligase (EC 6.2.1.12) [121].

Table 4.
Plant enzymes synthesizing dinucleoside polyphosphates (NpnN’s).
Enzymes degrading NpnN’s may be divided into substrate-specific and substrate non-specific. Substrate-specific enzymes degrading NpnN’s include, among others, asymmetrical Np4N hydrolase (EC 3.6.1.17), symmetrical Np4N hydrolase (EC 3.6.1.41), and dinucleoside triphosphate (Np3N) hydrolase (EC 3.6.1.29) [156]. In plants, the activity of asymmetrical Np4N hydrolase was detected in Lupinus luteus seeds [157], Helianthus annuus and marrow (Cucurbita pepo) seeds [158], tomato cells [159], Lupinus angustifolius seeds [160] and in Hordeum vulgare [161]. Among Np4N’s, only Ap4A can be degraded by the asymmetrical Np4N hydrolase giving ATP. The substrate non-specific enzymes in plants are phosphodiesterase I (EC 3.1.4.1) from Lupinus luteus [157] and nucleotide pyrophosphatase (EC 3.6.1.9) from potato tuber [162].
4.4. Function of Uncommon Nucleotides
The cellular level of NpnN’s is modified under various physiological and pathological conditions and the compounds have been suggested as intracellular messengers in diverse cellular processes [163,164]. The increased concentration of Ap4A was correlated with high cellular proliferation rates [165] or some phases of the cell cycle [165,166]. A dramatic increase of levels of Np4N and various NpnN’s has been observed in cells subjected to stresses, such as elevated temperature, ethanol or cadmium [143,144,145,167]. A very interesting function of uncommon nucleotides is their potential function as alarmones [168]. However, so far no clear metabolic or molecular targets or receptors of the postulated alarm signaled by the NpnN’s have been experimentally demonstrated. Alarmones are intracellular signaling molecules that are produced under adverse environmental factors. They regulate gene expression at the transcriptional level. Nucleotides (p)ppGpp and ppGpp have such a role in bacteria [169]. The results of recent studies on the role of uncommon nucleotides focus on microorganisms and animals, including man. It has been demonstrated that NpnN’s may accumulate in cells under stress conditions. In cells of Saccharomyces cerevisiae and Escherichia coli under elevated temperature and cadmium ions an increased level of NpnN’s was observed. The intracellular concentrations of these molecules vary from 10−9 M in a basal metabolic state to 10−4 M when the organisms are subjected to stresses, such as heat shock or exposure to cadmium [143,167]. In cyanobacteria the same effects are a result of thermal shock and heavy metals [145]. In Salmonella, the increased synthesis of NpnN’s was caused by ethanol [142]. Studies performed on orange fruit showed that under high temperature enzymes involved in NpnN’ metabolism were activated and an extremely large increase in bis(5′-adenosyl) triphosphatase protein content was observed. These results suggest intensive synthesis of Ap3A under high temperature [170]. Another group of enzymes involved in NpnN’ metabolism is Nudix (nucleoside diphosphate linked to x) hydrolases (NUDTs). An increase of transcript and protein NUDT7 levels was found in Arabidopsis thaliana plants under biotic stress evoked by Pseudomonas syringae avrRpt2 [171]. In Arabidopsis thaliana, there was also observed a quick increase in NUDT7 content under the effect of ozone and pathogen infection [172]. Recent studies on AtNUDT19 indicate the involvement of this hydrolase in the photo-oxidative stress response by regulating photosynthesis, the antioxidant system and the synthesis of salicylic acid—a signaling molecule involved in the response to biotic and abiotic stresses [173]. Additionally, it is suggested that barley NUDTs respond to abiotic stress [174]. It is also known that NUDTs bind RNA and participate in the regulation of gene expression in animals and plants [175,176,177].
Studies on the function of Ap4A in Escherichia coli have shown that it may be a damage metabolite and a few proteins binding Ap4A in these bacteria were identified [178]. Recent studies on the function of Ap4A have shown that aminoglycoside antibiotics induced synthesis of Ap4A in Escherichia coli and it caused bacterial cell killing by these antibiotics [179]. Lysyl-tRNA synthetase plays a key role in MITF (microphthalmia-associated transcription factor) transcriptional activity via Ap4A as an important signaling molecule in mast cells [180,181]. Studies on chronic myelogenous leukemia cells showed that NUDT2 disruption elevates Ap4A and down-regulates immune response and cancer promotion genes [182]. Ap3A and Ap4A induce activation of a signaling pathway that results in increase of proliferation of vascular smooth muscle cells by stimulation of ERK1/2(MAP kinase) [183]. It is known that NpnN’s can act as neurotransmitters and influence the vascular system through purinergic receptors. There is also evidence that Ap4A inhibits the initiation of DNA replication. It is also proposed that Ap4A acts as an important inducible ligand in the DNA damage response to prevent the replication of damaged DNA [184]. In contrast, the increase in the ratio of Ap3A/Ap4A in human HL60 cells can induce apoptosis [164].
So far there are only a few papers describing the role of uncommon nucleotides in plant cells. It was shown that the phenylpropanoid pathway may be affected by exogenous uncommon nucleotides [185,186,187,188]. The phenylpropanoid pathway is a source of secondary metabolites which are very important in plant responses to biotic and abiotic stresses [122]. The first data showed that Ap3A and Ap4A regulate the expression of genes and the activity of enzymes involved in the phenylpropanoid pathway in seven-day old Arabidopsis thaliana seedlings [185]. Ap3A and Ap4A caused an increase in the activity of phenylalanine ammonia-lyase (PAL) and 4:coumarate-CoA ligase (4CL) just 10 min after application. Analysis of PAL gene expression showed that there was strong induction (about 70-fold) of PAL2 gene expression by Ap3A and Ap4A. Additionally, PAL activity was significantly enhanced by Ap3A (about 9-fold). Moreover, it has been shown that the activity of 4CL and the 4CL gene expression level were higher in seedlings treated with Ap3A and Ap4A [185]. These results may indicate a dual role of 4CL in the plant response to stress factors. Firstly, this enzyme synthesizes NpnN’s, i.e., compounds that can play a function as alarmones [121], and secondly, it synthesizes secondary metabolites minimizing the adverse effects of stresses in plant cells [189]. Also, Ap3A caused changes in the gene expression level and the activity of chalcone synthase (CHS) in Arabidopsis thaliana seedlings. This enzyme catalyzes the synthesis of chalcone, which is the precursor of secondary metabolites—flavonoids [185]. These data strongly indicate the signaling role of NpnN’s in plants. The inductive effect of Ap3A on gene expression of the phenylpropanoid pathway proteins has also been confirmed in a cell suspension of Vitis vinifera cv. Monastrell. A synergistic effect evoked by Ap3A and cyclodextrins in trans-resveratrol biosynthesis has been demonstrated [186]. It may suggest the involvement of NpnN’s in the plant response to stress. Interestingly, cyclodextrins act as elicitors in their chemical similarity to the alkyl-derivatized pectic oligosaccharides, which are released from the cell wall during fungal infection [190]. It is known that one of the defense strategies of higher plants against biotic and abiotic stresses is activation of the phenylpropanoid pathway leading to enhanced production of various phenylpropanoid compounds, such as flavonoids [191,192], lignin [193,194], anthocyanins [194,195,196], and salicylic acid [197] (Figure 5). Recently the differences in the regulation of gene expression of the phenylpropanoid pathway enzymes and phenylpropanoid accumulation by purine or pyrimidine NpnN’s in Vitis vinifera suspension cell culture were described. The pyrimidine dinucleotides, such as Cp3C, Cp4C, and Ap4C, markedly (6- to 8-fold) induced the expression of the gene coding the cinnamoyl-CoA:NADP oxidoreductase (CCR) that controls lignin biosynthesis. The most effective in stilbene accumulation was Up4U, but other pyrimidine dinucleotides (Cp3C, Cp4C, and Ap4C) strongly inhibited the biosynthesis of these phenylpropanoids [188].
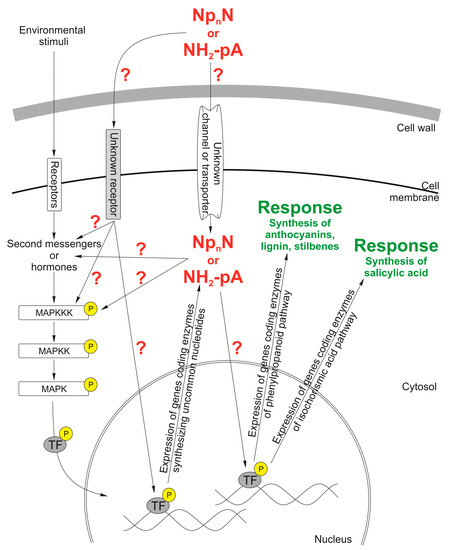
Figure 5.
Hypothetical working model of NpnN’ and NH2-pA signaling network in a plant cell. Dinucleoside polyphosphate (NpnN) and adenosine 5′-phosphoramidate (NH2-pA) trigger induction of the phenylpropanoid and the isochorismic acid pathways yielding metabolites protecting plant against various types of stresses. Plant cells respond to environmental stimuli by intensification of the production of various compounds, such as anthocyanins, lignin, stilbenes, and salicylic acid. Question marks indicate the hypothetical components of the signaling network.
As described above, in plants, the presence of enzymes degrading and synthesizing another uncommon nucleotide, NH2-pA, has been described [127,132]. It was evidenced that NH2-pA regulates the expression of genes coding enzymes of the phenylpropanoid pathway, such as PAL, cinnamate-4-hydroxylase (C4H), 4CL, CHS, CCR, and isochorismate synthase (ICS) in Arabidopsis thaliana seedlings. CCR is an enzyme involved in the biosynthesis of lignin, whereas ICS is involved in the biosynthesis of a signaling molecule—salicylic acid. Among the analyzed genes the strongest induction in gene expression caused by NH2-pA was observed for CCR2 (approx. 4-fold). This was also accompanied by an increased level of lignin in the seedlings. Another important effect caused by NH2-pA was an increase in the ICS gene expression and a significant increase (2-fold) in the level of free salicylic acid. In view of the fact that salicylic acid is one of the signaling molecules involved in the response of plants to biotic stress, the induction of the synthesis of salicylic acid by NH2-pA may suggest the interaction of both these compounds in plant responses to stresses [187].
Another uncommon nucleotide occurring in plants is nicotinic acid adenine dinucleotide phosphate (NAADP). The synthesis of NAADP is not fully understood. However, it is supposed that it can be synthesized by adenosine-5′- diphosphateribosyl-cyclase (EC 3.2.2.5) [198]. This nucleotide is involved in calcium signaling in many organisms including plants [199,200,201]. NAADP-mediated calcium release has been shown in the microsomal vesicles of red beets (Beta vulgaris) and cauliflower (Brassica oleracea). Analysis of sucrose gradient-separated cauliflower microsomes revealed that the NAADP-sensitive Ca2+ pool was derived from the endoplasmic reticulum [199].
4.5. Uncommon Nucleotide Signal Transduction Pathways in Plants
As yet there has been no information about signal transduction pathways or receptors activated by uncommon nucleotides. Based on the data existing in the literature about the involvement of these molecules in different biochemical and physiological processes in bacteria, fungi, animal, and plant organisms, we propose putative signaling pathways induced by uncommon nucleotides in plant cells (Figure 5). It is hypothesized that uncommon nucleotides may be signal molecules synthesized by pathogens outside the cell and interact with plant cells through unknown plasma membrane receptor(s). These nucleotides can also be transported inside the plant cell by unknown plasma membrane transporter(s). In addition, their level in the plant cell can be regulated by synthesizing and degrading enzymes. Both the extracellular and intracellular uncommon nucleotides may affect the biosynthesis of other plant cell signal molecules, e.g., secondary messengers and hormones that can engage MAPK cascades, which are involved in plant growth and development, cellular responses to hormones, regulation of the cell cycle, and responses to biotic and abiotic stresses, such as pathogen infection, wounding, low temperature, drought, high salinity, metals, and ROS. Moreover, it is known that MAPK cascades can regulate gene transcription by activation or repression of transcription factors. In addition, MAPK transcript levels in Arabidopsis thaliana seedlings were shown to increase in a time-dependent manner following exposure to Cu and Cd [202]. Many MAPK cascades respond to hormones such as abscisic acid, jasmonic acid, salicylic acid, ethylene, auxins, and brassinosteroids. Usually, signaling molecules participate in distinct signaling pathways, resulting in the formation of a cross-talking network that co-ordinates responses to different stresses [202,203,204]. We postulate that MAPK cascades can be involved in the signal transduction pathway triggered by uncommon nucleotides. So far, it is only known that extracellular uncommon nucleotides can induce gene expression of the phenylpropanoid pathway, the accumulation of phenylpropanoids, and the synthesis of salicylic acid. However, components of the nucleotide transduction pathway are still not known (Figure 5).
5. Conclusions
Plants are not able to escape from their abiotic (cold, heat, etc.) or their biotic (herbivores, insects) environment [205]. Furthermore, their food uptake and gas exchange take place through external surfaces (leaves: light and CO2, roots: ions and water). Plants must, therefore, possess systems to exchange information throughout the entire plant to ensure the coordination of plant development and defense. Evidence suggests that in plants information exchange relies on at least two different systems: one involving molecules that are transported within the plant and another that uses electrical and/or hydraulic signals to carry the information throughout the entire organism [205]. The systems for transmitting this information are complex and involve multiple components, which are far from being understood. For many years plant hormones were considered to be dominant molecules in plant signaling. Nowadays, this term is applied to many more compounds. It is suggested that dynamic changes in the level of second messengers, such as Ca2+, ROS, and NO, serve as signatures for both intracellular signaling and cell-to-cell communications. These second messenger signatures work in concert with physical signal signatures (such as electrical and hydraulic signals) to create a “lock and key” mechanism that triggers an appropriate response to various stresses [206]. Part of the system, new players in plant signaling, are chemical signals such as extracellular ATP and uncommon nucleotides described and discussed in this work. These molecules are essential for proper coordination of processes in particular organs as well as responses to internal and external signals playing analogical or similar functions in plants and in animals. The data obtained during the last few years suggest that in plants both purine and pyrimidine dinucleotides should be considered as members of a family of naturally occurring stress signaling molecules. So far, our knowledge of these signals in plants is still insufficient to clearly understand their signaling role. The mechanisms of signal perception and transduction are unknown. Further studies are required, which should aim at describing the signaling networks involving uncommon nucleotides, and also perform omic experiments to identify the network components and processes that are regulated by these signals.
Author Contributions
M.P.-B. and E.S.-N. conceived the topic of the article; M.P.-B. and J.D. wrote the majority of the manuscript and designed all figures; S.B. drew all the figures and was involved in the writing of the manuscript; E.S.-N. was involved in the discussion and writing of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
The publication was co-financed within the framework of the Ministry of Science and Higher Education program as “Regional Initiative Excellence” in years 2019-2022, Project No. 005/RID/2018/19.
Acknowledgments
Compliments to Andrzej Guranowski for his invaluable contribution to knowledge about the metabolism of uncommon nucleotides and special thanks to Professor from M.P.-B. for supervising Ph.D. thesis, many years of fruitful collaboration, inspiration in science and common scientific path. We thank Richard Ashcroft (Bioscience editor, www.anglopolonia.com/home.html) for the professional language editing of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| 1,3-ppGly | 1,3-Diphosphoglycerate |
| 3-pGly | 3-Phosphoglycerate |
| 4CL | 4-coumarate:CoA ligase |
| AARS | Aminoacyl t-RNA synthetase |
| ABC transporter | ATP-binding cassette ABC transporter |
| AMP, pA | Adenosine 5′-monophosphate |
| Ap3A, ApppA | Diadenosine 5′,5′′′-P1,P3-triphosphate |
| Ap4A, AppppA | Diadenosine 5′,5′’’-P1,P4-tetraphosphate |
| Ap4C | P1-(5’-Adenosyl)-P4-(5’-cytidyl)-tetraphosphate |
| ApU | Adenosine-uridine monophosphate |
| APY1 and APY2 | Apyrase 1 and 2 |
| C4H | Cinnamate-4-hydroxylase |
| cAMP | Adenosine 3′,5′-cyclic monophosphate |
| CAMTA3 | Calmodulin-binding transcription activator |
| CCR | Cinnamoyl-CoA:NADP oxidoreductase |
| cGMP | Guanosine 3′,5′-cyclic monophosphate |
| CHS | Chalcone synthase |
| Cp3C | Dicytosine 5′,5′′′-P1,P3-triphosphate |
| Cp4C | Dicytosine 5′,5′′′-P1,P4-tetraphosphate |
| DORN1 | Does not Respond to Nucleotide |
| E | Enzyme |
| eATP | Extracellular ATP |
| eNAD+ | Extracellular NAD+ |
| eNADP+ | Extracellular NADP+ |
| Fhit | Fragile histidine triad |
| F-pA | Adenosine 5′-O-phosphorofluoridate |
| Gp3G | Diguanosine 5′,5′′′-P1,P3-triphosphate |
| Gp4G | Diguanosine 5′,5′′′-P1,P4-tetraphosphate |
| HIT | Histidine triad |
| ICS | Isochorismate synthase |
| ISR | Induced systemic resistance |
| JAR1 | Jasmonate:amino acid synthetase |
| LecRK-1.9 | L-type lectin receptor-like kinase 1.9 |
| LysRS | Lysyl-tRNA synthetase |
| MAPK | Mitogen-activated protein kinases |
| MITF | Microphthalmia-associated transcription factor |
| MYC | Transcription factors |
| NAADP | Nicotinic acid adenine dinucleotide phosphate |
| NH2-pA | Adenosine 5′-phosphoramidate |
| NO | Nitric oxide |
| Non-AARS | Non-aminoacyl t-RNA synthetase |
| NpnN’ | Dinucleoside polyphosphate |
| NTP, pppN | Nucleoside 5′-triphosphate |
| NUDT | Nudix hydrolase |
