Long-Term Cd Exposure Alters the Metabolite Profile in Stem Tissue of Medicago sativa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Growth and Sampling
2.2. Measurement of Glutathione in Stem Tissue
2.3. Quantification of Primary Metabolites
2.3.1. HPLC-FLD Analysis of PAs in Stems
2.3.2. UHPLC–DAD Analysis of Amino Acids in Stems
2.3.3. GC–MS Analysis of Primary Metabolites in Leaves
2.4. Quantification of Secondary Metabolites
2.4.1. Extraction
2.4.2. UHPLC–MS Analysis
2.4.3. Data Processing
2.5. Gene Expression Analyses
3. Results
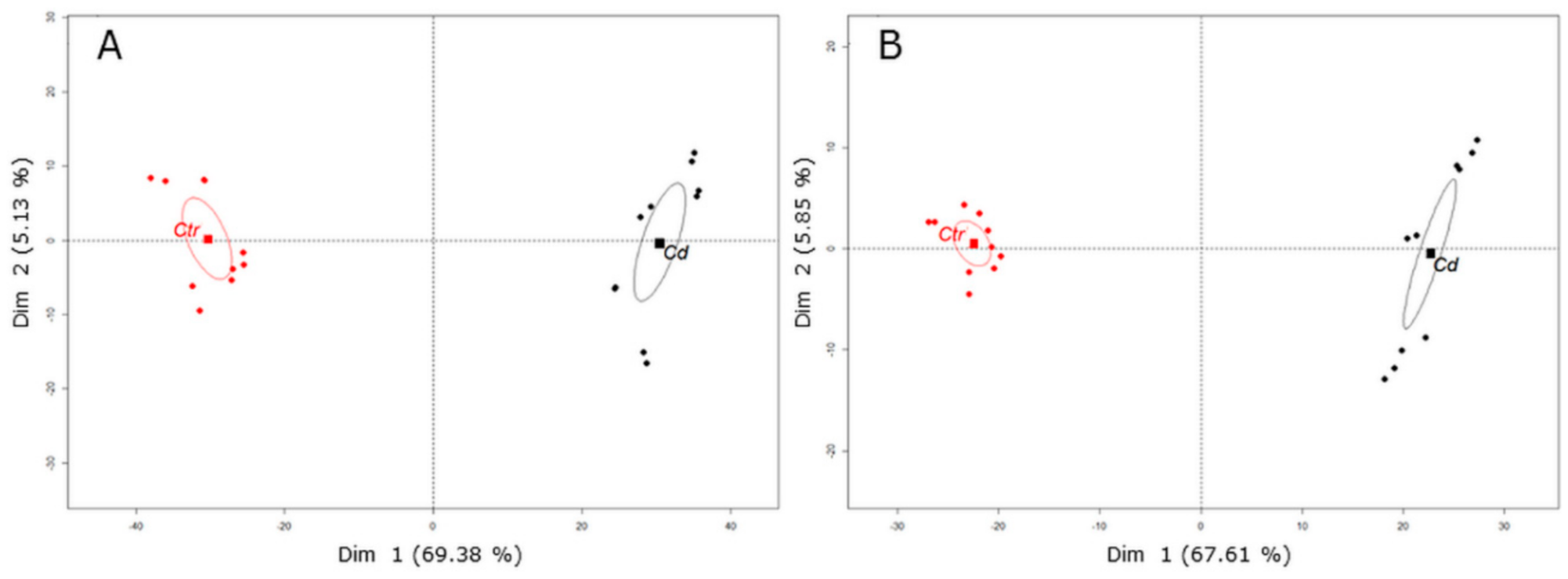
3.1. Cd Stress Provokes the Accumulation of (iso)Flavone Conjugates in Stem Tissue
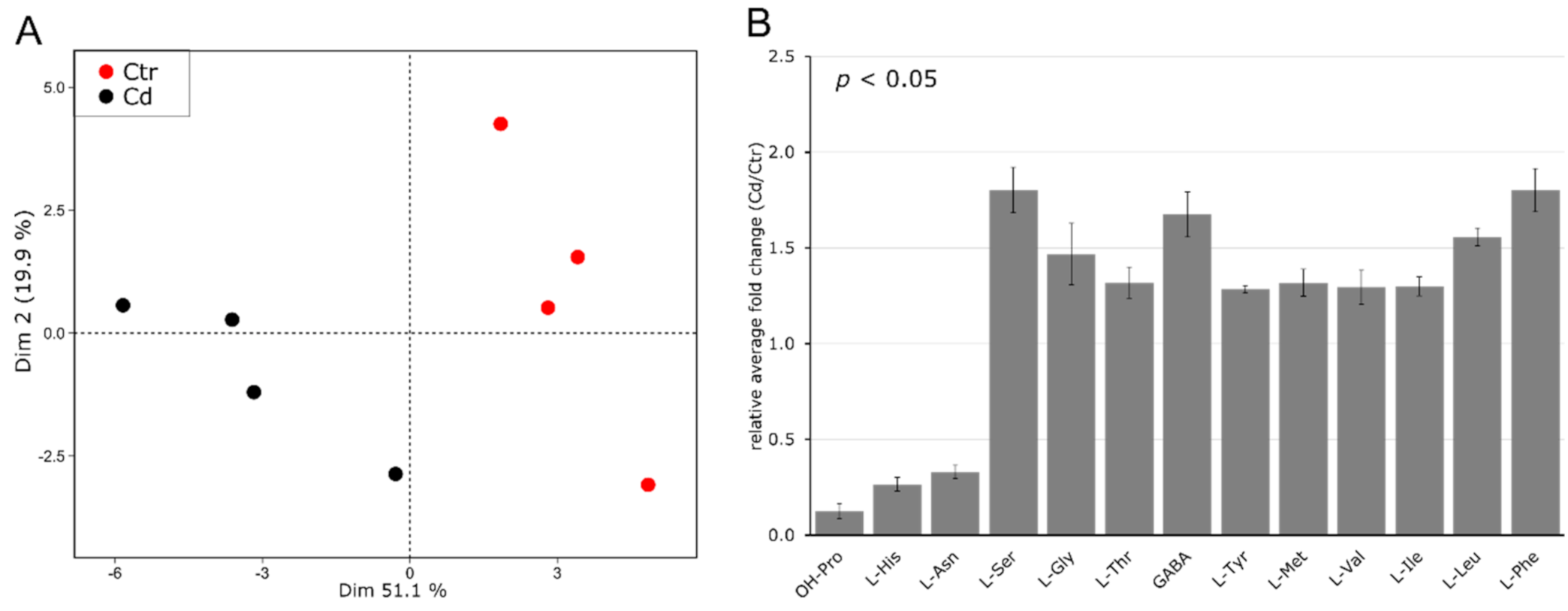
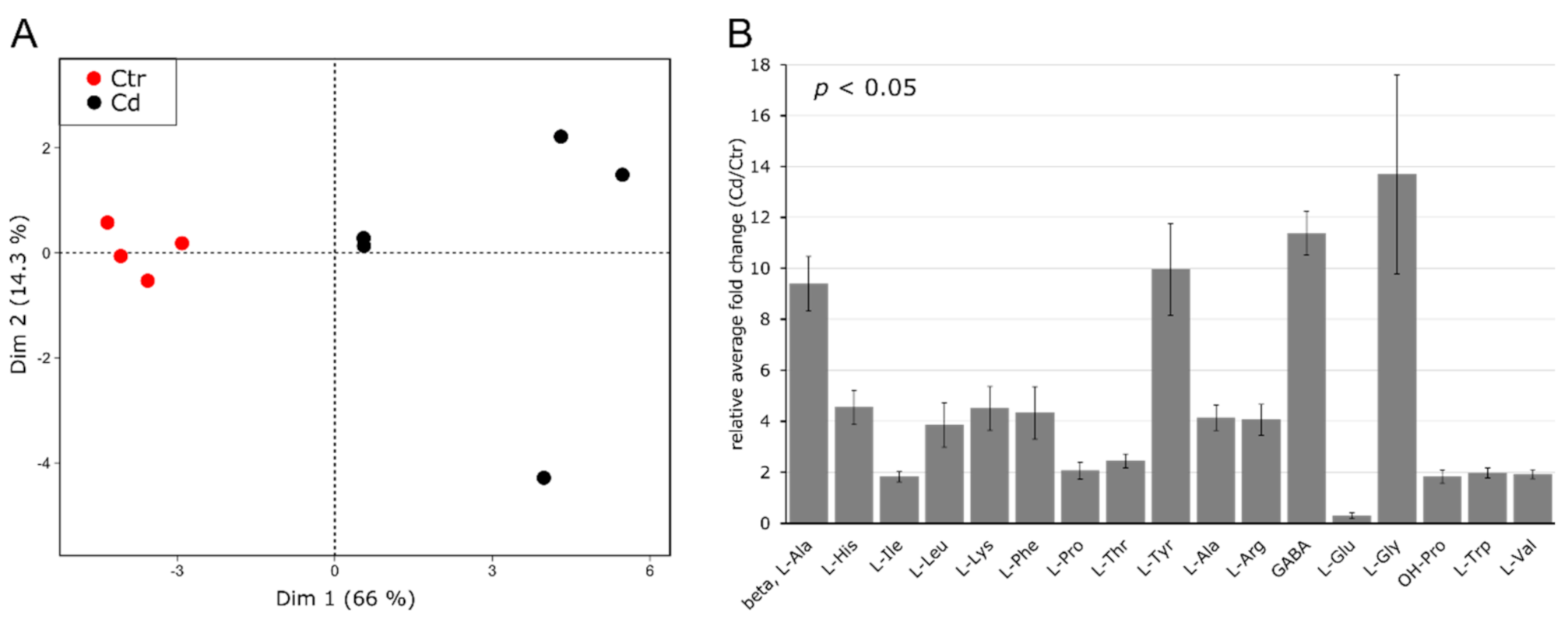
3.2. Cd Exposure Alters the Amino Acid Profile of Stems and Leaves Distinctively
3.3. Glutathione Plays a Minor Role during Long-Term Cd Exposure
4. Discussion
4.1. Cd Increases the Abundance of (iso)Flavones
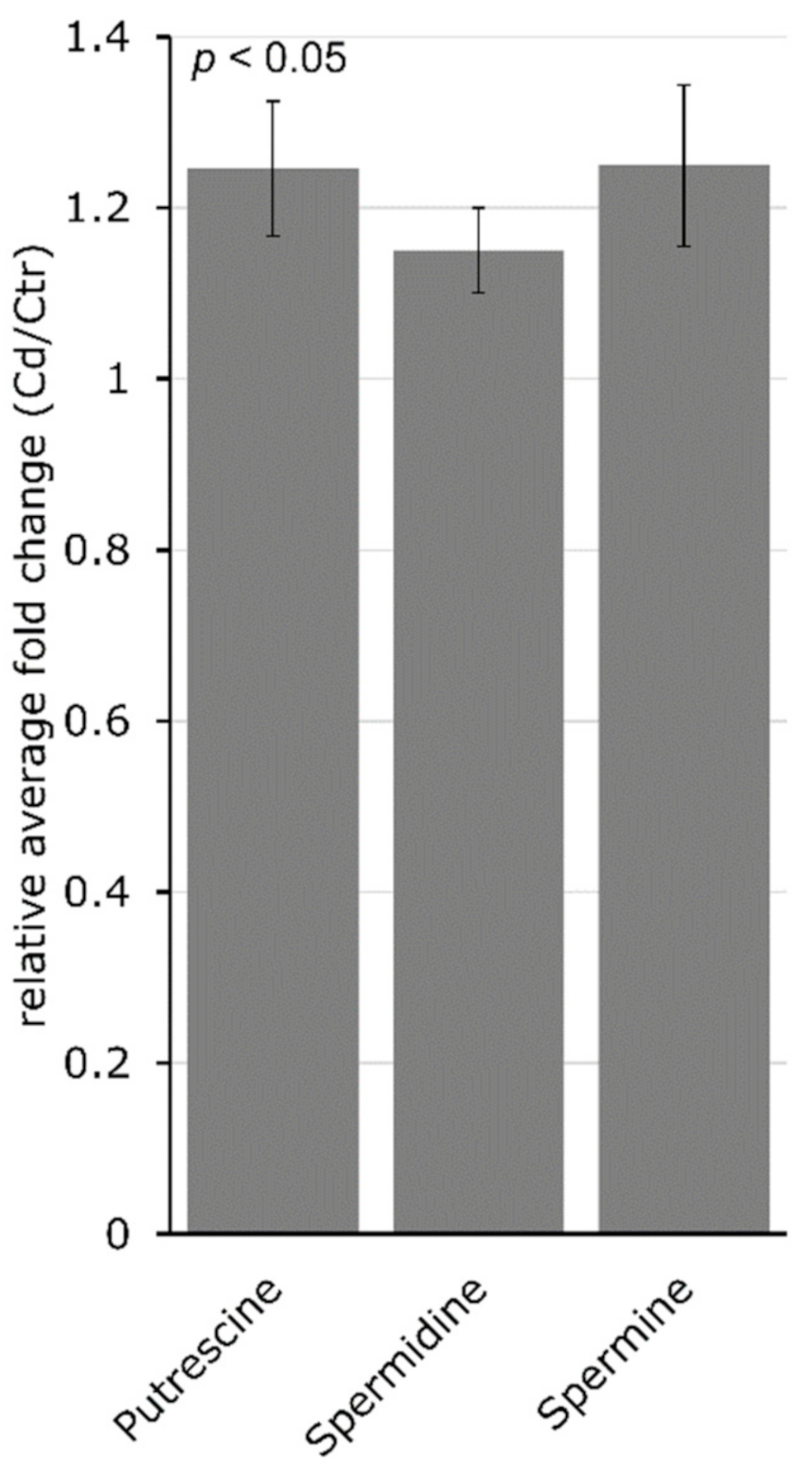
4.2. Cadmium Induces Changes in the Abundance of Amino Acids and Amino-Acid-Derived Molecules
4.3. Abscisic Acid Supports Tolerance Acquisition to Cd
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Clemens, S.; Ma, J.F. Toxic Heavy Metal and Metalloid Accumulation in Crop Plants and Foods. Annu. Rev. Plant Biol. 2016, 67, 489–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuypers, A.; Smeets, K.; Ruytinx, J.; Opdenakker, K.; Keunen, E.; Remans, T.; Horemans, N.; Vanhoudt, N.; Van Sanden, S.; Van Belleghem, F.; et al. The cellular redox state as a modulator in cadmium and copper responses in Arabidopsis thaliana seedlings. J. Plant Physiol. 2011, 168, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Semane, B.; Cuypers, A.; Smeets, K.; Van Belleghem, F.; Horemans, N.; Schat, H.; Vangronsveld, J. Cadmium responses in Arabidopsis thaliana: Glutathione metabolism and antioxidative defence system. Physiol. Plant. 2007, 129, 519–528. [Google Scholar] [CrossRef]
- Schützendübel, A.; Polle, A. Plant responses to abiotic stresses: Heavy metal-induced oxidative stress and protection by mycorrhization. J. Exp. Bot. 2002, 53, 1351–1365. [Google Scholar] [CrossRef]
- Perfus-Barbeoch, L.; Leonhardt, N.; Vavasseur, A.; Forestier, C. Heavy metal toxicity: Cadmium permeates through calcium channels and disturbs the plant water status. Plant J. 2002, 32, 539–548. [Google Scholar] [CrossRef]
- Sanità Di Toppi, L.; Gabbrielli, R. Response to cadmium in higher plants. Environ. Exp. Bot. 1999, 41, 105–130. [Google Scholar] [CrossRef]
