Identification of Novel Rare ABCC1 Transporter Mutations in Tumor Biopsies of Cancer Patients
Abstract
1. Introduction
2. Material and Methods
2.1. RNA Sequencing and Mutation Analysis
2.2. Homology Modeling, Protein Structure Preparation
2.3. Molecular Docking and Clustering
3. Results
3.1. ABC Transporters Mutation Analysis
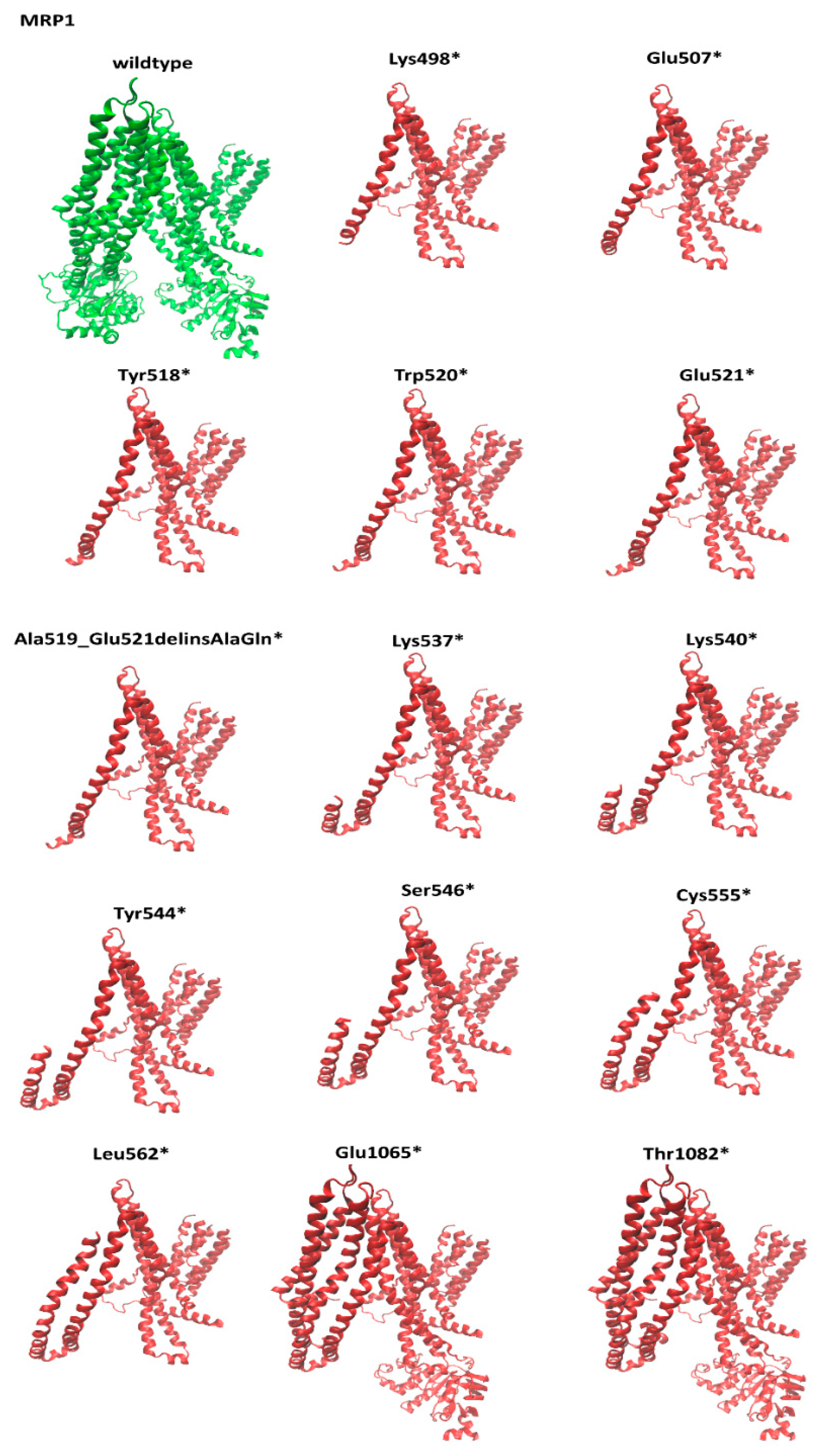
3.2. Structural Comparison of the Nonsense MRP1 Mutant Models
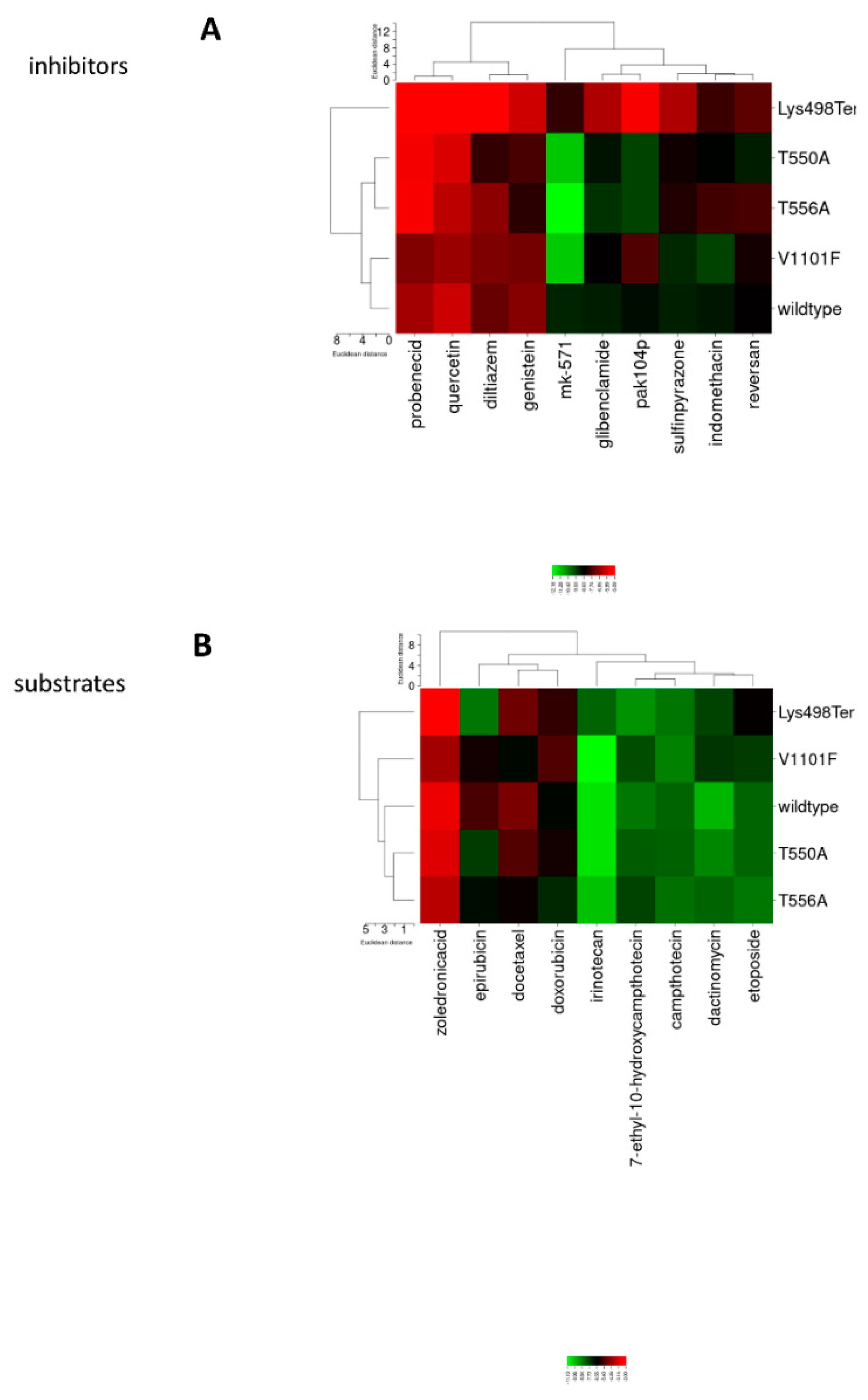
3.3. Molecular Docking Clustering
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ABC | ATP-binding cassette transporter |
| MRP | multidrug resistance-associated protein |
References
- Efferth, T.; Zeino, M.; Volm, M. Modulation of P-Glycoprotein-Mediated Multidrug Resistance by Synthetic and Phytochemical Small Molecules, Monoclonal Antibodies, and Therapeutic Nucleic Acids. In Resistance to Targeted ABC Transporters in Cancer; Efferth, T., Ed.; Springer: Cham, Switzerland, 2015; pp. 153–181. [Google Scholar] [CrossRef]
- Volm, M.; Efferth, T. Role of P-Glycoprotein for Resistance of Tumors to Anticancer Drugs: From Bench to Bedside. In Resistance to Targeted ABC Transporters in Cancer; Efferth, T., Ed.; Springer: Cham, Switzerland, 2015; pp. 1–26. [Google Scholar] [CrossRef]
- Roundhill, E.A.; Fletcher, J.I.; Haber, M.; Norris, M.D. Clinical Relevance of Multidrug-Resistance-Proteins (MRPs) for Anticancer Drug Resistance and Prognosis. In Resistance to Targeted ABC Transporters in Cancer; Efferth, T., Ed.; Springer: Cham, Switzerland, 2015; pp. 27–52. [Google Scholar] [CrossRef]
- Natarajan, K.; Baer, M.R.; Ross, D.D. Role of Breast Cancer Resistance Protein (BCRP, ABCG2) in Cancer Outcomes and Drug Resistance. In Resistance to Targeted ABC Transporters in Cancer; Efferth, T., Ed.; Springer: Cham, Switzerland, 2015; pp. 53–88. [Google Scholar] [CrossRef]
- Frampton, G.M.; Fichtenholtz, A.; Otto, G.A.; Wang, K.; Downing, S.R.; He, J.; Schnall-Levin, M.; White, J.; Sanford, E.M.; An, P.; et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. 2013, 31, 1023–1031. [Google Scholar] [CrossRef] [PubMed]
- Conley, B.A.; Doroshow, J.H. Molecular analysis for therapy choice: NCI MATCH. Semin. Oncol. 2014, 41, 297–299. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Little, P.; Hoyle, A.P.; Pegna, G.J.; Hayward, M.C.; Ivanova, A.; Parker, J.S.; Marron, D.L.; Soloway, M.G.; Jo, H.; et al. The Prognostic Significance of Low-Frequency Somatic Mutations in Metastatic Cutaneous Melanoma. Front. Oncol. 2018, 8, 584. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Dang, H.; Wang, X.W. The significance of intertumor and intratumor heterogeneity in liver cancer. Exp. Mol. Med. 2018, 50, e416. [Google Scholar] [CrossRef] [PubMed]
- Saunders, N.A.; Simpson, F.; Thompson, E.W.; Hill, M.M.; Endo-Munoz, L.; Leggatt, G.; Minchin, R.F.; Guminski, A. Role of intratumoral heterogeneity in cancer drug resistance: Molecular and clinical perspectives. EMBO Mol. Med. 2012, 4, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Lusito, E.; Felice, B.; D’Ario, G.; Ogier, A.; Montani, F.; Di Fiore, P.P.; Bianchi, F. Unraveling the role of low-frequency mutated genes in breast cancer. Bioinformatics 2019, 35, 36–46. [Google Scholar] [CrossRef]
- Leitner, H.M.; Kachadourian, R.; Day, B.J. Harnessing drug resistance: Using ABC transporter proteins to target cancer cells. Biochem. Pharm. 2007, 74, 1677–1685. [Google Scholar] [CrossRef]
- Dharmapuri, G.; Doneti, R.; Philip, G.H.; Kalle, A.M. Celecoxib sensitizes imatinib-resistant K562 cells to imatinib by inhibiting MRP1-5, ABCA2 and ABCG2 transporters via Wnt and Ras signaling pathways. Leuk. Res. 2015, 39, 696–701. [Google Scholar] [CrossRef]
- Steinbach, D.; Gillet, J.P.; Sauerbrey, A.; Gruhn, B.; Dawczynski, K.; Bertholet, V.; de Longueville, F.; Zintl, F.; Remacle, J.; Efferth, T. ABCA3 as a possible cause of drug resistance in childhood acute myeloid leukemia. Clin. Cancer Res. 2006, 12, 4357–4363. [Google Scholar] [CrossRef]
- Xu, M.; Li, L.; Liu, Z.Q.; Jiao, Z.J.; Xu, P.; Kong, X.Y.; Huang, H.J.; Zhang, Y.L. ABCB2 (TAP1) as the downstream target of SHH signaling enhances pancreatic ductal adenocarcinoma drug resistance. Cancer Lett. 2013, 333, 152–158. [Google Scholar] [CrossRef]
- Kawanobe, T.; Kogure, S.; Nakamura, S.; Sato, M.; Katayama, K.; Mitsuhashi, J.; Noguchi, K.; Sugimoto, Y. Expression of human ABCB5 confers resistance to taxanes and anthracyclines. Biochem. Biophys. Res. Commun. 2012, 418, 736–741. [Google Scholar] [CrossRef] [PubMed]
- Kadioglu, O.; Cao, J.; Kosyakova, N.; Mrasek, K.; Liehr, T.; Efferth, T. Genomic and transcriptomic profiling of resistant CEM/ADR-5000 and sensitive CCRF-CEM leukemia cells for unravelling the full complexity of multi-factorial multidrug resistance. Sci. Rep. 2016, 6, 36754. [Google Scholar] [CrossRef] [PubMed]
- Tajima, Y.; Nakagawa, H.; Tamura, A.; Kadioglu, O.; Satake, K.; Mitani, Y.; Murase, H.; Regasini, L.O.; Bolzani, V.D.; Ishikawa, T.; et al. Nitensidine A, a guanidine alkaloid from Pterogyne nitens, is a novel substrate for human ABC transporter ABCB1. Phytomed. Int. J. Phytother. Phytopharm. 2014, 21, 323–332. [Google Scholar] [CrossRef] [PubMed]
- de Groot, D.J.; van der Deen, M.; Le, T.K.; Regeling, A.; de Jong, S.; de Vries, E.G. Indomethacin induces apoptosis via a MRP1-dependent mechanism in doxorubicin-resistant small-cell lung cancer cells overexpressing MRP1. Br. J. Cancer 2007, 97, 1077–1083. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.R.; Jamieson, D.; Thomas, H.D.; Brown, C.D.; Boddy, A.V.; Veal, G.J. Characterization of the roles of ABCB1, ABCC1, ABCC2 and ABCG2 in the transport and pharmacokinetics of actinomycin D in vitro and in vivo. Biochem. Pharmacol. 2013, 85, 29–37. [Google Scholar] [CrossRef]
- Burkhart, C.A.; Watt, F.; Murray, J.; Pajic, M.; Prokvolit, A.; Xue, C.; Flemming, C.; Smith, J.; Purmal, A.; Isachenko, N.; et al. Small-molecule multidrug resistance-associated protein 1 inhibitor reversan increases the therapeutic index of chemotherapy in mouse models of neuroblastoma. Cancer Res. 2009, 69, 6573–6580. [Google Scholar] [CrossRef]
- Ebert, R.; Meissner-Weigl, J.; Zeck, S.; Maatta, J.; Auriola, S.; Coimbra de Sousa, S.; Mentrup, B.; Graser, S.; Rachner, T.D.; Hofbauer, L.C.; et al. Probenecid as a sensitizer of bisphosphonate-mediated effects in breast cancer cells. Mol. Cancer 2014, 13, 265. [Google Scholar] [CrossRef]
- Deeley, R.G.; Cole, S.P. Substrate recognition and transport by multidrug resistance protein 1 (ABCC1). FEBS Lett. 2006, 580, 1103–1111. [Google Scholar] [CrossRef]
- Van Zanden, J.J.; van der Woude, H.; Vaessen, J.; Usta, M.; Wortelboer, H.M.; Cnubben, N.H.; Rietjens, I.M. The effect of quercetin phase II metabolism on its MRP1 and MRP2 inhibiting potential. Biochem. Pharmacol. 2007, 74, 345–351. [Google Scholar] [CrossRef]
- Vulsteke, C.; Lambrechts, D.; Dieudonne, A.; Hatse, S.; Brouwers, B.; van Brussel, T.; Neven, P.; Belmans, A.; Schoffski, P.; Paridaens, R.; et al. Genetic variability in the multidrug resistance associated protein-1 (ABCC1/MRP1) predicts hematological toxicity in breast cancer patients receiving (neo-)adjuvant chemotherapy with 5-fluorouracil, epirubicin and cyclophosphamide (FEC). Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2013, 24, 1513–1525. [Google Scholar] [CrossRef]
- Tivnan, A.; Zakaria, Z.; O’Leary, C.; Kogel, D.; Pokorny, J.L.; Sarkaria, J.N.; Prehn, J.H. Inhibition of multidrug resistance protein 1 (MRP1) improves chemotherapy drug response in primary and recurrent glioblastoma multiforme. Front. Neurosci. 2015, 9, 218. [Google Scholar] [CrossRef] [PubMed]
- Brangi, M.; Litman, T.; Ciotti, M.; Nishiyama, K.; Kohlhagen, G.; Takimoto, C.; Robey, R.; Pommier, Y.; Fojo, T.; Bates, S.E. Camptothecin resistance: Role of the ATP-binding cassette (ABC), mitoxantrone-resistance half-transporter (MXR), and potential for glucuronidation in MXR-expressing cells. Cancer Res. 1999, 59, 5938–5946. [Google Scholar] [PubMed]
- Castro, A.F.; Altenberg, G.A. Inhibition of drug transport by genistein in multidrug-resistant cells expressing P-glycoprotein. Biochem. Pharmacol. 1997, 53, 89–93. [Google Scholar] [CrossRef]
- Marbeuf-Gueye, C.; Salerno, M.; Quidu, P.; Garnier-Suillerot, A. Inhibition of the P-glycoprotein- and multidrug resistance protein-mediated efflux of anthracyclines and calceinacetoxymethyl ester by PAK-104P. Eur. J. Pharmacol. 2000, 391, 207–216. [Google Scholar] [CrossRef]
- Zhang, D.W.; Nunoya, K.; Vasa, M.; Gu, H.M.; Cole, S.P.C.; Deeley, R.G. Mutational analysis of polar amino acid residues within predicted transmembrane helices 10 and 16 of multidrug resistance protein 1 (ABCC1): Effect on substrate specificity. Drug Metab. Dispos. 2006, 34, 539–546. [Google Scholar] [CrossRef]
- Kadioglu, O.; Saeed, M.E.M.; Valoti, M.; Frosini, M.; Sgaragli, G.; Efferth, T. Interactions of human P-glycoprotein transport substrates and inhibitors at the drug binding domain: Functional and molecular docking analyses. Biochem. Pharm. 2016, 104, 42–51. [Google Scholar] [CrossRef]
- Kadioglu, O.; Saeed, M.; Kuete, V.; Greten, H.J.; Efferth, T. Oridonin Targets Multiple Drug-Resistant Tumor Cells as Determined by in Silico and in Vitro Analyses. Front. Pharmacol. 2018, 9. [Google Scholar] [CrossRef]
- Ooko, E.; Kadioglu, O.; Greten, H.J.; Efferth, T. Pharmacogenomic Characterization and Isobologram Analysis of the Combination of Ascorbic Acid and Curcumin-Two Main Metabolites of Curcuma longa-in cancer Cells. Front. Pharmacol. 2017, 8. [Google Scholar] [CrossRef]
- Robey, R.W.; Pluchino, K.M.; Hall, M.D.; Fojo, A.T.; Bates, S.E.; Gottesman, M.M. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer 2018, 18, 452–464. [Google Scholar] [CrossRef]
- Roundhill, E.A.; Burchill, S.A. Detection and characterisation of multi-drug resistance protein 1 (MRP-1) in human mitochondria. Br. J. Cancer 2012, 106, 1224–1233. [Google Scholar] [CrossRef]
- Ji, L.L.; Li, H.; Gao, P.; Shang, G.G.; Zhang, D.N.D.; Zhang, N.; Jiang, T. Nrf2 Pathway Regulates Multidrug-Resistance-Associated Protein 1 in Small Cell Lung Cancer. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, Z.H.; Bi, L.K.; Li, K.Q.; Zhou, B.F.; Xu, C.; Huang, J.; Xu, K.W. NOTCH1 signaling promotes chemoresistance via regulating ABCC1 expression in prostate cancer stem cells. Mol. Cell. Biochem. 2014, 393, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.M.T.; Huynh, T.; Truong, A.M.; Haber, M.; Norris, M.D. ABC Transporters and Neuroblastoma. Adv. Cancer Res. 2015, 125, 139–170. [Google Scholar] [CrossRef] [PubMed]
- Bakos, E.; Homolya, L. Portrait of multifaceted transporter, the multidrug resistance-associated protein 1 (MRP1/ABCC1). Pflug. Arch. Eur. J. Physiol. 2007, 453, 621–641. [Google Scholar] [CrossRef] [PubMed]
- Abaan, O.D.; Mutlu, P.K.; Baran, Y.; Atalay, C.; Gunduz, U. Multidrug resistance mediated by MRP1 gene overexpression in breast cancer patients. Cancer Investig. 2009, 27, 201–205. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yin, J.; Zhang, J. Multidrug resistance-associated protein 1 (MRP1/ABCC1) polymorphism: From discovery to clinical application. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2011, 36, 927–938. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.; Iida, A.; Sekine, A.; Miura, Y.; Ogawa, C.; Kawauchi, S.; Higuchi, S.; Nakamura, Y. Identification of 779 genetic variations in eight genes encoding members of the ATP-binding cassette, subfamily C (ABCC/MRP/CFTR. J. Hum. Genet. 2002, 47, 147–171. [Google Scholar] [CrossRef]
- Fukushima-Uesaka, H.; Saito, Y.; Tohkin, M.; Maekawa, K.; Hasegawa, R.; Kawamoto, M.; Kamatani, N.; Suzuki, K.; Yanagawa, T.; Kajio, H.; et al. Genetic variations and haplotype structures of the ABC transporter gene ABCC1 in a Japanese population. Drug Metab. Pharm. 2007, 22, 48–60. [Google Scholar] [CrossRef]
- Leschziner, G.; Zabaneh, D.; Pirmohamed, M.; Owen, A.; Rogers, J.; Coffey, A.J.; Balding, D.J.; Bentley, D.B.; Johnson, M.R. Exon sequencing and high resolution haplotype analysis of ABC transporter genes implicated in drug resistance. Pharm. Genom. 2006, 16, 439–450. [Google Scholar] [CrossRef]
- Nussinov, R.; Tsai, C.J.; Jang, H. Why Are Some Driver Mutations Rare? Trends Pharmacol. Sci. 2019, 40, 919–929. [Google Scholar] [CrossRef]

| Patient ID | Tumor Type | Age | Gender |
|---|---|---|---|
| P01 | vulva cancer, colon cancer | 82 | Female |
| P02 | breast | 64 | Female |
| P03 | breast | 75 | Female |
| P04 | lung | 75 | Female |
| P05 | breast | 76 | Female |
| P06 | non-Hodgkin lymphoma | 56 | Male |
| P07 | non-Hodgkin lymphoma | 68 | Male |
| P08 | lymphoma | 43 | Female |
| P09 | breast | 50 | Female |
| P10 | cholangiocellular cancer of bile duct | 54 | Male |
| P11 | liver | 80 | Male |
| P12 | cervix | 54 | Female |
| P13 | invasive lobular breast cancer | 50 | Female |
| P14 | pancreas cancer | 62 | Male |
| P15 | squamous epithelium cancer of the base of the tongue | 57 | Male |
| P16 | acute myeloid leukemia | 64 | Female |
| P17 | serous adeno cancer of the tube | 57 | Female |
| P18 | acute myeloid leukemia | 82 | Male |
| Mutation | Patient |
|---|---|
| Ala519_Glu521delinsAlaGlnTer | P16, P03, P04, P17, P08 |
| Cys555Ter | P16, P03, P13, P08, P15 |
| Glu1065Ter | P16, P01, P02, P03, P05, P06, P13, P08, P11, P09, P10 |
| Glu507Ter | P03, P04, P17, P08 |
| Glu521Ter | P16, P03, P04, P17, P13, P08 |
| Leu562Ter | P16, P18, P02, P03, P05, P06, P17, P13, P08, P11, P12, P15, P14, P09 |
| Lys498Ter | P03, P04, P17, P08 |
| Lys537Ter | P16, P03, P17, P13, P08 |
| Lys540Ter | P16, P03, P17, P13, P08 |
| Ser546Ter | P16, P03, P13, P08 |
| Thr1082Ter | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 |
| Trp520Ter | P16, P03, P04, P17, P08 |
| Tyr518Ter | P16, P03, P04, P17, P08 |
| Tyr544Ter | P16, P03, P13, P08 |
| Mutation | Patient | Mutation | Patient | Mutation | Patient |
|---|---|---|---|---|---|
| Ala1104Asp | P16, P01, P03, P04, P07, P17, P13, P08, P11, P12 | His494Leu | P03, P04, P17, P08 | Phe1064Cys | P16, P01, P02, P03, P05, P06, P13, P08, P11, P09, P10 |
| Ala493Ser | P03, P04, P17, P08 | His494Phe | P03, P04, P17, P08 | Phe1064Val | P16, P01, P02, P03, P05, P06, P13, P08, P11, P09, P10 |
| Ala519Thr | P16, P03, P04, P17, P08 | His494Tyr | P03, P04, P17, P08 | Phe1076Leu | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 |
| Ala523_Asp526delinsValGluLeuSer | P16, P03, P04, P17, P13, P08 | His701Pro | P01, P01, P03, P03, P07, P07, P13, P13, P08, P08, P15, P15 | Phe1076Ser | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 |
| Ala523Val | P16, P03, P04, P17, P13, P08 | Ile1087Leu | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Phe1094_Gly1096delinsTrpSerGln | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 |
| Ala530_Ile531delinsGlyHis | P16, P03, P17, P13, P08 | Ile1087Thr | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Phe1094Cys | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 |
| Ala530Gly | P16, P03, P17, P13, P08 | Ile1091_Lys1092delinsMetGlu | P12 | Phe1094Leu | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P12, P09, P10 |
| Ala543Val | P16, P03, P13, P08 | Ile1091Asn | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Phe1099Leu | P16, P01, P03, P07, P13, P08, P11, P12 |
| Ala547Asp | P16, P03, P13, P08 | Ile1091Met | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P12, P09, P10 | Phe1099Ser | P16, P01, P03, P07, P13, P08, P11, P12 |
| Ala547Ser | P16, P03, P13, P08 | Ile1091Val | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Phe314Ala | P07, P13, P10 |
| Ala693_Glu694delinsMetLeu | P01, P03, P07, P13, P08, P15 | Ile1102Leu | P16, P01, P03, P04, P07, P17, P13, P08, P11, P12 | Phe314Ser | P07, P13, P10 |
| Ala693Thr | P01, P03, P07, P13, P08, P15 | Ile1102Pro | P16, P01, P03, P04, P07, P17, P13, P08, P11, P12 | Phe314Val | P07, P13, P10 |
| Ala693Val | P01, P03, P07, P13, P08, P15 | Ile1102Thr | P16, P01, P03, P04, P07, P17, P13, P08, P11, P12 | Phe321Val | P07, P13, P10 |
| Ala703Pro | P01, P03, P07, P13, P08, P15 | Ile345Leu | P04, P07, P13, P10 | Phe325Leu | P07, P13, P10 |
| Ala703Val | P01, P03, P07, P13, P08, P15 | Ile508Asn | P03, P04, P17, P08 | Phe524Leu | P16, P03, P04, P17, P13, P08 |
| Arg1066Pro | P16, P01, P02, P03, P05, P06, P13, P08, P11, P09, P10 | Ile508Leu | P03, P04, P17, P08 | Phe524Tyr | P16, P03, P04, P17, P13, P08 |
| Arg1075_Ser1077delinsLysProGly | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Ile512_Lys513delinsArgGlu | P16, P03, P04, P17, P08 | Phe524Val | P16, P03, P04, P17, P13, P08 |
| Arg1075His | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Ile512_Lys513delinsSerGlu | P03, P08 | Phe551Ser | P16, P03, P13, P08, P15 |
| Arg1075Ser | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Ile512Ser | P16, P03, P04, P17, P08 | Phe558Ile | P16, P03, P13, P08, P15 |
| Arg501Leu | P03, P04, P17, P08 | Ile531Asn | P16, P03, P17, P13, P08 | Phe558Ser | P16, P03, P13, P08, P15 |
| Arg532Met | P16, P03, P17, P13, P08 | Ile531Leu | P16, P03, P04, P17, P13, P08 | Phe565Leu | P16, P18, P02, P03, P05, P06, P17, P13, P08, P11, P12, P15, P14, P09 |
| Arg532Phe | P16, P03, P17, P13, P08 | Ile704Phe | P01, P03, P07, P13, P08, P15 | Phe565Ser | P16, P18, P02, P03, P05, P06, P17, P13, P08, P11, P12, P15, P14, P09 |
| Arg532Ser | P16, P03, P17, P13, P08 | Ile704Thr | P01, P03, P07, P13, P08, P15 | Phe565Val | P16, P18, P02, P03, P05, P06, P17, P13, P08, P11, P12, P15, P14, P09 |
| Arg532Trp | P16, P03, P17, P13, P08 | Leu1098_Val1101delinsProProValSer | P16, P01, P03, P07, P13, P08, P11, P12 | Pro1068Ala | P16, P01, P02, P03, P05, P06, P13, P08, P11, P09, P10 |
| Asn1071His | P16, P01, P02, P03, P05, P06, P07, P07, P13, P08, P11, P09, P10 | Leu1098Pro | P16, P01, P02, P02, P03, P05, P05, P06, P06, P07, P13, P08, P11, P12, P09, P09, P10, P10 | Pro1068Leu | P16, P01, P02, P03, P05, P06, P13, P08, P11, P09, P10 |
| Asn1071Lys | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Leu313Val | P07, P13, P10 | Pro1088Gln | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 |
| Asn1071Thr | P16, P01, P02, P03, P05, P06, P13, P08, P11, P09, P10 | Leu317Val | P07, P13, P10 | Pro1088Lys | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 |
| Asn1074Ile | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Leu504_Met505delinsHisLeu | P03, P04, P17, P08 | Pro1088Thr | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 |
| Asn1074Phe | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Leu504Gln | P03, P04, P17, P08 | Pro557_Phe558delinsArgThr | P16, P03, P13, P08, P15 |
| Asn1074Tyr | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Leu509Phe | P03, P04, P17, P08 | Pro557_Phe558delinsLeuThr | P16, P03, P13, P08, P15 |
| Asn1100Asp | P16, P01, P02, P03, P04, P06, P07, P17, P13, P08, P11, P12 | Leu515Ile | P16, P03, P04, P17, P08 | Pro557Leu | P16, P03, P13, P08, P15 |
| Asn1100Ile | P16, P01, P02, P03, P06, P07, P13, P08, P11, P12 | Leu515Pro | P16, P03, P04, P17, P08 | Ser1069Arg | P16, P01, P02, P03, P05, P06, P13, P08, P11, P09, P10 |
| Asn1100Lys | P16, P01, P03, P07, P13, P08, P11, P12 | Leu517Val | P16, P03, P04, P17, P08 | Ser1069Thr | P16, P01, P02, P03, P05, P06, P13, P08, P11, P09, P10 |
| Asn1100Val | P02, P06 | Leu522Arg | P16, P16, P03, P03, P04, P04, P17, P17, P13, P13, P08, P08 | Ser1077Ala | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 |
| Asn310Asp | P07, P13, P10 | Leu529Met | P16, P03, P04, P17, P13, P08 | Ser1077Cys | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 |
| Asn500Asp | P03, P04, P17, P08 | Leu529Pro | P16, P03, P04, P17, P13, P08 | Ser1085Phe | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 |
| Asn500Ile | P03, P04, P17, P08 | Leu536_Val538delinsHisCysArg | P16, P03, P17, P13, P08 | Ser1097Cys | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P12, P09, P10 |
| Asn500Lys | P03, P04, P17, P08 | Leu536Gln | P16, P03, P17, P13, P08 | Ser1097Trp | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P12, P09, P10 |
| Asn506His | P03, P04, P17, P08 | Leu559Pro | P16, P03, P13, P08, P15 | Ser497_Arg501delinsArgCysSerValIle | P03, P04, P17, P08 |
| Asn506Ile | P03, P04, P17, P08 | Leu562_Phe565delinsTerLysAspAla | P16, P18, P02, P03, P05, P06, P17, P13, P08, P11, P12, P15, P14, P09 | Ser497Arg | P03, P04, P17, P08 |
| Asn510His | P03, P04, P17, P08 | Leu691His | P01, P03, P07, P13, P08, P15 | Ser497Cys | P03, P04, P17, P08 |
| Asn510Pro | P03, P04, P17, P08 | Leu691Phe | P01, P03, P07, P13, P08, P15 | Ser542Leu | P16, P03, P17, P13, P08 |
| Asn510Thr | P03, P04, P17, P08 | Leu691Tyr | P01, P03, P07, P13, P08, P15 | Ser542Phe | P16, P03, P17, P13, P08 |
| Asp1081Phe | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Leu692Trp | P01, P03, P07, P13, P08, P15 | Ser542Pro | P16, P03, P17, P13, P08 |
| Asp1081Tyr | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Lys1078_Glu1079delinsAsnLys | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Ser546_Ala547delinsGlyTyr | P16, P03, P13, P08 |
| Asp1081Val | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Lys1078Asn | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10, P12 | Ser546Ala | P16, P03, P13, P08 |
| Asp1084Val | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Lys1092Asn | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P12, P09, P10 | Thr1067_Ser1069delinsThrValPro | P16, P01, P02, P03, P05, P06, P13, P08, P11, P09, P10 |
| Asp292His | P07, P13, P10 | Lys1092Glu | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Thr1067Ala | P16, P01, P02, P03, P05, P06, P13, P08, P11, P09, P10 |
| Asp499Ala | P03, P04, P17, P08 | Lys1092Met | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Thr1082Arg | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 |
| Asp499Tyr | P03, P04, P17, P08 | Lys315Thr | P07, P13, P10 | Thr1082Ser | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 |
| Asp526Ala | P16, P03, P04, P17, P13, P08 | Lys319_Phe321delinsAsnLeuVal | P07, P13, P10 | Thr1242Leu | P01, P02, P03, P13, P11, P14 |
| Asp526Glu | P16, P03, P04, P17, P13, P08 | Lys319Asn | P07, P13, P10 | Thr1242Met | P16, P01, P02, P03, P07, P17, P13, P08, P11, P14, P10 |
| Asp526Tyr | P16, P03, P04, P17, P13, P08, P01, P03, P07, P13, P08, P15 | Lys319Glu | P07, P13, P10 | Thr1242Pro | P01, P02, P03, P13, P11, P14 |
| Asp696Tyr | P01, P03, P07, P13, P08, P15 | Lys496_Ser497delinsLeuCys | P03, P04, P17, P08 | Thr320Ile | P07, P13, P10 |
| Asp696Val | P01, P03, P07, P13, P08, P15 | Lys496Asn | P03, P04, P17, P08 | Thr320Pro | P07, P13, P10 |
| Cys555Arg | P16, P03, P13, P08, P15 | Lys496Gln | P03, P04, P17, P08 | Thr550Ala | P16, P03, P13, P08 |
| Cys555Ser | P16, P03, P13, P08, P15 | Lys496Met | P03, P04, P17, P08 | Thr550Gly | P16, P03, P13, P08 |
| Cys563Ser | P16, P18, P02, P03, P05, P06, P17, P13, P08, P11, P12, P15, P14, P09 | Lys498Arg | P03, P04, P17, P08 | Thr550Ser | P16, P03, P13, P08, P15 |
| Cys563Trp | P16, P18, P02, P03, P05, P06, P17, P13, P08, P11, P12, P15, P14, P09 | Lys498Asn | P03, P04, P17, P08 | Thr552_Thr556delinsTrpLeuArgProGly | P16, P03, P13, P08, P15 |
| Cys563Tyr | P16, P18, P02, P03, P05, P06, P17, P13, P08, P11, P12, P15, P14, P09 | Lys503Arg | P03, P04, P17, P08 | Thr552Ser | P16, P16, P03, P03, P13, P13, P08, P08, P15, P15 |
| Gln344_Ile345delinsHisLeu | P04, P07, P13, P10 | Lys503Asn | P03, P04, P17, P08 | Thr556Ala | P16, P03, P13, P08, P15 |
| Gln344His | P04, P07, P13, P10 | Lys503Glu | P03, P04, P17, P08 | Thr556Arg | P16, P03, P13, P08, P15 |
| Gln533_Glu535delinsHisLeuGln | P16, P03, P17, P13, P08 | Lys503Gly | P03, P04, P17, P08 | Thr564Ala | P16, P18, P02, P03, P05, P06, P17, P13, P08, P11, P12, P15, P14, P09 |
| Gln533His | P16, P03, P17, P13, P08 | Lys513_Leu515delinsLysGlnThr | P16, P03, P04, P17, P08 | Thr564Lys | P16, P18, P02, P03, P05, P06, P17, P13, P08, P11, P12, P15, P14, P09 |
| Glu1065Asp | P16, P01, P02, P03, P05, P06, P13, P08, P11, P09, P10 | Lys513Glu | P16, P03, P04, P17, P08 | Trp309_Asn310delinsCysAsp | P07, P13, P10 |
| Glu1065Gly | P16, P01, P02, P03, P05, P06, P13, P08, P11, P09, P10 | Lys516Gln | P16, P03, P04, P17, P08 | Trp309Arg | P07, P13, P10 |
| Glu1079Lys | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Lys525Gln | P16, P03, P04, P17, P13, P08 | Trp309Cys | P07, P13, P10 |
| Glu1089_Met1093delinsArgProGluValLeu | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Lys525Met | P16, P03, P04, P17, P13, P08 | Trp520Arg | P16, P03, P04, P17, P08 |
| Glu1089Gln | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Lys527Met | P16, P03, P04, P17, P13, P08 | Trp553Arg | P16, P03, P13, P08, P15 |
| Glu1089Gly | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Lys537Arg | P16, P03, P17, P13, P08 | Trp553Cys | P16, P03, P13, P08, P15 |
| Glu308_Trp309delinsAspArg | P07, P13, P10 | Lys537Asn | P16, P03, P17, P13, P08 | Trp553Leu | P16, P03, P13, P08, P15 |
| Glu308Asp | P07, P13, P10 | Lys540_Lys541delinsCysGly | P16, P03, P17, P13, P08 | Tyr1243Arg | P16, P01, P02, P03, P07, P17, P13, P08, P11, P14, P10 |
| Glu507_Ile508delinsCysHis | P03, P04, P17, P08 | Lys540Arg | P16, P03, P17, P13, P08 | Tyr1243Cys | P16, P01, P02, P03, P07, P17, P13, P08, P11, P14, P10 |
| Glu507Asp | P03, P04, P17, P08 | Lys540Asn | P16, P03, P17, P13, P08 | Tyr1243His | P16, P01, P02, P03, P07, P17, P13, P08, P11, P14, P10 |
| Glu507Gly | P03, P04, P17, P08 | Lys541Arg | P16, P03, P17, P13, P08 | Tyr518_Ala519delinsTerThr | P16, P03, P04, P17, P08 |
| Glu521Asp | P16, P03, P04, P17, P13, P08 | Lys541Glu | P16, P03, P17, P13, P08 | Tyr544His | P16, P03, P13, P08 |
| Glu534Asp | P16, P03, P17, P13, P08 | Lys697Asn | P01, P03, P07, P13, P08, P15 | Tyr568Ser | P02, P03, P05, P13, P08, P09 |
| Glu534Gln | P16, P03, P17, P13, P08 | Lys697Gln | P01, P03, P07, P13, P08, P15 | Val1073Leu | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 |
| Glu534Val | P16, P03, P17, P13, P08 | Lys697Thr | P01, P03, P07, P13, P08, P15 | Val1083Gly | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 |
| Glu535Gln | P16, P03, P17, P13, P08 | Lys705Arg | P01, P03, P07, P13, P08, P15 | Val1090Ala | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 |
| Glu694Gln | P01, P03, P07, P13, P08, P15 | Lys705Glu | P01, P03, P07, P13, P08, P15 | Val1090Leu | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 |
| Glu694Val | P01, P03, P07, P13, P08, P15 | Met1086_Ile1087delinsSerPro | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Val1101Ala | P16, P01, P03, P04, P07, P17, P13, P08, P11, P12 |
| Glu699_Gly700delinsGlyHis | P01, P03, P07, P13, P08, P15 | Met1086Ile | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Val1101Phe | P16, P01, P03, P07, P13, P08, P11, P12 |
| Glu699Asp | P01, P03, P07, P13, P08, P15 | Met1086Leu | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Val1240Gly | P01, P02, P03, P13, P11, P14 |
| Glu699Gly | P01, P03, P07, P13, P08, P15 | Met1086Thr | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Val316Ala | P07, P13, P10 |
| Gly1070_Asn1071delinsHisPro | P16, P01, P02, P03, P05, P06, P13, P08, P11, P09, P10 | Met1093Ile | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Val492Gly | P03, P04, P17, P08 |
| Gly1070Arg | P16, P01, P02, P03, P05, P06, P13, P08, P11, P09, P10 | Met1093Leu | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Val492Met | P03, P04, P17, P08 |
| Gly1070Glu | P16, P01, P02, P03, P05, P06, P13, P08, P11, P09, P10 | Met1095Ile | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Val514Glu | P16, P03, P04, P17, P08 |
| Gly1096Arg | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P12, P09, P10 | Met1095Leu | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P09, P10 | Val514Leu | P16, P03, P04, P17, P08 |
| Gly1096Asp | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P12, P09, P10 | Met1095Thr | P16, P01, P02, P03, P05, P06, P07, P13, P08, P11, P12, P09, P10 | Val528_Leu529delinsThrThr | P16, P03, P04, P17, P13, P08 |
| Gly1096Gln | P12 | Met495Arg | P03, P03, P04, P04, P17, P17, P08, P08 | Val528Ala | P16, P03, P04, P17, P13, P08 |
| Gly1103Ala | P16, P01, P03, P04, P07, P17, P13, P08, P11, P12 | Met495Leu | P03, P04, P17, P08 | Val528Met | P16, P03, P04, P17, P13, P08 |
| Gly1103Arg | P16, P01, P03, P04, P07, P17, P13, P08, P11, P12 | Met505_Asn506delinsIleLeu | P03, P04, P17, P08 | Val538Gly | P16, P03, P17, P13, P08 |
| Gly1103Pro | P16, P01, P03, P04, P07, P17, P13, P08, P11, P12 | Met505Ile | P03, P04, P17, P08 | Val538Met | P16, P03, P17, P13, P08 |
| Gly511Ala | P03, P04, P17, P08 | Met505Leu | P03, P04, P17, P08 | Val548_Gly549delinsGlyCys | P16, P03, P13, P08 |
| Gly511Arg | P03, P04, P17, P08 | Met695_Lys697delinsSerPhePro | P01, P03, P07, P13, P08, P15 | Val548Gly | P16, P03, P13, P08 |
| Gly511Pro | P03, P03, P04, P17, P08, P08 | Met695Ile | P01, P03, P07, P13, P08, P15 | Val554Gly | P16, P03, P13, P08, P15 |
| Gly549Cys | P16, P03, P13, P08 | Met695Leu | P01, P03, P07, P13, P08, P15 | Val554Ile | P16, P03, P13, P08, P15 |
| Gly700Arg | P01, P03, P07, P13, P08, P15 | Met695Thr | P01, P03, P07, P13, P08, P15 | Val698Ala | P01, P03, P07, P13, P08, P15 |
| Gly700Glu | P01, P03, P07, P13, P08, P15 | Phe1064_Thr1067delinsGlyCysThrAla | P16, P01, P02, P03, P05, P06, P13, P08, P11, P09, P10 | Val702_Lys705delinsAlaLeuSerGly | P01, P03, P07, P13, P08, P15 |
| Val702Ala | P01, P03, P07, P13, P08, P15 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kadioglu, O.; Saeed, M.; Munder, M.; Spuller, A.; Greten, H.J.; Efferth, T. Identification of Novel Rare ABCC1 Transporter Mutations in Tumor Biopsies of Cancer Patients. Cells 2020, 9, 299. https://doi.org/10.3390/cells9020299
Kadioglu O, Saeed M, Munder M, Spuller A, Greten HJ, Efferth T. Identification of Novel Rare ABCC1 Transporter Mutations in Tumor Biopsies of Cancer Patients. Cells. 2020; 9(2):299. https://doi.org/10.3390/cells9020299
Chicago/Turabian StyleKadioglu, Onat, Mohamed Saeed, Markus Munder, Andreas Spuller, Henry Johannes Greten, and Thomas Efferth. 2020. "Identification of Novel Rare ABCC1 Transporter Mutations in Tumor Biopsies of Cancer Patients" Cells 9, no. 2: 299. https://doi.org/10.3390/cells9020299
APA StyleKadioglu, O., Saeed, M., Munder, M., Spuller, A., Greten, H. J., & Efferth, T. (2020). Identification of Novel Rare ABCC1 Transporter Mutations in Tumor Biopsies of Cancer Patients. Cells, 9(2), 299. https://doi.org/10.3390/cells9020299