| Nudix | Nucleoside diphosphate linked to x (x: any moiety) |
| P2K1 | P2 receptor kinase 1 |
| P2X | Ligand-gated ion channel |
| P2Y | G protein-coupled receptor |
| P3, ppp | Triphosphate |
| P4, pppp | Tetraphosphate |
| p4A, ppppA | Adenosine 5′-tetraphosphate |
| p4C | Cytosine 5′-tetraphosphate |
| p4G | Guanosine 5′-tetraphosphate |
| p4U | Uridine 5′-tetraphosphate |
| p5A, pppppA | Adenosine 5′-pentaphosphate |
| PAL | Phenylalanine ammonia-lyase |
| PGP1 | p-Glycoprotein 1 |
| PM-ANT1 | Plasma membrane-localized nucleotide transporters |
| pnN | Nucleoside 5′-polyphosphate |
| ppGpp | Guanosine 3′-diphosphate 5′-diphosphate |
| PPi | Pyrophosphate |
| (p)ppGpp | Guanosine 3′-diphosphate 5′-triphosphate |
| RBOHD | Respiratory burst oxidase homolog protein D |
| RNS | Reactive nitrogen species |
| ROS | Reactive oxygen species |
| SAR | Systemic acquired resistance |
| STS | Stilbene synthase |
| TF | Transcription factor |
| Up2U | Diuridine 5′,5′′′-P1,P2-diphosphate |
| UTP, pppU | Uridine 5′-triphosphate |
References
- Edel, K.H.; Marchadier, E.; Brownlee, C.; Kudla, J.; Hetherington, A.M. The evolution of calcium-based signalling in plants. Curr. Biol. 2017, 27, 667–679. [Google Scholar] [CrossRef]
- Del Rio, L.A. ROS and RNS in plant physiology: An overview. J. Exp. Bot. 2015, 66, 2827–2837. [Google Scholar] [CrossRef]
- Pietrowska-Borek, M.; Chadzinikolau, T.; Borek, S. Cyclic nucleotides and nucleotide cyclases in plants under stress. In Improvement of Crops in the Era of Climatic Changes; Ahmad, P., Wani, M.R., Azooz, M.M., Tran, L.S.P., Eds.; Springer Science and Business Media: New York, NY, USA, 2014; pp. 119–151. [Google Scholar]
- Isner, J.C.; Maathuis, F.J.M. cGMP signalling in plants: From enigma to main stream. Funct. Plant Biol. 2016, 45, 93–101. [Google Scholar] [CrossRef]
- Świeżawska, B.; Duszyn, M.; Jaworski, K.; Szmidt-Jaworska, A. Downstream targets of cyclic nucleotides in plants. Front. Plant Sci. 2018, 9, 1428. [Google Scholar] [CrossRef]
- Gehring, C.; Turek, I.S. Cyclic nucleotide monophosphates and their cyclases in plant signaling. Front. Plant Sci. 2017, 8, 1704. [Google Scholar] [CrossRef]
- Tripathi, D.; Tanaka, K. A crosstalk between extracellular ATP and jasmonate signaling pathways for plant defense. Plant Signal. Behav. 2018, 13, e1432229. [Google Scholar] [CrossRef] [PubMed]
- Burnstock, G. Purinergic nerves. Pharmacol. Rev. 1972, 24, 509–581. [Google Scholar]
- Drury, A.N.; Szent-Gyorgy, A. The physiological activity of adenine compounds with special reference to their action upon the mammalian heart. J. Physiol. 1929, 68, 213–237. [Google Scholar] [CrossRef] [PubMed]
- Cheung, K.K.; Chan, W.Y.; Burnstock, G. Expression of P2X purinoceptors during rat brain development and their inhibitory role on motor axon outgrowth in neural tube explant cultures. Neuroscience 2005, 133, 937–945. [Google Scholar] [CrossRef]
- Choi, J.; Tanaka, K.; Cao, Y.; Qi, Y.; Qiu, J.; Liang, Y.; Lee, S.Y.; Stacey, G. Identification of a plant receptor for extracellular ATP. Science 2014, 343, 290–294. [Google Scholar] [CrossRef]
- Kim, S.Y.; Sivaguru, M.; Stacey, G. Extracellular ATP in plants. Visualization, localization, and analysis of physiological significance in growth and signaling. Plant Physiol. 2006, 142, 984–992. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.J.; Liu, Y.S.; Wu, J.Y. The signaling role of extracellular ATP and its dependence on Ca2+ flux in elicitation of Salvia miltiorrhiza hairy root cultures. Plant Cell Physiol. 2008, 49, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Weerasinghe, R.R.; Swanson, S.J.; Okada, S.F.; Garrett, M.B.; Kim, S.Y.; Stacey, G.; Boucher, R.C.; Gilroy, S.; Jones, A.M. Touch induces ATP release in Arabidopsis roots that is modulated by the heterotrimeric G-protein complex. FEBS Lett. 2009, 583, 2521–2526. [Google Scholar] [CrossRef] [PubMed]
- Dark, A.; Demidchik, V.; Richards, S.L.; Shabala, S.; Davies, J.M. Release of extracellular purines from plant roots and effect on ion fluxes. Plant Signal. Behav. 2011, 6, 1855–1857. [Google Scholar] [CrossRef] [PubMed]
- Song, C.J.; Steinebrunner, I.; Wang, X.; Stout, S.C.; Roux, S.J. Extracellular ATP induces the accumulation of superoxide via NADPH oxidases in Arabidopsis. Plant Physiol. 2006, 140, 1222–1232. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.; Rajagopal, A.; Windsor, B.; Dudler, R.; Lloyd, A.; Roux, S.J. A role for ectophosphatase in xenobiotic resistance. Plant Cell 2000, 12, 519–533. [Google Scholar] [CrossRef] [PubMed]
- Rieder, B.; Neuhaus, H.E. Identification of an Arabidopsis plasma membrane-located ATP transporter important for anther development. Plant Cell 2011, 23, 1932–1944. [Google Scholar] [CrossRef]
- Lim, M.H.; Wu, J.; Clark, G.; Roux, S.J. Apyrase suppression raises extracellular ATP levels and induces gene expression and cell wall changes characteristic of stress responses. Plant Physiol. 2014, 164, 2054–2067. [Google Scholar] [CrossRef]
- Clark, G.; Roux, S.J. Apyrases, extracellular ATP and the regulation of growth. Curr. Opin. Plant Biol. 2011, 14, 700–706. [Google Scholar] [CrossRef]
- Parsons, H.T.; Christiansen, K.; Knierim, B.; Carroll, A.; Ito, J.; Batth, T.S.; Smith-Moritz, A.M.; Morrison, S.; McInerney, P.; Hadi, M.Z.; et al. Isolation and proteomic characterization of the Arabidopsis Golgi defines functional and novel components involved in plant cell wall biosynthesis. Plant Physiol. 2012, 159, 12–26. [Google Scholar] [CrossRef]
- Chiu, T.Y.; Christiansen, K.; Moreno, I.; Lao, J.; Loqué, D.; Orellana, A.; Heazlewood, J.L.; Clark, G.; Roux, S.J. AtAPY1 and AtAPY2 function as Golgi-localized nucleoside diphosphatases in Arabidopsis thaliana. Plant Cell Physiol. 2012, 53, 1913–1925. [Google Scholar] [CrossRef] [PubMed]
- Schiller, M.; Massalski, C.; Kurth, T.; Steinebrunner, I. The Arabidopsis apyrase AtAPY1 is localized in the Golgi instead of the extracellular space. BMC Plant Biol. 2012, 12, 123. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Steinebrunner, I.; Sun, Y.; Butterfield, T.; Torres, J.; Arnold, D.; Gonzalez, A.; Jacob, F.; Reichler, S.; Roux, S.J. Apyrases (nucleoside triphosphate-diphosphohydrolases) play a key role in growth control in Arabidopsis. Plant Physiol. 2007, 144, 961–975. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wu, J.; Clark, G.; Lundy, S.; Lim, M.; Arnold, D.; Chan, J.; Tang, W.Q.; Muday, G.K.; Gardner, G.; et al. Role for apyrases in polar auxin transport in Arabidopsis. Plant Physiol. 2012, 160, 1985–1995. [Google Scholar] [CrossRef] [PubMed]
- Day, R.B.; McAlvin, C.B.; Loh, J.T.; Denny, R.L.; Wood, T.C.; Young, N.D.; Stacey, G. Differential expression of two soybean apyrases, one of which is an early nodulin. Mol. Plant Microbe Interact. 2000, 13, 1053–1070. [Google Scholar] [CrossRef] [PubMed]
- Kiba, A.; Toyoda, K.; Yoshioka, K.; Tsujimura, K.; Takahashi, H.; Ichinose, Y.; Takeda, T.; Kato, T.; Shiraishi, T. A pea NTPase, PsAPY1, recognizes signal molecules from microorganisms. J. Gen. Plant Pathol. 2006, 72, 238–246. [Google Scholar] [CrossRef]
- Riewe, D.; Grosman, L.; Fernie, A.R.; Wucke, C.; Geigenberger, P. The potato-specific apyrase is apoplastically localized and has influence on gene expression, growth, and development. Plant Physiol. 2008, 147, 1092–1109. [Google Scholar] [CrossRef]
- Chiu, T.Y.; Lao, J.; Manalansan, B.; Loque, D.; Roux, S.J.; Heazlewood, J.L. Biochemical characterization of Arabidopsis APYRASE family reveals their roles in regulating endomembrane NDP/NMP homoeostasis. Biochem. J. 2015, 472, 43–54. [Google Scholar] [CrossRef]
- Burnstock, G. Purinergic receptors. J. Theor. Biol. 1976, 62, 491–503. [Google Scholar] [CrossRef]
- Burnstock, G. Purine and pyrimidine receptors. Cell Mol. Life Sci. 2007, 64, 1471–1483. [Google Scholar] [CrossRef]
- Burnstock, G. A basis for distinguishing two types of purinergic receptor. In Cell Membrane Receptors for Drugs and Hormones: A Multidisciplinary Approach; Bolis, L., Straub, R.C.O., Eds.; Raven Press: New York, NY, USA, 1978; pp. 107–118. [Google Scholar]
- Möhlmann, T.; Steinebrunner, I.; Neuhaus, E. Nucleotides and nucleosides: Transport, metabolism, and signaling function of extracellular ATP. In Progress in Botany; Lüttge, U., Beyschlag, W., Cushman, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; Volume 75, pp. 119–144. [Google Scholar]
- Tanaka, K.; Gilroy, S.; Jones, A.M.; Stacey, G. Extracellular ATP signaling in plants. Trends Cell Biol. 2010, 20, 601–608. [Google Scholar] [CrossRef] [PubMed]
- Jewell, J.B.; Tanaka, K. Transcriptomic perspective on extracellular ATP signaling: A few curious trifles. Plant Signal. Behav. 2019, 14, 1659079. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Shang, Z. Extracellular ATP: An essential apoplastic messenger in plants. In Progress in Botany; Cánovas, F.M., Lüttge, U., Matyssek, R., Eds.; Springer: Cham, Switzerland, 2016; pp. 121–144. [Google Scholar]
- Cho, S.H.; Nguyen, C.T.; Choi, J.; Stacey, G. Molecular mechanism of plant recognition of extracellular ATP. Adv. Exp. Med. Biol. 2017, 1051, 233–253. [Google Scholar]
- Jewell, J.B.; Sowders, J.M.; He, R.; Willis, M.A.; Gang, D.R.; Tanaka, K. Extracellular ATP shapes a defense-related transcriptome both independently and along with other defense signaling pathways. Plant Physiol. 2019, 179, 1144–1158. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.J.; Dong, X.X.; Hao, W.W.; Gao, W.; Zhang, W.Z.; Xia, S.Y.; Liu, T.; Shang, Z.L. Heterotrimeric G protein-regulated Ca2+ influx and PIN2 asymmetric distribution are involved in Arabidopsis thaliana roots’ avoidance response to extracellular ATP. Front. Plant Sci. 2017, 8, 1522. [Google Scholar] [CrossRef] [PubMed]
- Matthus, E.; Sun, J.; Wang, L.; Bhat, M.G.; Mohammad-Sidik, A.B.; Wilkins, K.A.; Leblanc-Fournier, N.; Legué, V.; Moulia, B.; Stacey, G.; et al. DORN1/P2K1 and purino-calcium signalling in plants: Making waves with extracellular ATP. Ann. Bot. 2020, 124, 1227–1242. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Choi, J.; Cao, Y.; Stacey, G. Extracellular ATP acts as a damage-associated molecular pattern (DAMP) signalin plants. Front. Plant Sci. 2014, 5, 446. [Google Scholar] [CrossRef]
- Demidchik, V.; Nichols, C.; Oliynyk, M.; Dark, A.; Glover, B.J.; Davies, J.M. Is ATP a signaling agent in plants? Plant Physiol. 2003, 133, 456–461. [Google Scholar] [CrossRef]
- Wu, S.; Wu, J. Extracellular ATP-induced NO production and its dependence on membrane Ca2+ flux in Salvia miltiorrhiza hairy roots. J. Exp. Bot. 2008, 59, 4007–4016. [Google Scholar] [CrossRef]
- Rodrigues, O.; Reshetnyak, G.; Grondin, A.; Saijo, Y.; Leonhardt, N.; Maurel, C.; Verdoucq, L. Aquaporins facilitate hydrogen peroxide entry into guard cells to mediate ABA- and pathogen-triggered stomatal closure. Proc. Natl. Acad. Sci. USA 2017, 114, 9200–9205. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhang, C.L.; Deng, S.R.; Lu, C.F.; Shen, X.; Zhou, X.Y.; Zheng, X.J.; Hu, Z.M.; Chen, S.L. An ATP signalling pathway in plant cells: Extracellular ATP triggers programmed cell death in Populus euphratica. Plant Cell Environ. 2012, 35, 893–916. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.X.; Ahammed, G.J.; Wu, C.; Fan, S.Y.; Zhou, Y.H. Crosstalk among jasmonate, salicylate and ethylene signaling pathways in plant disease and immune responses. Curr. Protein Pept. Sci. 2015, 16, 450–461. [Google Scholar] [CrossRef] [PubMed]
- Bouwmeester, K.; de Sain, M.; Weide, R.; Gouget, A.; Klamer, S.; Canut, H.; Govers, F. The lectin receptor kinase LecRK-I.9 is a novel Phytophthora resistance component and a potential host target for a RXLR effector. PLoS Pathog. 2011, 7, e1001327. [Google Scholar] [CrossRef] [PubMed]
- Balague, C.; Gouget, A.; Bouchez, O.; Souriac, C.; Haget, N.; Boutet-Mercey, S.; Govers, F.; Roby, D.; Canut, H. The Arabidopsis thaliana lectin receptor kinase LecRK-I.9 is required for full resistance to Pseudomonas syringae and affects jasmonate signalling. Mol. Plant Pathol. 2017, 18, 937–948. [Google Scholar] [CrossRef] [PubMed]
- Vahabi, K.; Sherameti, I.; Bakshi, M.; Mrozinska, A.; Ludwig, A.; Reichelt, M.; Oelmüller, R. The interaction of Arabidopsis with Piriformospora indica shifts from initial transient stress induced by fungus-released chemical mediators to a mutualistic interaction after physical contact of the two symbionts. BMC Plant Biol. 2015, 15, 58. [Google Scholar] [CrossRef]
- Nizam, S.; Qiang, X.; Wawra, S.; Nostadt, R.; Getzke, F.; Schwanke, F.; Dreyer, I.; Langen, G.; Zuccaro, A. Serendipita indica E5′NT modulates extracellular nucleotide levels in the plant apoplast and affects fungal colonisation. EMBO Rep. 2019, 20, e47430. [Google Scholar] [CrossRef]
- Tanaka, K.; Tóth, K.; Stacey, G. Role of ectoapyrases in nodulation. In Biological Nitrogen Fixation, 1st ed.; de Bruijn Frans, J., Ed.; John Wiley & Sons, Inc.: New York, NY, USA, 2015; pp. 517–524. [Google Scholar]
- Clark, G.; Fraley, D.; Steinebrunner, I.; Cervantes, A.; Onyirimba, J.; Liu, A.; Torres, J.; Tang, W.; Kim, J.; Roux, S.J. Extracellular nucleotides and apyrases regulate stomatal aperture in Arabidopsis. Plant Physiol. 2011, 156, 1740–1753. [Google Scholar] [CrossRef]
- Jeter, C.R.; Tang, W.; Henaff, E.; Butterfield, T.; Roux, S.J. Evidence of a novel cell signaling role for extracellular adenosine triphosphates and diphosphates in Arabidopsis. Plant Cell 2004, 16, 2652–2664. [Google Scholar] [CrossRef]
- Hou, Q.Z.; Ye, G.J.; Wang, R.F.; Jia, L.Y.; Liang, J.Y.; Feng, H.Q.; Wen, J.; Shi, D.L.; Wang, Q.W. Changes by cadmium stress in lipid peroxidation and activities of lipoxygenase and antioxidant enzymes in Arabidopsis are associated with extracellular ATP. Biologia 2017, 72, 1467–1474. [Google Scholar] [CrossRef]
- Chen, D.; Cao, Y.; Li, H.; Kim, D.; Ahsan, N.; Thelen, J.; Stacey, G. Extracellular ATP elicits DORN1-mediated RBOHD phosphorylation to regulate stomatal aperture. Nat. Commun. 2017, 8, 2265. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Yang, S.H.; Kim, T.J.; Han, J.S.; Suh, J.W. Hypertonic stress increased extracellular ATP levels and the expression of stress-responsive genes in Arabidopsis thaliana seedlings. Biosci. Biotechnol. Biochem. 2009, 73, 1252–1256. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Strnad, M. Jasmonates: News on occurrence, biosynthesis, metabolism and action of an ancient group of signaling compounds. Int. J. Mol. Sci. 2018, 19, 2539. [Google Scholar] [CrossRef] [PubMed]
- Rizwan, M.; Ali, S.; Adrees, M.; Ibrahim, M.; Tsang, D.; Zia-ur-Rehman, M.; Zahir, Z.; Rinklebe, J.; Tack, F.; Ok, Y. A critical review on effects, tolerance mechanisms and management of cadmium in vegetables. Chemosphere 2017, 182, 90–105. [Google Scholar] [CrossRef]
- Fargašová, A.; Pastierová, J.; Svetková, K. Effect of Se-metal pair combinations (Cd, Zn, Cu, Pb) on photosynthetic pigments production and metal accumulation in Sinapis alba L. seedlings. Plant Soil Environ. 2006, 52, 8–15. [Google Scholar] [CrossRef]
- Sarry, J.E.; Kuhn, L.; Ducruix, C.; Lafaye, A. The early responses of Arabidopsis thaliana cells to cadmium exposure explored by protein and metabolite profiling analyses. Proteomics 2006, 6, 2180–2198. [Google Scholar] [CrossRef]
- Herbette, S.; Taconnat, L.; Hugouvieux, V.; Piette, L.; Magniette, M.L.M.; Cuine, S. Genome-wide transcriptome profiling of the early cadmium response of Arabidopsis roots and shoots. Biochimie 2006, 88, 1751–1765. [Google Scholar] [CrossRef]
- Sanità di Toppi, L.; Gabbrielli, R. Response to cadmium in higher plants. Environ. Exp. Bot. 1999, 41, 105–130. [Google Scholar] [CrossRef]
- Berger, F.; Ramirez-Hernandez, M.H.; Ziegler, M. The new life of a centenarian: Signalling functions of NAD(P). Trends Biochem. Sci. 2004, 29, 111–118. [Google Scholar] [CrossRef]
- Noctor, G. Metabolic signalling in defence and stress: The central roles of soluble redox couples. Plant Cell Environ. 2006, 29, 409–425. [Google Scholar] [CrossRef]
- Billington, R.A.; Bruzzone, S.; De Flora, A.; Genazzani, A.A.; Koch-Nolte, F.; Ziegler, M.; Zocchi, E. Emerging functions of extracellular pyridine nucleotides. Mol. Med. 2006, 12, 324–327. [Google Scholar] [CrossRef]
- Zhang, X.; Mou, Z. Extracellular pyridine nucleotides induce PR gene expression and disease resistance in Arabidopsis. Plant J. 2009, 57, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Mou, Z. Expression of the human NAD(P)-metabolizing ectoenzyme CD38 compromises systemic acquired resistance in Arabidopsis. Mol Plant Microbe Interact. 2012, 25, 1209–1218. [Google Scholar] [CrossRef]
- Wang, C.; Zhou, M.; Zhang, X.; Yao, J.; Zhang, Y.; Mou, Z. A lectin receptor kinase as a potential sensor for extracellular nicotinamide adenine dinucleotide in Arabidopsis thaliana. eLife 2017, 6, e25474. [Google Scholar] [CrossRef]
- Wang, C.; Huang, X.; Li, Q.; Zhang, Y.; Li, J.L.; Mou, Z. Extracellular pyridine nucleotides trigger plant systemic immunity through a lectin receptor kinase/BAK1 complex. Nat. Commun. 2019, 10, 4810. [Google Scholar] [CrossRef]
- Marrian, D.H. A new adenine nucleotide. Biochim. Biophys. Acta 1953, 12, 492. [Google Scholar] [CrossRef]
- Marrian, D.H. A new adenine nucleotide. Biochim. Biophys. Acta 1954, 13, 278–281. [Google Scholar] [CrossRef]
- Lieberman, I. Identification of adenosine tetraphosphate from horse muscle. J. Am. Chem. Soc. 1955, 77, 3373–3375. [Google Scholar] [CrossRef]
- Sacks, J. Adenosine pentaphosphate from commercial ATP. Biochim. Biophys. Acta 1955, 16, 436. [Google Scholar] [CrossRef]
- Gardner, J.A.A.; Hoagland, M.B. The isolation of guanosine tetraphosphate from commercially available preparations of guanosine triphosphate. J. Biol. Chem. 1965, 240, 1244–1246. [Google Scholar]
- Moreno, A.; Vallejo, C.G.; Sillero, A.; Sillero, M.A.G. Wide occurrence of guanosine 5′-tetraphosphate in commercial preparations of GTP. Anal. Biochem. 1975, 68, 648–650. [Google Scholar] [CrossRef]
- Moreno, A.; Lobatón, C.D.; Günther Sillero, M.A.; Sillero, A. Dinucleosidetetraphosphatase from Ehrlich ascites tumourcells: Inhibition by adenosine, guanosine and uridine 5′-tetraphosphates. Int. J. Biochem. 1982, 14, 629–634. [Google Scholar] [CrossRef]
- Costas, M.J.; Cameselle, J.C.; Günther Sillero, M.A.; Sillero, A. Presence of cytidine 5′-tetraphosphate in commercial samples of cytidine 5′-triphosphate. Anal. Biochem. 1983, 134, 455–458. [Google Scholar] [CrossRef]
- Heldt, H.W.; Klingenberg, M. Endogenous nucleotides of mitochondria participating in phosphate transfer reactions as studied with 32P labelled orthophosphate and ultramicroscale ion exchange chromatography. Biochem. Z. 1965, 343, 433–451. [Google Scholar] [PubMed]
- Zamecnik, P.C.; Stephenson, M.L. A possible regulatory site located at the gateway to protein synthesis. In Regulatory Mechanisms for Protein Synthesis in Mammalian Cells; Lamborg, M.R., Kenney, P.T., Eds.; Academic Press Inc.: San Pietro, NY, USA, 1968; pp. 3–16. [Google Scholar]
- Small, G.; Cooper, C. Studies on the occurrence and biosynthesis of adenosine tetraphosphate. Biochemistry 1966, 5, 26–33. [Google Scholar] [CrossRef]
- Van Dyke, K.; Robinson, R.; Urquilla, P.; Smith, D.; Taylor, M.; Trush, M.; Wilson, M. An analysis of nucleotides and catecholamines in bovine medullary granules by anion exchange high pressure liquid chromatography and fluorescence. Evidence that most of catecholamines in chromaffin granules are stored without associated ATP. Pharmacology 1977, 15, 377–391. [Google Scholar] [CrossRef]
- Sillero, M.A.; Del Valle, M.; Zaera, E.; Michelena, P.; García, A.G.; Sillero, A. Diadenosine 5′,5′′′-P1,P4-tetraphosphate (Ap4A), ATP and catecholamine content in bovine adrenal medulla, chromaffin granules and chromaffin cells. Biochimie 1994, 76, 404–409. [Google Scholar] [CrossRef]
- Gualix, J.; Abal, M.; Pintor, J.; Miras-Portugal, M.T. Presence of ε-adenosine tetraphosphate in chromaffin granules after transport of ε-ATP. FEBS Lett. 1996, 391, 195–198. [Google Scholar] [CrossRef]
- Lee, J.W.; Jeon, S.J.; Kong, I.D.; Jeong, S.W. Identification of adenosine 5′-tetraphosphate in rabbit platelets and its metabolism in blood. Korean J. Physiol. 1995, 29, 217–223. [Google Scholar]
- Jakubowski, H. Sporulation of the yeast Saccharomyces cerevisiae is accompanied by synthesis of adenosine 5’-tetraphosphate and adenosine 5’-pentaphosphate. Proc. Natl. Acad. Sci. USA 1986, 83, 2378–2382. [Google Scholar] [CrossRef]
- Fraga, H.; Fontes, R. Enzymatic synthesis of mono and dinucleoside polyphosphates. Biochim. Biophys. Acta 2011, 1810, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Zamecnik, P.C.; Stephenson, M.L. Nucleoside pyrophosphate compounds related to the first step in protein synthesis. In Alfred Benzon Symposium I: The Role of Nucleotides for the Function and Conformation of Enzymes; Kalckar, H.M., Klenow, H., Munch-Peterson, G., Ottesen, M., Thuysen, J.M., Eds.; Munksguard: Copenhagen, Denmark, 1969; pp. 276–291. [Google Scholar]
- Theoclitou, M.E.; El-Thaher, T.S.H.; Miller, A.D. Enzymatic synthesis of diadenosine 5′,5‴-P1,P4-tetraphosphate (Ap4A) analogues by stress protein LysU. J. Chem. Soc. Chem. Commun. 1994, 5, 659–661. [Google Scholar] [CrossRef]
- Zamecnik, P.C.; Stephenson, M.L.; Janeway, C.M.; Randerath, K. Enzymatic synthesis of diadenosine tetraphosphate and diadenosine triphosphate with a purified lysyl-sRNA synthetase. Biochem. Biophys. Res. Commun. 1966, 24, 91–97. [Google Scholar] [CrossRef]
- Guranowski, A. Studies on dinucleoside polyphosphates: Some intriguing biochemical, physiological, and medical aspects. J. Clin. Biochem. 2000, 28, 177–189. [Google Scholar] [CrossRef]
- McLennan, A.G.; Zamecnik, P.C. Dinucleoside polyphosphates—An introduction. In Ap4A and Other Dinucleoside Polyphosphates; McLennan, A.G., Ed.; CRC Press: Boca Raton, FL, USA, 1992; pp. 1–7. [Google Scholar]
- Goerlich, O.; Foeckler, R.; Holler, E. Mechanism of synthesis of adenosine(5′)tetraphospho(5′)adenosine (AppppA) by aminoacyl-tRNA synthetases. Eur. J. Biochem. 1982, 126, 135–142. [Google Scholar] [CrossRef]
- Blanquet, S.; Plateau, P.; Brevet, A. The role of zinc in 5′,5′-diadenosine tetraphosphate production by aminoacyl-transfer RNA synthetases. Mol. Cell. Biochem. 1983, 52, 3–11. [Google Scholar] [CrossRef]
- Biryukov, A.I.; Zhukov, Y.N.; Lavrik, O.I.; Khomutov, R.M. Influence of the aminoacyl-tRNA synthetase inhibitors and the diadenosine-5′-tetraphosphate phosphonat analogues on the catalysis of diadenosyl oligophosphates formation. FEBS Lett. 1990, 273, 208–210. [Google Scholar] [CrossRef]
- Charlier, J.; Sanchez, R. Lysyl-tRNA synthetase from Escherichia coli K12. Chromatographic heterogeneity and the lysU-gene product. Biochem. J. 1987, 248, 43–51. [Google Scholar] [CrossRef]
- Lazewska, D.; Guranowski, A. P alpha-chiral phosphorothioate analogues of bis (5′-adenosyl)tetraphosphate (Ap4A); their enzymatic synthesis and degradation. Nucleic Acid Res. 1990, 18, 6083–6088. [Google Scholar] [CrossRef]
- Brevet, A.; Chen, J.; Lévêque, F.; Blanquet, S.; Plateau, P. Comparison of the enzymatic properties of the two Escherichia coli Lysyl-tRNA synthetase species. J. Biol. Chem. 1995, 270, 14439–14444. [Google Scholar] [CrossRef]
- Wright, M.; Boonyalai, N.; Tanner, J.A.; Hindley, A.D.; Miller, A.D. The duality of LysU, a catalyst for both Ap4A and Ap3A formation. FEBS J. 2006, 273, 3534–3544. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, B.; Sillero, A.; Sillero, M.A.G. Specific synthesis of adenosine(5′)tetraphospho(5′)nucleoside and adenosine(5′)oligophospho(5′)adenosine (n greater than 4) catalyzed by firefly luciferase. Eur. J. Biochem. 1993, 215, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Guranowski, A.; de Diego, A.; Sillero, A.; Sillero, M.A.G. Uridine 5 ′-polyphosphates (p4U and p5U) and uridine(5′)polyphospho(5′)nucleosides (UpnNs) can be synthesized by UTP:glucose-1-phosphate uridylyltransferase from Saccharomyces cerevisiae. FEBS Lett. 2004, 561, 83–88. [Google Scholar] [CrossRef]
- García-Diaz, M.; Canales, J.; Günther Sillero, M.A.; Sillero, A.; Cameselle, J.C. Phosphoghlycerate kinase from yeast synthesizes guanosine 5′-tetraphosphate. Biochem. Int. 1989, 19, 1253–1264. [Google Scholar]
- Kupriyanov, V.V.; Ferretti, J.A.; Balaban, R.S. Muscle adenylate kinase catalyzes adenosine 5′-tetraphosphate synthesis from ATP and ADP. Biochim. Biophys. Acta 1986, 869, 107–111. [Google Scholar] [CrossRef]
- Luo, G.; Nishimura, J.S. Adenosine 5′-tetraphosphate is synthesized by the histidine a142—asparagine mutant of Escherichia coli succinyl-CoA synthetase. J. Biol. Chem. 1992, 267, 9516–9520. [Google Scholar]
- Guranowski, A.; Günther Sillero, M.A.; Sillero, A. Adenosine 5′-tetraphosphate and adenosine 5′-pentaphosphate are synthesized by yeast acetyl coenzyme A synthetase. J. Bacteriol. 1994, 176, 2986–2990. [Google Scholar] [CrossRef]
- Sillero, M.A.; de Diego, A.; Pérez-Zúñiga, F.; Sillero, A. Synthesis of biphosphonate derivatives of ATP by T4 DNA ligase, ubiquitin activating enzyme (E1) and other ligases. Biochem. Pharmacol. 2008, 75, 1959–1965. [Google Scholar] [CrossRef]
- Fontes, R.; Sillero, M.A.; Sillero, A. Acyl coenzyme A synthetase from Pseudomonas fragi catalyzes the synthesis of adenosine 5′-polyphosphates and dinucleoside polyphosphates. J. Bacteriol. 1998, 180, 3152–3158. [Google Scholar] [CrossRef]
- Fontes, R.; Günther Sillero, M.A.; Sillero, A. Acyl-CoA synthetase catalyzes the synthesis of diadenosine hexaphosphate (Ap6A). Biochimie 1999, 81, 229–233. [Google Scholar] [CrossRef]
- Madrid, O.; Martin, D.; Atencia, E.A.; Sillero, A.; Sillero, M.A.G. T4 DNA ligase synthesizes dinucleoside polyphosphates. FEBS Lett. 1998, 433, 283–286. [Google Scholar] [CrossRef]
- Günther Sillero, M.A.; Montes, M.; de Diego, A.; del Valle, M.; Atencia, E.; Sillero, A. Thermostable Pyrococcus furiosus DNA ligase catalyzes the synthesis of (di)nucleoside polyphosphates. Extremophiles 2002, 6, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Sillero, M.A.; de Diego, A.; Tavares, J.E.F.; Silva, C.; Pérés-Zúñiga, J.; Sillero, A. Synthesis of ATP derivatives of compounds of the mevalonate pathway (isopentenyl di- and triphosphate; geranyl di- and triphosphate, farnesyl di- and triphosphate, and dimethylallyl diphosphate) catalyzed by T4 RNA ligase, T4 DNA ligase and other ligases potential relationship with the effect of bisphosphonates on osteoclasts. Biochem. Pharmacol. 2009, 78, 335–343. [Google Scholar] [PubMed]
- Atencia, E.A.; Madrid, O.; Sillero, M.A.G.; Sillero, A. T4 RNA ligase catalyzes thesynthesis of dinucleoside polyphosphates. Eur. J. Biochem. 1999, 261, 802–811. [Google Scholar] [CrossRef] [PubMed]
- Bouhss, A.; Dementin, S.; van Heijenoort, J.; Parquet, C.; Blanot, D. Formation of adenosine 5′-tetraphosphate from the acyl phosphate intermediate: A difference between the MurC and MurD synthetases of Escherichia coli. FEBS Lett. 1999, 453, 15–19. [Google Scholar] [CrossRef]
- Guranowski, A.; Starzyńska, E.; Rataj-Guranowska, M. Purification of apyrase from yellow lupin cotyledons after extraction with perchloric acid. Protein. Expr. Purif. 1991, 4, 235–239. [Google Scholar] [CrossRef]
- Guranowski, A.; Starzyńska, E.; Sillero, M.A.; Sillero, A. Conversion of adenosine(5′) oligophospho(5′) adenosines into inosine(5′)oligophospho(5′)inosines by non-specific adenylate deaminase from the snail Helix pomatia. Biochim. Biophys. Acta 1995, 1243, 78–84. [Google Scholar] [CrossRef]
- Mateo, J.; Miras-Portugal, M.T.; Rotllán, P. Ecto-enzymatic hydrolysis of diadenosine polyphosphates by cultured adrenomedullary vascular endothelial cells. Am. J. Physiol. 1997, 273, 6918–6927. [Google Scholar] [CrossRef]
- Mateo, J.; Rotllán, P.; Marti, E.; Gomez de Aranda, I.; Solsona, C.; Miras-Portugal, M.T. Diadenosine polyphosphate hydrolase from presynaptic plasma membranes of Torpedo electric organ. Biochem. J. 1997, 323, 677–684. [Google Scholar] [CrossRef]
- Plateau, P.; Fromant, M.; Brevet, A.; Gesquière, A.; Blanquet, S. Catabolism of bis(5′-nucleosidyl) oligophosphates in Escherichia coli; metal requirements and substrate specificity of homogeneous diadenosine-5′,5′′′-tetraphosphate (symmetrical) pyrophosphohydrolase. Biochemistry 1985, 24, 914–922. [Google Scholar] [CrossRef]
- Guranowski, A.; Blanquet, S. Phosphorolytic cleavage of diadenosine 5’,5’’’-P1,P4-tetraphosphate; properties of homogeneous diadenosine 5’,5’’’-P1,P4-tetraphosphate α,β-phosphorylase from Saccharomyces cerevisiae. J. Biol. Chem. 1985, 260, 3542–3547. [Google Scholar] [PubMed]
- Guranowski, A.; Just, G.; Holler, E.; Jakubowski, H. Synthesis of diadenosine 5’, 5’’’-P1,P4-tetraphosphate (AppppA) from adenosine 5’- phosphosulfate and adenosine 5’-triphosphate catalyzed by yeast AppppA phosphorylase. Biochemistry 1988, 27, 2959–2964. [Google Scholar] [CrossRef]
- Guranowski, A.; Starzyńska, E.; Brown, P.; Blackburn, G.M. Adenosine 5′-tetraphosphate phosphohydrolase from yellow lupin seeds: Purification to homogeneity and some properties. Biochem. J. 1997, 328, 257–262. [Google Scholar] [CrossRef]
- Pietrowska-Borek, M.; Stuible, H.P.; Kombrink, E.; Guranowski, A. 4-Coumarate:coenzyme A ligase has the catalytic capacity to synthesize and reuse various (di)adenosine polyphosphates. Plant Physiol. 2003, 131, 1401–1410. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.A.; Paiva, N.L. Stress-induced phenylpropanoid metabolism. Plant Cell. 1995, 7, 1085–1097. [Google Scholar] [CrossRef] [PubMed]
- Guranowski, A.; Miersch, O.; Staswick, P.E.; Suza, W.; Wasternack, C. Substrate specificty and products of side-reactions catalyzed by jasmonate:amino acid synthetase (JAR1). FEBS Lett. 2007, 581, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Staswick, P.E.; Tiryaki, I.; Rowe, M.L. Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole 3-acetic acids in an assay for adenylation. Plant Cell 2002, 14, 1405–1415. [Google Scholar] [CrossRef]
- Frankhauser, H.; Berkovitz, G.A.; Schiff, J.A. A nucleotide with the properties of adenosine 5′-phosphoramidate from Chlorella cells. Biochem. Biophys. Res. Commun. 1981, 101, 524–532. [Google Scholar] [CrossRef]
- Frankhauser, H.; Schiff, J.A.; Garber, L.J. Purification and properties of adenylyl sulfate: Ammonia adenylyltransferase from Chlorella catalyzing the formation of adenosine 5′-phosphoramidate from adenosine 5′-phosphosulfate and ammonia. Biochem. J. 1981, 195, 545–560. [Google Scholar] [CrossRef]
- Wojdyła-Mamoń, A.M.; Guranowski, A. Adenylylsulfate:ammonia adenylyltransferase activity is another inherent property of Fhit proteins. Bioscence Rep. 2015, 35, e00235. [Google Scholar] [CrossRef]
- Kuba, M.; Okizaki, T.; Ohmori, H.; Kumon, A. Nucleoside monophosphoramidate hydrolase from rat liver: Purification and characterization. Int. J. Biochem. Cell Biol. 1994, 26, 235–245. [Google Scholar]
- Bieganowski, P.; Garrison, P.N.; Hodawadekar, S.C.; Faye, G.; Barnes, L.D.; Brenner, C. Adenosine monophosphoramidase activity of Hint and Hnt1 supports function of Kin28, Ccl1, and Tfb3. J. Biol. Chem. 2002, 13, 10852–10860. [Google Scholar] [CrossRef] [PubMed]
- Guranowski, A.; Wojdyła, A.M.; Zimny, J.; Wypijewska, A.; Kowalska, J.; Jemielity, J.; Davis, R.E.; Bieganowski, P. Dual activity of certain HIT-proteins: A. thaliana Hint4 and C. elegans DcpS act on adenosine 5′-phosphosulfate as hydrolases (forming AMP) and as phosphorylases (forming ADP). FEBS Lett. 2010, 584, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Guranowski, A.; Wojdyła, A.M.; Zimny, J.; Wypijewska, A.; Kowalska, J.; Łukaszewicz, M.; Jemielity, J.; Darżynkiewicz, E.; Jagiełło, A.; Bieganowski, P. Recognition of different nucleotidyl-derivatives as substrates of re-actions catalyzed by various HIT-proteins. New J. Chem. 2010, 34, 888–893. [Google Scholar] [CrossRef]
- Guranowski, A.; Wojdyła, A.M.; Rydzik, A.M.; Stępiński, J.; Jemielity, J. Plant nucleoside 5’-phosphoramidate hydrolase; simple purification from yellow lupin (Lupinus luteus) seeds and properties of homogeneous enzyme. Acta Biochim. Pol. 2011, 58, 131–136. [Google Scholar] [CrossRef]
- Bretes, E.; Wojdyła-Mamoń, A.M.; Kowalska, J.; Jemielity, J.; Kaczmarek, R.; Baraniak, J.; Guranowski, A. Hint2, the mitochondrial nucleoside 5′-phosphoramidate hydrolase; properties of the homogeneous protein from sheep (Ovis aries) liver. Acta Biochim. Pol. 2013, 60, 249–254. [Google Scholar] [CrossRef]
- Huang, K.; Arabshahi, A.; Wei, Y.; Frey, P.A. The mechanism of action of the fragile histidine triad, Fhit: Isolation of a covalent adenylyl enzyme and chemical rescue of H96G-Fhit. Biochemistry 2004, 43, 7637–7642. [Google Scholar] [CrossRef]
- Brenner, C. Hint, Fhit, and GalT: Function, structure, evolution, and mechanism of three branches of the histidine triad superfamily of nucleotide hydrolases and transferases. Biochemistry 2002, 41, 9003–9014. [Google Scholar] [CrossRef]
- Guranowski, A.; Wojdyła, A.M.; Pietrowska-Borek, M.; Bieganowski, P.; Khurs, E.N.; Cliff, M.J.; Blackburn, G.M.; Błaziak, D.; Stec, W.J. Fhit proteins can also recognize substrates other than dinucleoside polyphosphates. FEBS Lett. 2008, 582, 3152–3158. [Google Scholar] [CrossRef]
- Christie, S.M.H.; Elmore, D.T.; Kenner, G.W.; Todd, A.R.; Weymouth, F.J. Nucleotides, Part XXII. Syntheses of P1,P2-diadenosine-5′ and P1,P2-diuridine-5′-pyrophosphates. J. Chem. Soc. 1953, 3, 2947–2953. [Google Scholar] [CrossRef]
- Finamore, F.J.; Warner, A.H. The occurrence of P1,P4-diguanosine 5′-tetraphosphate in brine shrimp eggs. J. Biol. Chem. 1963, 238, 344–348. [Google Scholar] [PubMed]
- Warner, A.H.; Finamore, F.J. Isolation, purification, and characterization of P1,P4-diguanosine 5′-tetraphosphatea asymmetrical-pyrophosphohydrolase from brine shrimp eggs. Biochemistry 1965, 4, 1568–1575. [Google Scholar] [CrossRef] [PubMed]
- Garrison, P.N.; Barnes, L.D. Determination of dinucleoside polyphosphates. In Ap4A and Other Dinucleoside Polyphosphates; McLennan, A.G., Ed.; CRC Press: Boca Raton, FL, USA, 1992; pp. 29–61. [Google Scholar]
- Plateau, P.; Blanquet, S. Dinucleoside oligophosphates in micro-organisms. Adv. Microb. Physiol. 1994, 36, 81–109. [Google Scholar] [PubMed]
- Lee, P.C.; Barry, R.; Ames, B.N. Diadenosine 5’,5’’’-P1,P4-tetraphosphate and related adenylylated nucleotides in Salmonella typhimurium. J. Biol. Chem. 1983, 258, 6827–6834. [Google Scholar] [PubMed]
- Coste, H.; Brevet, A.; Plateau, P.; Blanquet, S. Non-adenylylated bis(5’-nucleosidyl) tetraphosphates occur in Saccharomyces cerevisiae and in Escherichia coli and accumulate upon temperature shift or exposure to cadmium. J. Biol. Chem. 1987, 262, 12096–12103. [Google Scholar] [PubMed]
- Baltzinger, M.; Ebel, J.P.; Remy, P. Accumulation of dinucleoside polyphosphates in Saccharomyces cerevisiae under stress conditions. High levels are associated with cell death. Biochimie 1986, 68, 1231–1236. [Google Scholar] [CrossRef]
- Palfi, Z.; Suranyi, G.; Borbely, G. Alterations in the accumulation of adenylated nucleotides in heavy-metal-ion-stressed and heat-stressed Synechococcus sp. strain PCC 6301, cyanobacterium, in light and dark. Biochem. J. 1991, 276, 487–491. [Google Scholar] [CrossRef][Green Version]
- Lüthje, J.; Olivie, A. The presence of diadenosine 5’, 5’’’-P1,P3- triphosphate (Ap3A) in human platelets. Biochem. Biophys. Res. Commun. 1983, 115, 253–260. [Google Scholar] [CrossRef]
- Flodgaard, H.; Klenow, H. Abundant amounts of diadenosine 5’, 5’’’-P1,P4-tetraphosphate are present and releasable, but metabolically inactive, in human platelets. Biochem. J. 1982, 208, 737–742. [Google Scholar] [CrossRef]
- Morioka, M.; Shimada, H. Synthesis of diadenosine 5’, 5’’’-P1,P4-tetraphosphate (Ap4A) in sea urchin embryos. Cell Differ. 1984, 14, 53–58. [Google Scholar] [CrossRef]
- Miller, D.; McLennan, A.G. Changes in intracellular levels of Ap3A and Ap4A in cysts and larvae of Artemia do not correlate with changes in protein synthesis after heat-shock. Nucleic Acids Res. 1986, 14, 6031–6040. [Google Scholar] [CrossRef] [PubMed]
- Brevet, A.; Plateau, P.; Best-Belpomme, M.; Blanquet, S. Variation of Ap4A and other dinucleoside polyphosphates in stressed Drosophila cells. J. Biol. Chem. 1985, 260, 15566–15570. [Google Scholar] [PubMed]
- Plateau, P.; Mayaux, J.F.; Blanquet, S. Zinc (II)-dependent synthesis of diadenosine 5’,5’’’-P1,P4-tetraphosphate by Escherichia coli and yeast phenylalanyl transfer ribonucleic acid synthetases. Biochemistry 1981, 20, 4654–4662. [Google Scholar] [CrossRef] [PubMed]
- Jakubowski, H. Synthesis of diadenosine 5’,5’’’-P1,P4-tetraphosphate and related compounds by plant (Lupinus luteus) seryl-tRNA and phenylalanyl-tRNA synthetases. Acta Biochim. Pol. 1983, 30, 51–69. [Google Scholar]
- Guranowski, A.; Günther Sillero, M.A.; Sillero, A. Firefly luciferase synthesizes P1,P4-bis(5’-adenosyl) tetraphosphate (Ap4A) and other dinucleoside polyphosphates. FEBS Lett. 1990, 271, 215–218. [Google Scholar] [CrossRef]
- Wang, D.; Shatkin, A.J. Synthesis of Gp4N and Gp3N compounds by guanylyltransferase purified from yeast. Nucleic Acids Res. 1984, 12, 2303–2315. [Google Scholar] [CrossRef]
- Jin, Z.; Leveque, V.; Ma, H.; Johnson, K.A.; Klumpp, K. NTP-mediated nucleotide excision activity of hepatitis C virus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA 2013, 110, 348–357. [Google Scholar] [CrossRef]
- Guranowski, A. Specific and non-specific enzymes involved in the catabolism of mononucleoside and dinucleoside polyphosphates. Pharma Ther. 2000, 87, 117–139. [Google Scholar] [CrossRef]
- Jakubowski, H.; Guranowski, A. Enzymes hydrolyzing ApppA and/or AppppA in higher plants purification and some properties of diadenosine triphosphatase, diadenosine tetraphosphatase, and phosphodiesterase from yellow lupin (Lupinus luteus) seeds. J. Biol. Chem. 1983, 258, 9982–9989. [Google Scholar]
- Guranowski, A. Fluoride is a strong and specific inhibitor of (asymmetrical) Ap4A hydrolases. FEBS Lett. 1990, 262, 205–208. [Google Scholar] [CrossRef]
- Feussner, K.; Guranowski, A.; Kostka, S.; Wasternack, C. Diadenosine 5′,5′′′-P1,P4-tetraphosphate (Ap4A) hydrolase from tomato (Lycopersicon esculentum cv. Lukullus)–Purification, biochemical Ap4A and related nucleotides in plants properties and behaviour during stress. Z. Naturforsch. C J. Biosci. 1996, 51, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Maksel, D.; Guranowski, A.; Ilgoutz, S.C.; Moir, A.; Blackburn, M.G.; Gayler, K.R. Cloning and expression of diadenosine 5′,5′′′-P1,P4-tetraphosphate hydrolase from Lupinus angustifolius L. Biochem. J. 1998, 329, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Churin, J.; Hause, B.; Feussner, I.; Maucher, H.P.; Feussner, K.; Börner, T.; Wasternack, C. Cloning and expression of a new cDNA from monocotyledonous plants coding for a diadenosine 5′,5′′′-P1,P4-tetraphosphate hydrolase from barley (Hordeum vulgare). FEBS Lett. 1998, 431, 481–485. [Google Scholar] [CrossRef]
- Bartkiewicz, M.; Sierakowska, H.; Shugar, D. Nucleotide pyrophosphatase from potato tubers; purification and properties. Eur. J. Biochem. 1984, 143, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Zamecnik, P. Diadenosine 5′,5‴-P1,P4-tetraphosphate (Ap4A): Its role in cellular metabolism. Anal. Biochem. 1983, 134, 1–10. [Google Scholar] [CrossRef]
- McLennan, A.G. Dinucleoside polyphosphates - friend or foe? Pharmacol. Ther. 2000, 87, 73–89. [Google Scholar] [CrossRef]
- Rapaport, E.; Zamecnik, P.C. Presence of diadenosine 5′,5‴-P1,P4-tetraphosphate (Ap4A) in mamalian cells in levels varying widely with proliferative activity of the tissue: A possible positive “pleiotypic activator”. Proc. Natl. Acad. Sci. USA 1976, 73, 3984–3988. [Google Scholar] [CrossRef]
- Weinmann-Dorsch, C.; Pierron, G.; Wick, R.; Sauer, H.; Grummt, F. High diadenosine tetraphosphate (Ap4A) level at initiation of S phase in the naturally synchronous mitotic cycle of Physarum polycephalum. Exp. Cell Res. 1984, 155, 171–177. [Google Scholar] [CrossRef]
- Lee, P.C.; Bochner, B.R.; Ames, B.N. AppppA, heat-shock stress, and cell oxidation. Proc. Natl. Acad. Sci. USA 1983, 80, 7496–7500. [Google Scholar] [CrossRef]
- Bochner, B.R.; Lee, P.C.; Wilson, S.W.; Cutler, C.W.; Ames, B.N. Ap4A and related nucleotides are synthesized as consequence of oxidation stress. Cell 1984, 37, 225–232. [Google Scholar] [CrossRef]
- Hauryliuk, V.; Atkinson, G.C.; Murakami, K.S.; Tenson, T.; Gerdes, K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 2015, 13, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Perotti, V.E.; Del Vecchio, H.A.; Sansevich, A.; Meier, G.; Bello, F.; Cocco, M.; Garrán, S.M.; Anderson, C.; Vázquez, D.; Podestá, F.E. Proteomic, metabalomic, and biochemical analysis of heat treated Valencia oranges during storage. Postharv. Biol. Technol. 2011, 62, 97–114. [Google Scholar] [CrossRef]
- Jambunathan, N.; Penaganti, A.; Tang, Y.; Mahalingam, R. Modulation of redox homeostasis under suboptimal conditions by Arabidopsis nudix hydrolase 7. BMC Plant Biol. 2010, 10, 173. [Google Scholar] [CrossRef] [PubMed]
- Muthuramalingam, M.; Zeng, X.; Iyer, N.J.; Klein, P.; Mahalingam, R. A GCC-box motif in the promoter of nudix hydrolase 7 (AtNUDT7) gene plays a role in ozone response of Arabidopsis ecotypes. Genomics 2015, 105, 31–38. [Google Scholar] [CrossRef]
- Maruta, T.; Ogawa, T.; Tsujimura, M.; Ikemoto, K.; Yoshida, T.; Takahashi, H.; Yoshimura, K.; Shigeoka, S. Loss-of-function of an Arabidopsis NADPH pyrophosphohydrolase, AtNUDX19, impacts on the pyridine nucleotides status and confers photooxidative stress tolerance. Sci. Rep. 2016, 6, 37432. [Google Scholar] [CrossRef]
- Tanaka, S.; Kihara, M.; Sugimoto, M. Structure and molecular characterization of barley nudix hydrolase genes. Biosci. Biotechnol. Biochem. 2015, 79, 394–401. [Google Scholar] [CrossRef]
- Xu, J.; Yang, J.Y.; Niu, Q.W.; Chua, N.H. Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell 2006, 18, 3386–3398. [Google Scholar] [CrossRef]
- Gunawardana, D.; Cheng, H.C.; Gayler, K.R. Identification of functional domains in Arabidopsis thaliana mRNA decapping enzyme (AtDcp2). Nucleic Acids Res. 2008, 36, 203–216. [Google Scholar] [CrossRef]
- Song, M.G.; Bail, S.; Kiledjian, M. Multiple Nudix family proteins possess mRNA decapping activity. RNA 2013, 19, 390–399. [Google Scholar]
- Despotović, D.; Brandis, A.; Savidor, A.; Levin, Y.; Fumagalli, L.; Tawfik, D.S. Diadenosine tetraphosphate (Ap4A) – an E. coli alarmone or a demage metabolite? FEBS J. 2017, 284, 2194–2215. [Google Scholar] [CrossRef]
- Ji, X.; Zou, J.; Peng, H.; Stolle, A.S.; Xie, R.; Zhang, H.; Peng, B.; Mekalanos, J.J.; Zheng, J. Alarmone Ap4A is elevated by aminoglycoside antibiotics and enhances their bactericidal activity. Proc. Nat. Acad. Sci. USA 2019, 116, 9578–9585. [Google Scholar] [CrossRef]
- Lee, Y.-N.; Nechushtan, H.; Figov, N.; Razin, E. The function of lysyl-tRNA synthetase and Ap4A as signaling regulators of MITF activity in FcϵRI-activated mast cells. Immunity 2004, 20, 145–151. [Google Scholar] [CrossRef]
- Carmi-Levy, I.; Yannay-Cohen, N.; Kay, G.; Razin, E.; Nechushtan, H. Diadenosine tetraphosphate hydrolase is part of the transcriptional regulation network in immunologically activated mast cells. Mol. Cell. Biol. 2008, 28, 5777–5784. [Google Scholar] [CrossRef]
- Marriott, A.S.; Vasieva, O.; Fang, Y.; Copeland, N.A.; McLennan, A.G.; Jones, N.J. NUDT2 disruption elevates diadenosine tetraphosphate (Ap4A) and down-regulates immune response and cancer promotion genes. PLoS ONE 2016, 11, e0154674. [Google Scholar] [CrossRef]
- Bobert, P.; Schlüter, H.; Schultheiss, H.P.; Reusch, H.P. Diadenosine polyphosphates Ap3A and Ap4A, but not Ap5A or Ap6A, induce proliferation of vascular smooth muscle cells. Biochem. Pharmacl. 2008, 75, 1966–1973. [Google Scholar] [CrossRef]
- Marriott, A.S.; Copeland, N.A.; Cunningham, R.; Wilkinson, M.C.; McLennan, A.G.; Jones, N.J. Diadenosine 5′, 5′′′-P1,P4-tetraphosphate (Ap4A) is synthesized in response to DNA damage and inhibits the initiation of DNA replication. DNA Repair. 2015, 33, 90–100. [Google Scholar] [CrossRef]
- Pietrowska-Borek, M.; Nuc, K.; Zielezińska, M.; Guranowski, A. Diadenosine polyphosphates (Ap3A and Ap4A) behave as alarmones triggering the synthesis of enzymes of the phenylpropanoid pathway in Arabidopsis thaliana. FEBS Open Bio 2011, 1, 1–6. [Google Scholar] [CrossRef]
- Pietrowska-Borek, M.; Czekała, Ł.; Belchí-Navarro, S.; Pedreño, M.A.; Guranowski, A. Diadenosine triphosphate is a novel factor which in combination with cyclodextrins synergistically enhances the biosynthesis of trans-resveratrol in Vitis vinifera cv. Monastrell suspension cultured cells. Plant Physiol. Biochem. 2014, 84, 271–276. [Google Scholar] [CrossRef]
- Pietrowska-Borek, M.; Nuc, K.; Guranowski, A. Exogenous adenosine 5′-phosphoramidate behaves as a signal molecule in plants; it augments metabolism of phenylpropanoids and salicylic acid in Arabidopsis thaliana seedlings. Plant Physiol. Biochem. 2015, 94, 144–152. [Google Scholar] [CrossRef]
- Pietrowska-Borek, M.; Wojdyła-Mamoń, A.; Dobrogojski, J.; Młynarska-Cieślak, A.; Baranowski, M.R.; Dąbrowski, J.M.; Kowalska, J.; Jemielity, J.; Borek, S.; Pedreño, M.A.; et al. Purine and pyrimidine dinucleoside polyphosphates differentially affect the phenylpropanoid pathway in Vitis vinifera L. cv. Monastrell suspension cultured cells. Plant Physiol. Biochem. 2020, 147, 125–132. [Google Scholar] [CrossRef]
- Ehlting, J.; Büttner, D.; Wang, Q.; Douglas, C.J.; Somssich, I.E.; Kombrink, E. Three 4-coumarate:coenzyme A ligases in A. thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 1999, 19, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Bru, R.; Sellés, S.; Casado-Vela, J.; Belchí-Navarro, S.; Pedreño, M.A. Modified cyclodextrins are chemically defined glucan inducers of defense responses in grapevine cell cultures. J. Agric. Food Chem. 2006, 54, 66–71. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 2002, 5, 218–223. [Google Scholar] [CrossRef]
- Olsen, M.K.; Lea, U.S.; Slimestad, R.; Verheul, M.; Lillo, C. Differential expression of four Arabidopsis PAL genes; PAL1 and PAL2 have functional specialization in abiotic environmental-triggered flavonoid synthesis. J. Plant Physiol. 2008, 165, 1491–1499. [Google Scholar] [CrossRef] [PubMed]
- Moura, J.C.M.S.; Bonine, C.A.V.; Viana, J.O.F.; Dornelas, M.C.; Mazzafera, P. Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 2010, 52, 360–376. [Google Scholar] [CrossRef]
- Pietrowska-Borek, M.; Nuc, K. Stimulating effect of cadmium on phenylpropanoid pathway in Arabidopsis thaliana seedlings. Acta Soc. Bot. Polon. 2010, 79 (Suppl. 1), 90. [Google Scholar]
- Steyn, W.J.; Wand, S.J.E.; Holcroft, D.M.; Jacobs, G. Anthocyanins in vegetative tissues: A proposed unified function in photoprotection. New Phytol. 2002, 55, 349–361. [Google Scholar] [CrossRef]
- Bandurska, H.; Pietrowska-Borek, M.; Cieślak, M. Response of barley seedlings to water deficit and enhanced UV-B irradiation acting alone and in combination. Acta Physiol. Plant. 2012, 34, 161–171. [Google Scholar] [CrossRef][Green Version]
- Hayat, Q.; Hayat, S.; Irfan, M.; Ahmad, A. Effect of exogenous salicylic acid under changing environment: A review. Environ. Exp. Bot. 2010, 68, 14–25. [Google Scholar] [CrossRef]
- Gakière, B.; Hao, J.; de Bont, L.; Pétriacq, P.; Nunes-Nesi, A.; Fernie, A.R. NAD+ biosynthesis and signaling in plants. Crit. Rev. Plant Sci. 2018, 3, 259–307. [Google Scholar] [CrossRef]
- Navazio, L.; Bewell, M.A.; Siddiqua, A.; Dickinson, G.D.; Galione, A.; Sanders, D. Calcium release from the endoplasmic reticulum of higher plants elicited by the NADP metabolite nicotinic acid adenine dinucleotide phosphate. Proc. Natl. Acad. Sci. USA 2000, 97, 8693–8698. [Google Scholar] [CrossRef] [PubMed]
- Navazio, L.; Mariani, P.; Sanders, D. Mobilization of Ca2+ by cyclic ADP-ribose from the endoplasmic reticulum of cauliflower florets. Plant Physiol. 2001, 125, 2129–2138. [Google Scholar] [CrossRef] [PubMed]
- Guse, A.H.; Lee, H.C. NAADP: A universal Ca2+ trigger. Sci. Signal. 2008, 1, re10. [Google Scholar] [CrossRef]
- Opdenakker, K.; Remans, T.; Vangronsveld, J.; Cuypers, A. Mitogen-activated protein (MAP) kinases in plant metal stress: Regulation and responses in comparison to other biotic and abiotic stresses. Int. J. Mol. Sci. 2012, 13, 7828–7853. [Google Scholar] [CrossRef]
- Devoto, A.; Turner, J.G. Regulation of jasmonate-mediated plant responses in Arabidopsis. Ann. Bot. 2003, 92, 329–337. [Google Scholar] [CrossRef]
- Mine, A.; Berens, M.L.; Nobori, T.; Anver, S.; Fukumoto, K.; Winkelmüller, T.M.; Takeda, A.; Becker, D.; Tsuda, K. Pathogen exploitation of an abscisic acid- and jasmonate-inducible MAPK phosphatase and its interception by Arabidopsis immunity. Proc. Natl. Acad. Sci. USA 2017, 114, 7456–7461. [Google Scholar] [CrossRef]
- Vian, A.; Stankovic, B.; Davies, E. Signalomics: Diversity and methods of analysis of systemic signals in plants. In Plant Omics: The Omics of Plant Science; Barh, D., Khan, M.S., Davies, E., Eds.; Springer: New Delhi, India, 2015; pp. 459–489. [Google Scholar]
- Marcec, M.J.; Gilroy, S.; Poovaiah, B.W.; Tanaka, K. Mutual interplay of Ca2+ and ROS signaling in plant immune response. Plant Sci. 2019, 283, 343–354. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).