- Yang, X.; Baligar, V.C.; Martens, D.C.; Clark, R.B. Cadmium effects on influx and transport of mineral nutrients in plant species. J. Plant Nutr. 1996, 19, 643–656. [Google Scholar] [CrossRef]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive Oxygen Species, Oxidative Damage, and Antioxidative Defense Mechanism in Plants under Stressful Conditions. J. Bot. 2012, 2012, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Sandalio, L.M.; Dalurzo, H.C.; Gómez, M.; Romero-Puertas, M.C.; del Río, L.A. Cadmium-induced changes in the growth and oxidative metabolism of pea plants. J. Exp. Bot. 2001, 52, 2115–2126. [Google Scholar] [CrossRef]
- Romero-Puertas, M.C.; Palma, J.M.; Gómez, M.; Río, L.A.D.E.L.; Sandalio, L.M. Cadmium causes the oxidative modification of proteins in pea plants. Plant Cell. Environ. 2002, 25, 677–686. [Google Scholar] [CrossRef]
- Berni, R.; Luyckx, M.; Xu, X.; Legay, S.; Sergeant, K.; Hausman, J.F.; Lutts, S.; Cai, G.; Guerriero, G. Reactive oxygen species and heavy metal stress in plants: Impact on the cell wall and secondary metabolism. Environ. Exp. Bot. 2019, 161, 98–106. [Google Scholar] [CrossRef]
- Fernie, A.R.; Pichersky, E. Focus issue on metabolism: Metabolites, metabolites everywhere. Plant Physiol. 2015, 169, 1421–1423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirai, M.Y.; Yano, M.; Goodenowe, D.B.; Kanaya, S.; Kimura, T.; Awazuhara, M.; Arita, M.; Fujiwara, T.; Saito, K. From the cover: Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2004, 101, 10205–10210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goossens, A.; Häkkinen, S.T.; Laakso, I.; Seppänen-Laakso, T.; Biondi, S.; De Sutter, V.; Lammertyn, F.; Nuutila, A.M.; Söderlund, H.; Zabeau, M.; et al. A functional genomics approach toward the understanding of secondary metabolism in plant cells. Proc. Natl. Acad. Sci. USA. 2003, 100, 8595–8600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obata, T.; Fernie, A.R. The use of metabolomics to dissect plant responses to abiotic stresses. Cell. Mol. Life Sci. 2012, 69, 3225–3243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kroymann, J. Natural diversity and adaptation in plant secondary metabolism. Curr. Opin. Plant Biol. 2011, 14, 246–251. [Google Scholar] [CrossRef]
- Yang, L.; Wen, K.-S.; Ruan, X.; Zhao, Y.-X.; Wei, F.; Wang, Q. Response of Plant Secondary Metabolites to Environmental Factors. Molecules 2018, 23, 762. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, E.; Arshad, M.; Zakriyya Khan, M.; Shoaib Amjad, M.; Mehreen Sadaf, H.; Riaz, I.; Sidra Sabir, P.; Ahmad, N. Sabaoon Secondary metabolites and their multidimensional prospective in plant life. J. Pharmacogn. Phytochem. 2017, 6, 205–214. [Google Scholar]
- Winkel-Shirley, B. Flavonoid Biosynthesis. A Colorful Model for Genetics, Biochemistry, Cell Biology, and Biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [Green Version]
- Erb, M.; Kliebenstein, D.J. Plant Secondary Metabolites as Defenses, Regulators, and Primary Metabolites: The Blurred Functional Trichotomy. Plant Physiol. 2020, 184, 39–52. [Google Scholar] [CrossRef]
- Ghosson, H.; Schwarzenberg, A.; Jamois, F.; Yvin, J.C. Simultaneous untargeted and targeted metabolomics profiling of underivatized primary metabolites in sulfur-deficient barley by ultra-high performance liquid chromatography-quadrupole/time-of-flight mass spectrometry. Plant Methods 2018, 14, 62. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Wei, S.; Liu, B.; Guo, D.; Zheng, B.; Feng, L.; Liu, Y.; Tomás-Barberán, F.A.; Luo, L.; Huang, D. A novel integrated non-targeted metabolomic analysis reveals significant metabolite variations between different lettuce (Lactuca sativa. L.) varieties. Hortic. Res. 2018, 5, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernie, A.R.; Trethewey, R.N.; Krotzky, A.J. Metabolite profiling: From diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 2004, 5, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Jorge, T.F.; Rodrigues, J.A.; Caldana, C.; Schmidt, R.; van Dongen, J.T.; Thomas-Oates, J.; Antonio, C. Mass spectrometry-based plant metabolomics: Metabolite responses to abiotic stress. Mass Spectrom. Rev. 2016, 35, 620–649. [Google Scholar] [CrossRef] [PubMed]
- Rogachev, I.; Aharoni, A. UPLC-MS-Based Metabolite Analysis in Tomato. In Plant Metabolomics; Methods in Molecular Biology (Methods and Protocols), Hardy, N., Hall, R., Eds.; Humana Press: Clifton, NJ, USA, 2011; Volume 860, pp. 129–144. [Google Scholar]
- Zhang, Y.; Zhang, A.; Zhang, Y.; Sun, H.; Meng, X.; Yan, G.; Wang, X. Application of ultra-performance liquid chromatography with time-of-flight mass spectrometry for the rapid analysis of constituents and metabolites from the extracts of Acanthopanax senticosus Harms leaf. Pharmacogn. Mag. 2016, 12, 145–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakabayashi, R.; Saito, K. Integrated metabolomics for abiotic stress responses in plants. Curr. Opin. Plant Biol. 2015, 24, 10–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scalabrin, E.; Radaelli, M.; Rizzato, G.; Bogani, P.; Buiatti, M.; Gambaro, A.; Capodaglio, G. Metabolomic analysis of wild and transgenic Nicotiana langsdorffii plants exposed to abiotic stresses: Unraveling metabolic responses. Anal. Bioanal. Chem. 2015, 407, 6357–6368. [Google Scholar] [CrossRef]
- Keunen, E.; Florez-Sarasa, I.; Obata, T.; Jozefczak, M.; Remans, T.; Vangronsveld, J.; Fernie, A.R.; Cuypers, A. Metabolic responses of Arabidopsis thaliana roots and leaves to sublethal cadmium exposure are differentially influenced by alternative oxidase 1a. Environ. Exp. Bot. 2016, 124, 64–78. [Google Scholar] [CrossRef]
- Pavlíková, D.; Zemanová, V.; Procházková, D.; Pavlík, M.; Száková, J.; Wilhelmová, N. The long-term effect of zinc soil contamination on selected free amino acids playing an important role in plant adaptation to stress and senescence. Ecotoxicol. Environ. Saf. 2014, 100, 166–170. [Google Scholar] [CrossRef]
- Hédiji, H.; Djebali, W.; Cabasson, C.; Maucourt, M.; Baldet, P.; Bertrand, A.; Boulila Zoghlami, L.; Deborde, C.; Moing, A.; Brouquisse, R.; et al. Effects of long-term cadmium exposure on growth and metabolomic profile of tomato plants. Ecotoxicol. Environ. Saf. 2010, 73, 1965–1974. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, L.; Shen, H.; Wang, J.; Liu, W.; Zhu, X. Metabolomic analysis with GC-MS to reveal potential metabolites and biological pathways involved in Pb & Cd stress response of radish roots. Sci. Rep. 2015, 5, 18296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skorzynska-Polit, E.; Drazkiewicz, M.; Wianowska, D.; Maksymiec, W.; Dawidowicz, A.L.; Tukiendorf, A. The influence of heavy metal stress on the level of some flavonols in the primary leaves of Phaseolus coccineus. Acta Physiol. Plant. 2004, 26, 247–254. [Google Scholar] [CrossRef]
- Pawlak-Sprada, S.; Arasimowicz-Jelonek, M.; Podgórska, M.; Deckert, J. Activation of phenylpropanoid pathway in legume plants exposed to heavy metals. Part I. Effects of cadmium and lead on phenylalanine ammonia-lyase gene expression, enzyme activity and lignin content. Acta Biochim. Pol. 2011, 58, 211–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bali, S.; Jamwal, V.L.; Kohli, S.K.; Kaur, P.; Tejpal, R.; Bhalla, V.; Ohri, P.; Gandhi, S.G.; Bhardwaj, R.; Al-Huqail, A.A.; et al. Jasmonic acid application triggers detoxification of lead (Pb) toxicity in tomato through the modifications of secondary metabolites and gene expression. Chemosphere 2019, 235, 734–748. [Google Scholar] [CrossRef]
- Kısa, D.; Elmastaş, M.; Öztürk, L.; Kayır, Ö. Responses of the phenolic compounds of Zea mays under heavy metal stress. Appl. Biol. Chem. 2016, 59, 813–820. [Google Scholar] [CrossRef]
- Karimi, E.; Oskoueian, E.; Oskoueian, A.; Omidvar, V.; Hendra, R.; Nazeran, H. Insight into the functional and medicinal properties of Medicago sativa (Alfalfa) leaves extract. J. Med. Plants Res. 2013, 7, 290–297. [Google Scholar] [CrossRef]
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattini, M. Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 2012, 196, 67–76. [Google Scholar] [CrossRef]
- Karamian, R.; Asadbegy, M. Antioxidant activity, total phenolic and flavonoid contents of three Onobrychis species from Iran. Pharm. Sci. 2016, 22, 112–119. [Google Scholar] [CrossRef] [Green Version]
- Olennikov, D.; Kashchenko, N.; Chirikova, N.; Olennikov, D.N.; Kashchenko, N.I.; Chirikova, N.K. A Novel HPLC-Assisted Method for Investigation of the Fe2+-Chelating Activity of Flavonoids and Plant Extracts. Molecules 2014, 19, 18296–18316. [Google Scholar] [CrossRef] [Green Version]
- Kidd, P.S.; Llugany, M.; Poschenrieder, C.; Gunsé, B.; Barceló, J. The role of root exudates in aluminium resistance and silicon-induced amelioration of aluminium toxicity in three varieties of maize (Zea mays L.). J. Exp. Bot. 2001, 52, 1339–1352. [Google Scholar] [CrossRef] [Green Version]
- Veronesi, F. Fodder Crops and Amenity Grasses; Springer: Heidelberg, Germany, 2010; ISBN 9781441907592. [Google Scholar]
- Messina, M.J. Legumes and Soybeans: Overview of their Nutritional Profiles and Health Effects. Am. J Clin. Nutr. 1999, 70, 439s–450s. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Printz, B.; Guerriero, G.; Sergeant, K.; Audinot, J.-N.; Guignard, C.; Renaut, J.; Lutts, S.; Hausman, J.-F. Combining -Omics to Unravel the Impact of Copper Nutrition on Alfalfa (Medicago sativa) Stem Metabolism. Plant Cell Physiol. 2016, 57, 407–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verdonk, J.C.; Hatfield, R.D.; Sullivan, M.L. Proteomic analysis of cell walls of two developmental stages of alfalfa stems. Front. Plant Sci. 2012, 3, 279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samac, D.A.; Jung, H.; Lamb, J.F.S. Development of alfalfa (Medicago sativa L.) as a feedstock for production of ethanol and other bioproducts. In Alcoholic Fuels; Minteer, S., Ed.; CRC Press: Boca Raton, FL, USA, 2006; p. 79. ISBN 0737-8025. [Google Scholar]
- Sanderson, M.A.; Martin, N.P.; Adler, P. Biomass, Energy, and Industrial Uses of Forages. In Forages: The Science of Grassland Agriculture; Moore, K.J., Barnes, R.F., Nelson, C.J., Collins, M., Eds.; Blackwell: Ames, IA, USA, 2006; pp. 635–647. [Google Scholar]
- Rafińska, K.; Pomastowski, P.; Wrona, O.; Górecki, R.; Buszewski, B. Medicago sativa as a source of secondary metabolites for agriculture and pharmaceutical industry. Phytochem. Lett. 2017, 20, 520–539. [Google Scholar] [CrossRef]
- Pan, J.; Plant, J.A.; Voulvoulis, N.; Oates, C.J.; Ihlenfeld, C. Cadmium levels in Europe: Implications for human health. Environ. Geochem. Health 2010, 32, 1–12. [Google Scholar] [CrossRef]
- Gutsch, A.; Keunen, E.; Guerriero, G.; Renaut, J.; Cuypers, A.; Hausman, J.-F.; Sergeant, K. Long-term cadmium exposure influences the abundance of proteins that impact the cell wall structure in Medicago sativa stems. Plant Biol. J. 2018, 20, 1023–1035. [Google Scholar] [CrossRef] [Green Version]
- Gutsch, A.; Zouaghi, S.; Renaut, J.; Cuypers, A.; Hausman, J.-F.; Sergeant, K. Changes in the Proteome of Medicago sativa Leaves in Response to Long-Term Cadmium Exposure Using a Cell-Wall Targeted Approach. Int. J. Mol. Sci. 2018, 19, 2498. [Google Scholar] [CrossRef] [Green Version]
- Gutsch, A.; Sergeant, K.; Keunen, E.; Prinsen, E.; Guerriero, G.; Renaut, J.; Hausman, J.F.; Cuypers, A. Does long-term cadmium exposure influence the composition of pectic polysaccharides in the cell wall of Medicago sativa stems? BMC Plant Biol. 2019, 19, 271. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Serrano, M.; Romero-Puertas, M.C.; Zabalza, A.; Corpas, F.J.; Gómez, M.; Del Río, L.A.; Sandalio, L.M. Cadmium effect on oxidative metabolism of pea (Pisum sativum L.) roots. Imaging of reactive oxygen species and nitric oxide accumulation in vivo. Plant. Cell Environ. 2006, 29, 1532–1544. [Google Scholar] [CrossRef]
- Passardi, F.; Penel, C.; Dunand, C. Performing the paradoxical: How plant peroxidases modify the cell wall. Trends Plant Sci. 2004, 9, 534–540. [Google Scholar] [CrossRef]
- Queval, G.; Noctor, G. A plate reader method for the measurement of NAD, NADP, glutathione, and ascorbate in tissue extracts: Application to redox profiling during Arabidopsis rosette development. Anal. Biochem. 2007, 363, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Lefèvre, I.; Gratia, E.; Lutts, S. Discrimination between the ionic and osmotic components of salt stress in relation to free polyamine level in rice (Oryza sativa). Plant Sci. 2001, 161, 943–952. [Google Scholar] [CrossRef]
- Lisec, J.; Schauer, N.; Kopka, J.; Willmitzer, L.; Fernie, A.R. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protoc. 2006, 1, 387–396. [Google Scholar] [CrossRef]
- Schauer, N.; Steinhauser, D.; Strelkov, S.; Schomburg, D.; Allison, G.; Moritz, T.; Lundgren, K.; Roessner-Tunali, U.; Forbes, M.G.; Willmitzer, L.; et al. GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett. 2005, 579, 1332–1337. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Guerriero, G.; Legay, S.; Hausman, J.F. Alfalfa cellulose synthase gene expression under abiotic stress: A hitchhiker’s guide to RT-qPCR normalization. PLoS ONE 2014, 9, e103808. [Google Scholar] [CrossRef] [Green Version]
- Elobeid, M.; Göbel, C.; Feussner, I.; Polle, A. Cadmium interferes with auxin physiology and lignification in poplar. J. Exp. Bot. 2012, 63, 1413–1421. [Google Scholar] [CrossRef] [Green Version]
- Chaoui, A.; El Ferjani, E. Effects of cadmium and copper on antioxidant capacities, lignification and auxin degradation in leaves of pea (Pisum sativum L.) seedlings. C. R. Biol. 2005, 328, 23–31. [Google Scholar] [CrossRef]
- Cui, W.; Gao, Z.; Wu, H.; Xie, Y.; Shen, W. Haem oxygenase-1 is involved in salicylic acid-induced methylation and chromatin patterning alleviation of oxidative stress due to cadmium stress in Medicago sativa. J. Exp. Bot. 2012, 63, 5521–5534. [Google Scholar] [CrossRef] [Green Version]
- Staszków, A.; Swarcewicz, B.; Banasiak, J.; Muth, D.; Jasiński, M.; Stobiecki, M. LC/MS profiling of flavonoid glycoconjugates isolated from hairy roots, suspension root cell cultures and seedling roots of Medicago truncatula. Metabolomics 2011, 7, 604–613. [Google Scholar] [CrossRef] [Green Version]
- De Rijke, E.; Out, P.; Niessen, W.M.A.; Ariese, F.; Gooijer, C.; Brinkman, U.A.T. Analytical separation and detection methods for flavonoids. J. Chromatogr. A 2006, 1112, 31–63. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.Z.; He, X.G.; Lindenmaier, M.; Yang, J.; Cleary, M.; Qiu, S.X.; Cordell, G.A. LC-ESI-MS study of the flavonoid glycoside malonates of red clover (Trifolium pratense). J. Agric. Food Chem. 2000, 48, 354–365. [Google Scholar] [CrossRef] [PubMed]
- Farag, M.A.; Huhman, D.V.; Dixon, R.A.; Sumner, L.W. Metabolomics Reveals Novel Pathways and Differential Mechanistic and Elicitor-Specific Responses in Phenylpropanoid and Isoflavonoid Biosynthesis in Medicago truncatula Cell Cultures. Plant Physiol. 2008, 146, 387–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wojakowska, A.; Piasecka, A.; García-López, P.M.; Zamora-Natera, F.; Krajewski, P.; Marczak, Ł.; Kachlicki, P.; Stobiecki, M. Structural analysis and profiling of phenolic secondary metabolites of Mexican lupine species using LC-MS techniques. Phytochemistry 2013, 92, 71–86. [Google Scholar] [CrossRef]
- Farag, M.A.; Huhman, D.V.; Lei, Z.; Sumner, L.W. Metabolic profiling and systematic identification of flavonoids and isoflavonoids in roots and cell suspension cultures of Medicago truncatula using HPLC-UV-ESI-MS and GC-MS. Phytochemistry 2007, 68, 342–354. [Google Scholar] [CrossRef]
- Prasain, J.K.; Jones, K.; Kirk, M.; Wilson, L.; Smith-Johnson, M.; Weaver, C.; Barnes, S. Profiling and quantification of isoflavonoids in kudzu dietary supplements by high-performance liquid chromatography and electrospray ionization tandem mass spectrometry. J. Agric. Food Chem. 2003, 51, 4213–4218. [Google Scholar] [CrossRef]
- Sumner, L.W.; Paiva, N.L.; Dixon, R.A.; Geno, P.W. High-performance liquid chromatography/continuous-flow liquid secondary ion mass spectrometry of flavonoid glycosides in leguminous plant extracts. J. Mass Spectrom. 1996, 31, 472–485. [Google Scholar] [CrossRef]
- Xu, J.; Yin, H.; Li, X. Protective effects of proline against cadmium toxicity in micropropagated hyperaccumulator, Solanum nigrum L. Plant Cell Rep. 2009, 28, 325–333. [Google Scholar] [CrossRef]
- Tian, B.; Qiao, Z.; Zhang, L.; Li, H.; Pei, Y. Hydrogen sulfide and proline cooperate to alleviate cadmium stress in foxtail millet seedlings. Plant Physiol. Biochem. 2016, 109, 293–299. [Google Scholar] [CrossRef]
- Chou, T.-S.; Chao, Y.Y.; Huang, W.D.; Kao, C.H. Effect of Magnesium deficiency on antioxidant status and Cadmium toxicity in rice seedlings. J. Plant Physiol. 2011, 168, 1021–1030. [Google Scholar] [CrossRef]
- Wang, C.Q.; Song, H. Calcium protects Trifolium repens L. seedlings against cadmium stress. Plant Cell Rep. 2009, 28, 1341–1349. [Google Scholar] [CrossRef] [PubMed]
- Peralta-Videa, J.R.; de la Rosa, G.; Gonzalez, J.H.; Gardea-Torresdey, J.L. Effects of the growth stage on the heavy metal tolerance of alfalfa plants. Adv. Environ. Res. 2004, 8, 679–685. [Google Scholar] [CrossRef]
- Jozefczak, M.; Keunen, E.; Schat, H.; Bliek, M.; Hernández, L.E.; Carleer, R.; Remans, T.; Bohler, S.; Vangronsveld, J.; Cuypers, A. Differential response of Arabidopsis leaves and roots to cadmium: Glutathione-related chelating capacity vs antioxidant capacity. Plant Physiol. Biochem. 2014, 83, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Delhaize, E.; Jackson, P.J.; