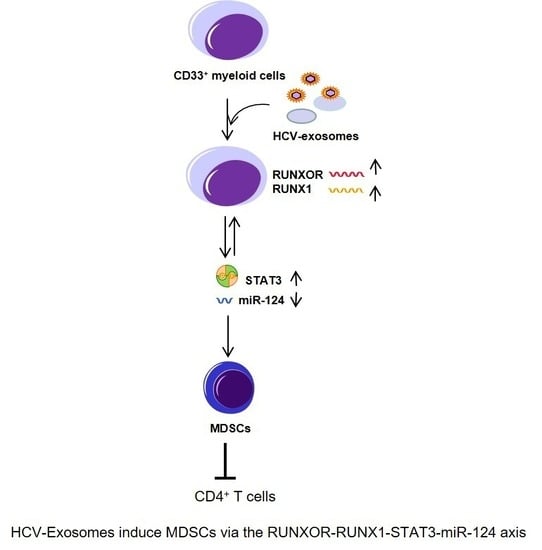
HCV-Associated Exosomes Upregulate RUNXOR and RUNX1 Expressions to Promote MDSC Expansion and Suppressive Functions through STAT3–miR124 Axis
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Cell Isolation, Culture, and Flow Cytometric Analysis
2.3. Exosome Isolation and Purification
2.4. lncRNA and miRNA Arrays and RT-qPCR Validation
2.5. Transfection and Co-Culture Experiments
2.6. Statistical Analysis
3. Results
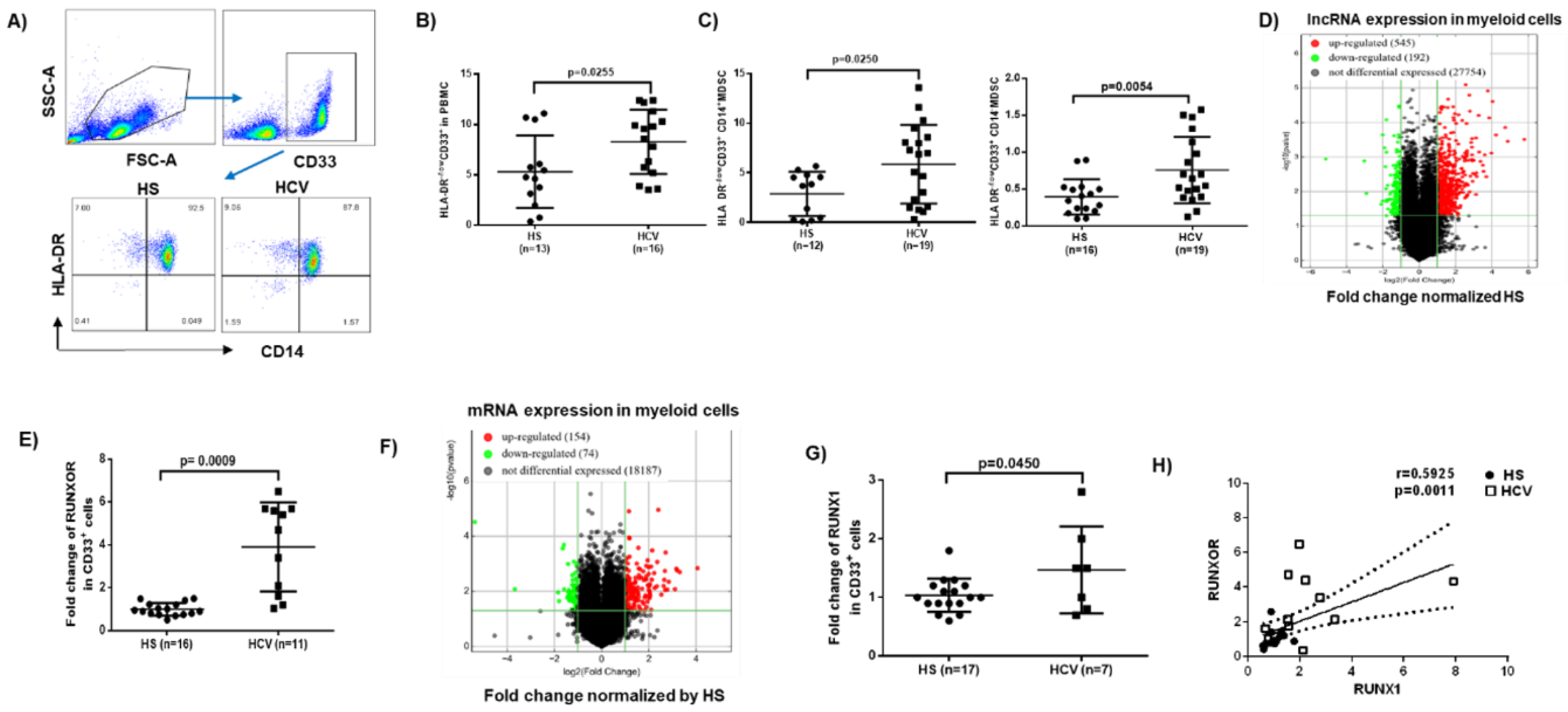
3.1. MDSCs Accumulate in Peripheral Blood during Chronic HCV Infection
3.2. RUNXOR and RUNX1 Are Upregulated in MDSCs during Chronic HCV Infection
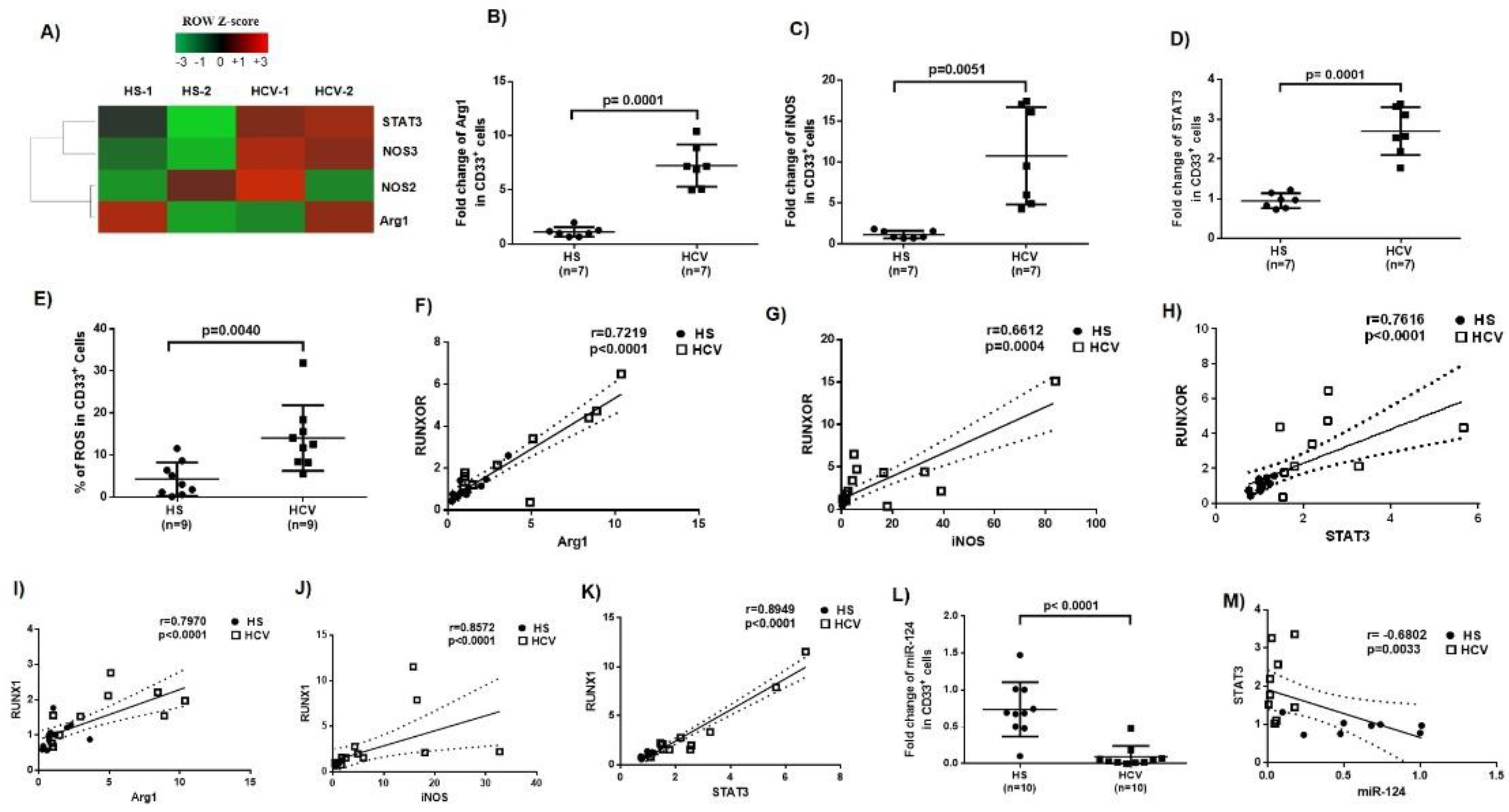
3.3. Immunosuppressive Molecules Are Elevated in MDSCs during Chronic HCV Infection
3.4. MiR-124 Expression Negatively Correlates with STAT3 Levels in MDSCs during HCV Infection
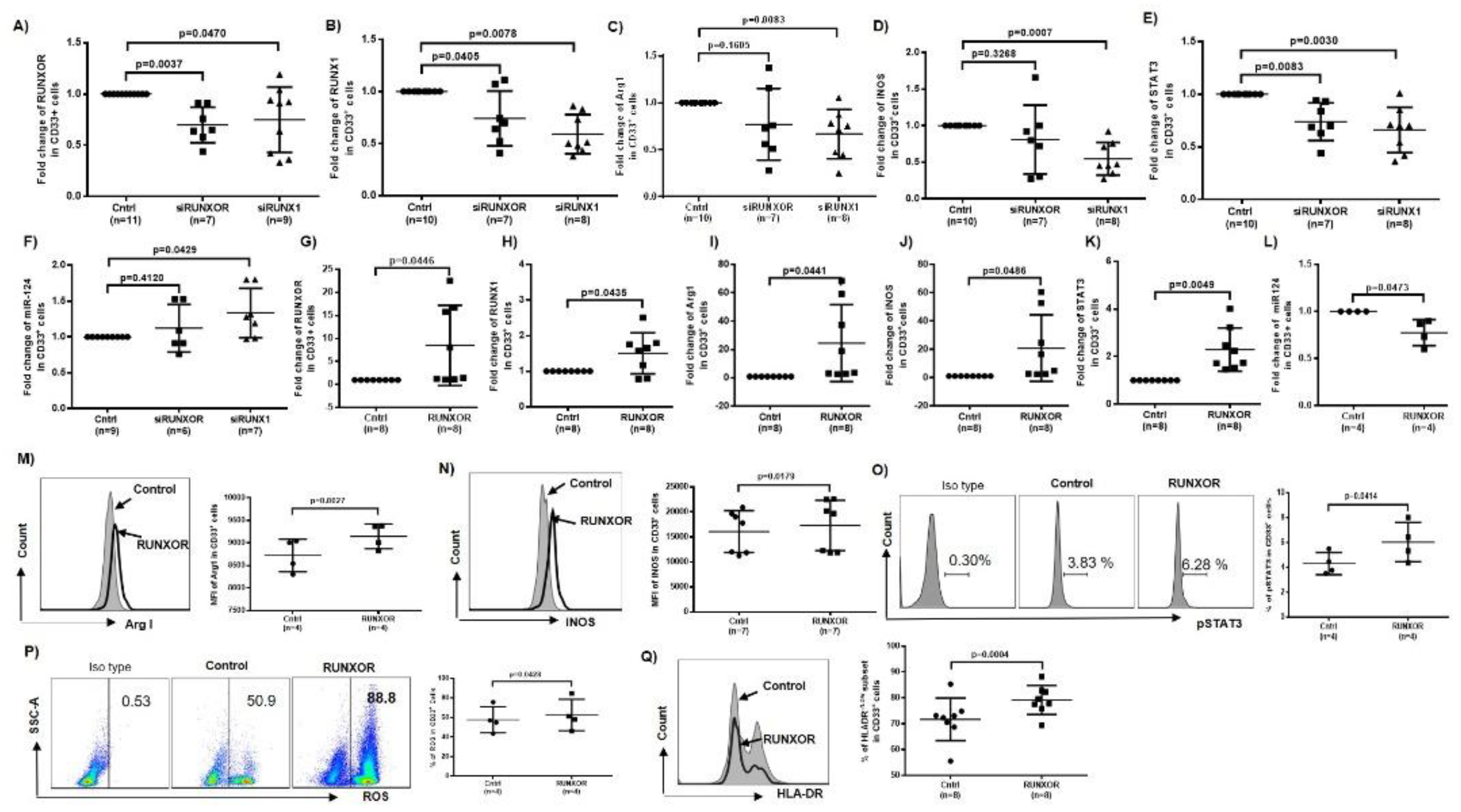
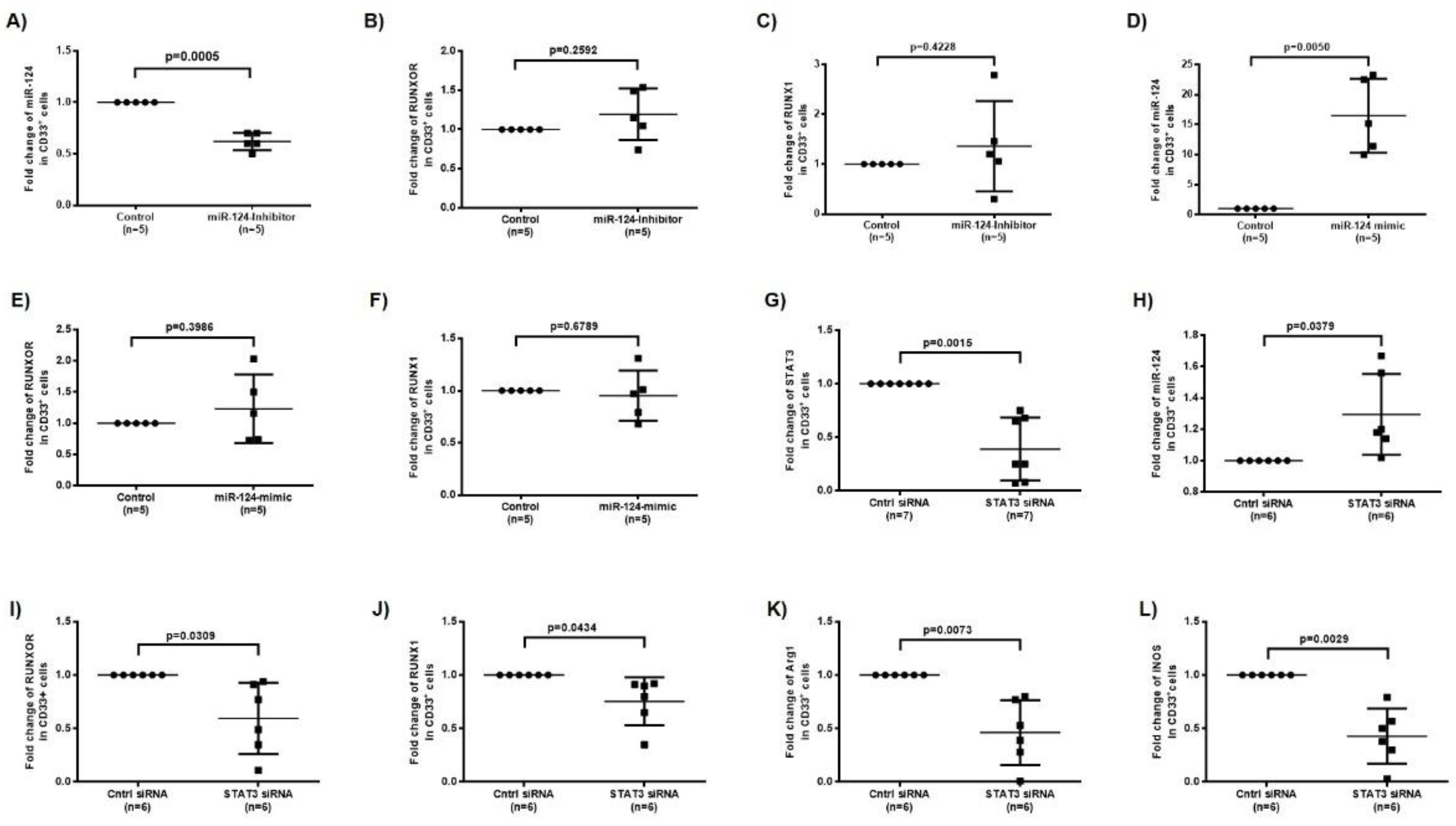
3.5. RUNXOR and RUNX1 Regulate Each Other’s and Control miR-124 Expression via the STAT3 Signaling in MDSCs during HCV Infection
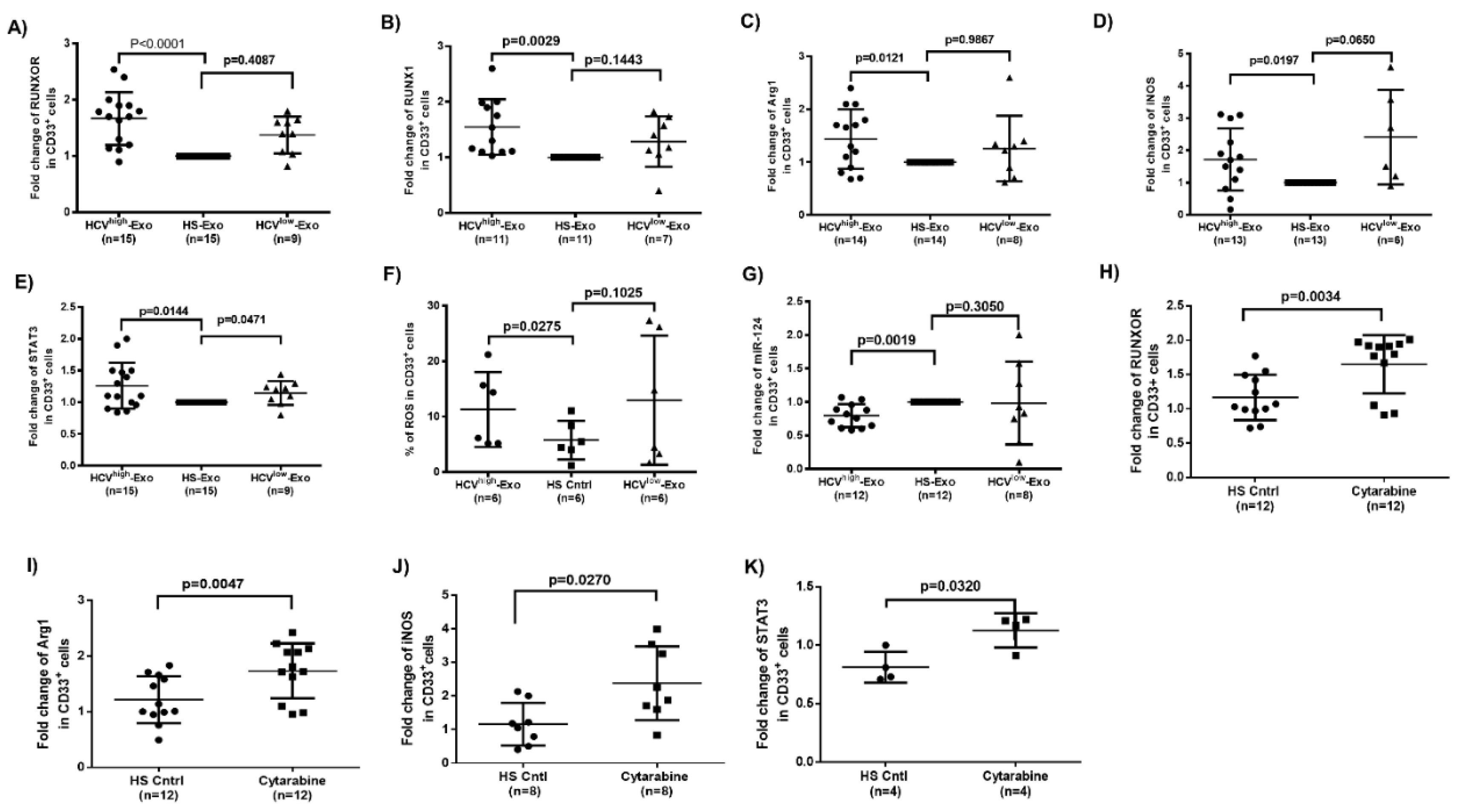
3.6. HCV-Associated Exosomes Regulate RUNXOR, RUNX1, and Suppressive Molecule Expressions in MDSCs
3.7. Cytarabine Regulates RUNXOR and Immunosuppressive Molecules in Healthy CD33+ Cells
3.8. Silencing RUNXOR and RUNX1 Expressions Reduce MDSC Frequencies and Suppressive Functions
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rosen, H.R. Emerging concepts in immunity to hepatitis C virus infection. J. Clin. Investig. 2013, 123, 4121–4130. [Google Scholar] [CrossRef]
- Park, S.-H.; Rehermann, B. Immune Responses to HCV and Other Hepatitis Viruses. Immunity 2014, 40, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Manns, M.P.; Von Hahn, T. Novel therapies for hepatitis C—One pill fits all? Nat. Rev. Drug Discov. 2013, 12, 595–610. [Google Scholar] [CrossRef] [PubMed]
- Pawlotski, J.-M. Hepatitis C Virus Resistance to Direct-Acting Antiviral Drugs in Interferon-Free Regimens. Gastroenterology 2016, 151, 70–86. [Google Scholar] [CrossRef]
- Vranjkovic, A.; Deonarine, F.; Kaka, S.; Angel, J.B.; Cooper, C.L.; Crawley, A.M. Direct-Acting Antiviral Treatment of HCV Infection Does Not Resolve the Dysfunction of Circulating CD8+ T-Cells in Advanced Liver Disease. Front. Immunol. 2019, 10, 1926. [Google Scholar] [CrossRef] [PubMed]
- Ostrand-Rosenberg, S.; Sinha, P. Myeloid-Derived Suppressor Cells: Linking Inflammation and Cancer. J. Immunol. 2009, 182, 4499–4506. [Google Scholar] [CrossRef]
- Gabrilovich, D.I.; Nagaraj, S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009, 9, 162–174. [Google Scholar] [CrossRef]
- Dai, J.; El Gazzar, M.; Li, G.Y.; Moorman, J.P.; Yao, Z.Q. Myeloid-Derived Suppressor Cells: Paradoxical Roles in Infection and Immunity. J. Innate Immun. 2015, 7, 116–126. [Google Scholar] [CrossRef]
- Huang, A.; Zhang, B.; Yan, W.; Wang, B.; Wei, H.; Zhang, F.; Wu, L.; Fan, K.-X.; Guo, Y. Myeloid-Derived Suppressor Cells Regulate Immune Response in Patients with Chronic Hepatitis B Virus Infection through PD-1–Induced IL-10. J. Immunol. 2014, 193, 5461–5469. [Google Scholar] [CrossRef]
- Vollbrecht, T.; Stirner, R.; Tufman, A.; Roider, J.; Huber, R.M.; Bogner, J.R.; Lechner, A.; Bourquin, C.; Draenert, R. Chronic progressive HIV-1 infection is associated with elevated levels of myeloid-derived suppressor cells. AIDS 2012, 26, F31–F37. [Google Scholar] [CrossRef]
- Bowers, N.L.; Helton, E.S.; Huijbregts, R.P.H.; Goepfert, P.A.; Heath, S.L.; Hel, Z. Immune Suppression by Neutrophils in HIV-1 Infection: Role of PD-L1/PD-1 Pathway. PLoS Pathog. 2014, 10, e1003993. [Google Scholar] [CrossRef]
- Norris, B.A.; Uebelhoer, L.S.; Nakaya, H.I.; Price, A.A.; Grakoui, A.; Pulendran, B. Chronic but Not Acute Virus Infection Induces Sustained Expansion of Myeloid Suppressor Cell Numbers that Inhibit Viral-Specific T Cell Immunity. Immunity 2013, 38, 309–321. [Google Scholar] [CrossRef]
- Tacke, R.S.; Lee, H.-C.; Goh, C.; Courtney, J.; Polyak, S.J.; Rosen, H.R.; Hahn, Y.S. Myeloid suppressor cells induced by hepatitis C virus suppress T-cell responses through the production of reactive oxygen species. Hepatology 2011, 55, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Qin, A.; Guo, P.; Yan, D.; Hu, F.; Yang, Q.; Xu, M.; Fu, Y.; Zhou, J.; Tang, X. Clinical Significance and Functional Studies of Myeloid-Derived Suppressor Cells in Chronic Hepatitis C Patients. J. Clin. Immunol. 2013, 33, 798–808. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhao, J.; Ren, J.P.; Wu, X.Y.; Morrison, Z.D.; A Elgazzar, M.; Ning, S.B.; Moorman, J.P.; Yao, Z.Q. Expansion of myeloid-derived suppressor cells promotes differentiation of regulatory T cells in HIV-1+ individuals. AIDS 2016, 30, 1521–1531. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.P.; Zhao, J.; Dai, J.; Griffin, J.W.D.; Wang, L.; Wu, X.Y.; Morrison, Z.D.; Li, G.Y.; El Gazzar, M.; Ning, S.; et al. Hepatitis C virus-induced myeloid-derived suppressor cells regulate T-cell differentiation and function via the signal transducer and activator of transcription 3 pathway. Immunology 2016, 148, 377–386. [Google Scholar] [CrossRef]
- Ren, J.P.; Wang, L.; Zhao, J.; Ning, S.; El Gazzar, M.; Moorman, J.P.; Yao, Z.Q.; Wang, L. Decline of miR-124 in myeloid cells promotes regulatory T-cell development in hepatitis C virus infection. Immunology 2016, 150, 213–220. [Google Scholar] [CrossRef]
- Wang, L.; Cao, D.; Zhao, J.; Nguyen, L.N.; Dang, X.; Ji, Y.; Wu, X.Y.; Morrison, Z.D.; Xie, Q.; El Gazzar, M.; et al. HCV-associated exosomes promote myeloid-derived suppressor cell expansion via inhibiting miR-124 to regulate T follicular cell differentiation and function. Cell Discov. 2018, 4, 1–15. [Google Scholar] [CrossRef]
- Jandura, A.; Krause, H.M. The New RNA World: Growing Evidence for Long Noncoding RNA Functionality. Trends Genet. 2017, 33, 665–676. [Google Scholar] [CrossRef]
- Morceau, F.; Chateauvieux, S.; Gaigneaux, A.; Dicato, M.; Diederich, M. Long and Short Non-Coding RNAs as Regulators of Hematopoietic Differentiation. Int. J. Mol. Sci. 2013, 14, 14744–14770. [Google Scholar] [CrossRef]
- Zhang, X.; Hong, R.; Chen, W.; Xu, M.; Wang, L. The role of long noncoding RNA in major human disease. Bioorg. Chem. 2019, 92, 103214. [Google Scholar] [CrossRef] [PubMed]
- The ENCODE Project Consortium Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nat. Cell Biol. 2007, 447, 799–816. [CrossRef]
- Tian, X.; Tian, J.; Tang, X.; Ma, J.; Wang, S. Long non-coding RNAs in the regulation of myeloid cells. J. Hematol. Oncol. 2016, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Zhao, Z.; Wu, T.; Yin, X.; Zhou, Y.; Wang, Z.; Shen, S.; Qiu, Y. Roles of noncoding RNAs in metastasis of nonsmall cell lung cancer: A mini review. J. Cancer Res. Ther. 2015, 11, 7. [Google Scholar] [CrossRef]
- Wan, L.; Sun, M.; Liu, G.-J.; Wei, C.-C.; Zhang, E.-B.; Kong, R.; Xu, T.-P.; Huang, M.-D.; Wang, Z.-X. Long Noncoding RNA PVT1 Promotes Non-Small Cell Lung Cancer Cell Proliferation through Epigenetically Regulating LATS2 Expression. Mol. Cancer Ther. 2016, 15, 1082–1094. [Google Scholar] [CrossRef]
- Peng, H.; Liu, Y.; Tian, J.; Ma, J.; Tang, X.; Rui, K.; Tian, X.; Mao, C.; Lu, L.; Xu, H.; et al. The Long Noncoding RNA IFNG-AS1 Promotes T Helper Type 1 Cells Response in Patients with Hashimoto’s Thyroiditis. Sci. Rep. 2015, 5, 17702. [Google Scholar] [CrossRef]
- Reddy, M.A.; Chen, Z.; Park, J.T.; Wang, M.; Lanting, L.; Zhang, Q.; Bhatt, K.; Leung, A.; Wu, X.; Putta, S.; et al. Regulation of Inflammatory Phenotype in Macrophages by a Diabetes-Induced Long Noncoding RNA. Diabetes 2014, 63, 4249–4261. [Google Scholar] [CrossRef]
- Nie, Y.; Zhou, L.; Wang, H.; Chen, N.; Jia, L.; Wang, C.; Wang, Y.; Chen, J.; Wen, X.; Niu, C.; et al. Profiling the epigenetic interplay of lncRNA RUNXOR and oncogenic RUNX1 in breast cancer cells by gene in situ cis-activation. Am. J. Cancer Res. 2019, 9, 1635–1649. [Google Scholar]
- Wang, H.; Li, W.; Guo, R.; Sun, J.; Cui, J.; Wang, G.; Hoffman, A.R.; Hu, J.-F. An intragenic long noncoding RNA interacts epigenetically with theRUNX1promoter and enhancer chromatin DNA in hematopoietic malignancies. Int. J. Cancer 2014, 135, 2783–2794. [Google Scholar] [CrossRef]
- Tian, X.; Ma, J.; Wang, T.; Tian, J.; Zheng, Y.; Peng, R.; Wang, Y.; Zhang, Y.; Mao, L.; Xu, H.; et al. Long non-coding RNA RUNXOR accelerates MDSC-mediated immunosuppression in lung cancer. BMC Cancer 2018, 18, 1–10. [Google Scholar] [CrossRef]
- Keller, S.; Sanderson, M.P.; Stoeck, A.; Altevogt, P. Exosomes: From biogenesis and secretion to biological function. Immunol. Lett. 2006, 107, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Pegtel, D.M.; Cosmopoulos, K.; Thorley-Lawson, D.A.; van Eijndhoven, M.A.; Hopmans, E.S.; Lindenberg, J.L.; de Gruijl, T.D.; Würdinger, T.; Middeldorp, J.M. Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. USA 2010, 107, 6328–6333. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Tvall, J.O.L.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Masciopinto, F.; Giovani, C.; Campagnoli, S.; Galli-Stampino, L.; Colombatto, P.; Brunetto, M.; Yen, T.S.B.; Houghton, M.; Pileri, P.; Abrignani, S. Association of hepatitis C virus envelope proteins with exosomes. Eur. J. Immunol. 2004, 34, 2834–2842. [Google Scholar] [CrossRef]
- Bukong, T.N.; Momen-Heravi, F.; Kodys, K.; Bala, S.; Szabo, G. Exosomes from Hepatitis C Infected Patients Transmit HCV Infection and Contain Replication Competent Viral RNA in Complex with Ago2-miR122-HSP90. PLoS Pathog. 2014, 10, e1004424. [Google Scholar] [CrossRef]
- Cosset, F.-L.; Dreux, M. HCV transmission by hepatic exosomes establishes a productive infection. J. Hepatol. 2014, 60, 674–675. [Google Scholar] [CrossRef]
- Dreux, M.; Garaigorta, U.; Boyd, B.; Décembre, E.; Chung, J.; Whitten-Bauer, C.; Wieland, S.; Chisari, F.V. Short-Range Exosomal Transfer of Viral RNA from Infected Cells to Plasmacytoid Dendritic Cells Triggers Innate Immunity. Cell Host Microbe 2012, 12, 558–570. [Google Scholar] [CrossRef]
- Achyut, B.R.; Arbab, A.S. Taming immune suppressor: Application of myeloid-derived suppressor cells in anti-cancer gene therapy. Transl. Cancer Res. 2017, 6, S160–S162. [Google Scholar] [CrossRef]
- Bizymi, N.; Bjelica, S.; Kittang, A.O.; Mojsilovic, S.; Velegraki, M.; Pontikoglou, C.; Roussel, M.; Ersvær, E.; Santibañez, J.F.; Lipoldová, M.; et al. Myeloid-Derived Suppressor Cells in Hematologic Diseases: Promising Biomarkers and Treatment Targets. HemaSphere 2019, 3, e168. [Google Scholar] [CrossRef]
- Pawelec, G.; Verschoor, C.P.; Ostrand-Rosenberg, S. Myeloid-Derived Suppressor Cells: Not Only in Tumor Immunity. Front. Immunol. 2019, 10, 1099. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.; Fritz, A.J.; Finstad, K.H.; Fitzgerald, M.P.; Weinheimer, A.; Viens, A.L.; Ramsey, J.; Stein, J.L.; Lian, J.B.; Stein, G.S. Suppression of Breast Cancer Stem Cells and Tumor Growth by the RUNX1 Transcription Factor. Mol. Cancer Res. 2018, 16, 1952–1964. [Google Scholar] [CrossRef]
- Van Bragt, M.P.; Hu, X.; Xie, Y.; Li, Z. RUNX1, a transcription factor mutated in breast cancer, controls the fate of ER-positive mammary luminal cells. eLife 2014, 3, 03881. [Google Scholar] [CrossRef]
- Jeong, M.; Goodell, M.A. Noncoding Regulatory RNAs in Hematopoiesis. Curr. Top. Dev. Biol. 2016, 118, 245–270. [Google Scholar] [CrossRef] [PubMed]
- El Gazzar, M. microRNAs as potential regulators of myeloid-derived suppressor cell expansion. Innate Immun. 2013, 20, 227–238. [Google Scholar] [CrossRef] [PubMed]
- McCartney, E.M.; Helbig, K.J.; Narayana, S.K.; Eyre, N.S.; Aloia, A.L.; Beard, M.R. Signal transducer and activator of transcription 3 is a proviral host factor for hepatitis C virus. Hepatology 2013, 58, 1558–1568. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Hanada, T.; Tokuhisa, T.; Kosai, K.-I.; Sata, M.; Kohara, M.; Yoshimura, A. Activation of STAT3 by the Hepatitis C Virus Core Protein Leads to Cellular Transformation. J. Exp. Med. 2002, 196, 641–653. [Google Scholar] [CrossRef]
- Gong, G.; Waris, G.; Tanveer, R.; Siddiqui, A. Human hepatitis C virus NS5A protein alters intracellular calcium levels, induces oxidative stress, and activates STAT-3 and NF- B. Proc. Natl. Acad. Sci. USA 2001, 98, 9599–9604. [Google Scholar] [CrossRef]
- Zeng, B.; Li, Z.; Chen, R.; Guo, N.; Zhou, J.; Zhou, Q.; Lin, Q.; Cheng, D.; Liao, Q.; Zheng, L.; et al. Epigenetic regulation of miR-124 by Hepatitis C Virus core protein promotes migration and invasion of intrahepatic cholangiocarcinoma cells by targeting SMYD3. FEBS Lett. 2012, 586, 3271–3278. [Google Scholar] [CrossRef]
- Peterson, L.F.; Zhang, D.-E. The 8;21 translocation in leukemogenesis. Oncogene 2004, 23, 4255–4262. [Google Scholar] [CrossRef]
- Speck, N.A.; Gilliland, D.G. Core-binding factors in haematopoiesis and leukaemia. Nat. Rev. Cancer 2002, 2, 502–513. [Google Scholar] [CrossRef]
- Lam, K. RUNX1 and RUNX1-ETO: Roles in hematopoiesis and leukemogenesis. Front. Biosci. 2012, 17, 1120. [Google Scholar] [CrossRef] [PubMed]
- Lichtinger, M.; Hoogenkamp, M.; Krysinska, H.; Ingram, R.; Bonifer, C. Chromatin regulation by RUNX1. Blood Cells Mol. Dis. 2010, 44, 287–290. [Google Scholar] [CrossRef] [PubMed]
- Link, K.A.; Chou, F.-S.; Mulloy, J.C. Core binding factor at the crossroads: Determining the fate of the HSC. J. Cell. Physiol. 2009, 222, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Dowdy, C.R.; Frederick, D.; Zaidi, S.K.; Colby, J.L.; Lian, J.B.; Van Wijnen, A.J.; Gerstein, R.M.; Stein, J.L.; Stein, G.S. A germline point mutation in Runx1 uncouples its role in definitive hematopoiesis from differentiation. Exp. Hematol. 2013, 41, 980–991.e1. [Google Scholar] [CrossRef]
- Guo, H.; Ma, O.; Speck, N.A.; Friedman, A.D. Runx1 deletion or dominant inhibition reduces Cebpa transcription via conserved promoter and distal enhancer sites to favor monopoiesis over granulopoiesis. Blood 2012, 119, 4408–4418. [Google Scholar] [CrossRef]
- Sood, R.; Kamikubo, Y.; Liu, P.P. Role of RUNX1 in hematological malignancies. Blood 2017, 129, 2070–2082. [Google Scholar] [CrossRef]
- Osato, M.; Asou, N.; Abdalla, E.; Hoshino, K.; Yamasaki, H.; Okubo, T.; Suzushima, H.; Takatsuki, K.; Kanno, T.; Shigesada, K.; et al. Biallelic and Heterozygous Point Mutations in the Runt Domain of theAML1/PEBP2alphaB Gene Associated with Myeloblastic Leukemias. Blood 1999, 93, 1817–1824. [Google Scholar] [CrossRef]
- Osato, M. Point mutations in the RUNX1/AML1 gene: Another actor in RUNX leukemia. Oncogene 2004, 23, 4284–4296. [Google Scholar] [CrossRef]
- Preudhomme, C.; Warot-Loze, D.; Roumier, C.; Grardel-Duflos, N.; Garand, R.; Lai, J.L.; Dastugue, N.; Macintyre, E.; Denis, C.; Bauters, F.; et al. High incidence of biallelic point mutations in the Runt domain of the AML1/PEBP2 alpha B gene in Mo acute myeloid leukemia and in myeloid malignancies with acquired trisomy 21. Blood 2000, 96, 2862–2869. [Google Scholar] [CrossRef]
- Tang, J.-L.; Hou, H.-A.; Chen, C.-Y.; Liu, C.-Y.; Chou, W.-C.; Tseng, M.-H.; Huang, C.-F.; Lee, F.-Y.; Liu, M.-C.; Yao, M.; et al. AML1/RUNX1 mutations in 470 adult patients with de novo acute myeloid leukemia: Prognostic implication and interaction with other gene alterations. Blood 2009, 114, 5352–5361. [Google Scholar] [CrossRef] [PubMed]
- Lam, K.; Muselman, A.; Du, R.; Harada, Y.; Scholl, A.G.; Yan, M.; Matsuura, S.; Weng, S.; Harada, H.; Zhang, D.-E. Hmga2 is a direct target gene of RUNX1 and regulates expansion of myeloid progenitors in mice. Blood 2014, 124, 2203–2212. [Google Scholar] [CrossRef] [PubMed]
- Nishimoto, N.; Arai, S.; Ichikawa, M.; Nakagawa, M.; Goyama, S.; Kumano, K.; Takahashi, T.; Kamikubo, Y.; Imai, Y.; Kurokawa, M. Loss of AML1/Runx1 accelerates the development of MLL-ENL leukemia through down-regulation of p19ARF. Blood 2011, 118, 2541–2550. [Google Scholar] [CrossRef] [PubMed]
- Motoda, L.; Osato, M.; Yamashita, N.; Jacob, B.; Chen, L.Q.; Yanagida, M.; Ida, H.; Wee, H.-J.; Sun, A.X.; Taniuchi, I.; et al. Runx1Protects Hematopoietic Stem/Progenitor Cells from Oncogenic Insult. Stem Cells 2007, 25, 2976–2986. [Google Scholar] [CrossRef]
- Goyama, S.; Schibler, J.; Cunningham, L.; Zhang, Y.; Rao, Y.; Nishimoto, N.; Nakagawa, M.; Olsson, A.; Wunderlich, M.; Link, K.A.; et al. Transcription factor RUNX1 promotes survival of acute myeloid leukemia cells. J. Clin. Investig. 2013, 123, 3876–3888. [Google Scholar] [CrossRef]
- Scheitz, C.J.F.; Tumbar, T. New insights into the role of Runx1 in epithelial stem cell biology and pathology. J. Cell. Biochem. 2013, 114, 985–993. [Google Scholar] [CrossRef]
- Blyth, K.; Cameron, E.R.; Neil, J.C. The runx genes: Gain or loss of function in cancer. Nat. Rev. Cancer 2005, 5, 376–387. [Google Scholar] [CrossRef]
- Chimge, N.-O.; Frenkel, B. The RUNX family in breast cancer: Relationships with estrogen signaling. Oncogene 2013, 32, 2121–2130. [Google Scholar] [CrossRef]
- Kadota, M.; Yang, H.H.; Gomez, B.; Sato, M.; Clifford, R.J.; Meerzaman, D.; Dunn, B.K.; Wakefield, L.M.; Lee, M.P. Delineating Genetic Alterations for Tumor Progression in the MCF10A Series of Breast Cancer Cell Lines. PLoS ONE 2010, 5, e9201. [Google Scholar] [CrossRef]
- Wang, L.; Brugge, J.S.; Janes, K.A. Intersection of FOXO- and RUNX1-mediated gene expression programs in single breast epithelial cells during morphogenesis and tumor progression. Proc. Natl. Acad. Sci. USA 2011, 108, E803–E812. [Google Scholar] [CrossRef]
- Janes, K.A. RUNX1 and its understudied role in breast cancer. Cell Cycle 2011, 10, 3461–3465. [Google Scholar] [CrossRef] [PubMed]
- Chimge, N.-O.; Ahmed-Alnassar, S.; Frenkel, B. Relationship between RUNX1 and AXIN1 in ER-negative versus ER-positive Breast Cancer. Cell Cycle 2017, 16, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, M.; Tan, T.Z.; Sulaiman, N.B.S.; Lamar, J.M.; Bansal, P.; Cui, J.; Qiao, Y.; Ito, Y. RUNX1 and RUNX3 protect against YAP-mediated EMT, stem-ness and shorter survival outcomes in breast cancer. Oncotarget 2018, 9, 14175–14192. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, N.; Mohammed, Z.M.A.; Nixon, C.; Mason, S.; Mallon, E.; McMillan, D.C.; Morris, J.S.; Cameron, E.R.; Edwards, J.; Blyth, K. Expression of RUNX1 Correlates with Poor Patient Prognosis in Triple Negative Breast Cancer. PLoS ONE 2014, 9, e100759. [Google Scholar] [CrossRef]
- Becker, H.; Marcucci, G.; Maharry, K.; Radmacher, M.D.; Mrózek, K.; Margeson, D.; Whitman, S.P.; Wu, Y.-Z.; Schwind, S.; Paschka, P.; et al. Favorable prognostic impact of NPM1 mutations in older patients with cytogenetically normal de novo acute myeloid leukemia and associated gene-and microRNA-expression signatures: A Cancer and Leukemia Group B study. J. Clin. Oncol. 2010, 28, 596–604. [Google Scholar] [CrossRef]
- Ulitsky, I.; Bartel, D.P. lincRNAs: Genomics, Evolution, and Mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef]
- Cartwright, P.; McLean, C.; Sheppard, A.; Rivett, D.; Jones, K.; Dalton, S. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development 2005, 132, 885–896. [Google Scholar] [CrossRef]
- Sarper, S.E.; Inubushi, T.; Kurosaka, H.; Minagi, H.O.; Kuremoto, K.-I.; Sakai, T.; Taniuchi, I.; Yamashiro, T. Runx1-Stat3 signaling regulates the epithelial stem cells in continuously growing incisors. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]


| Subjects | Numbers | Age (Mean) | Gender (M/F) | Viral Load and Other Characteristics |
|---|---|---|---|---|
| HCV | 50 | 29–65 (46) | 39/11 | 17,000–17,000,000 IU/mL, 36GT1, 8GT2, 6GT3 |
| HS | 54 | 22–64 (33) | 41/13 | All the tested negative for HCV, HBV and HIV |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thakuri, B.K.C.; Zhang, J.; Zhao, J.; Nguyen, L.N.; Nguyen, L.N.T.; Schank, M.; Khanal, S.; Dang, X.; Cao, D.; Lu, Z.; et al. HCV-Associated Exosomes Upregulate RUNXOR and RUNX1 Expressions to Promote MDSC Expansion and Suppressive Functions through STAT3–miR124 Axis. Cells 2020, 9, 2715. https://doi.org/10.3390/cells9122715
Thakuri BKC, Zhang J, Zhao J, Nguyen LN, Nguyen LNT, Schank M, Khanal S, Dang X, Cao D, Lu Z, et al. HCV-Associated Exosomes Upregulate RUNXOR and RUNX1 Expressions to Promote MDSC Expansion and Suppressive Functions through STAT3–miR124 Axis. Cells. 2020; 9(12):2715. https://doi.org/10.3390/cells9122715
Chicago/Turabian StyleThakuri, Bal Krishna Chand, Jinyu Zhang, Juan Zhao, Lam N. Nguyen, Lam N. T. Nguyen, Madison Schank, Sushant Khanal, Xindi Dang, Dechao Cao, Zeyuan Lu, and et al. 2020. "HCV-Associated Exosomes Upregulate RUNXOR and RUNX1 Expressions to Promote MDSC Expansion and Suppressive Functions through STAT3–miR124 Axis" Cells 9, no. 12: 2715. https://doi.org/10.3390/cells9122715
APA StyleThakuri, B. K. C., Zhang, J., Zhao, J., Nguyen, L. N., Nguyen, L. N. T., Schank, M., Khanal, S., Dang, X., Cao, D., Lu, Z., Wu, X. Y., Jiang, Y., El Gazzar, M., Ning, S., Wang, L., Moorman, J. P., & Yao, Z. Q. (2020). HCV-Associated Exosomes Upregulate RUNXOR and RUNX1 Expressions to Promote MDSC Expansion and Suppressive Functions through STAT3–miR124 Axis. Cells, 9(12), 2715. https://doi.org/10.3390/cells9122715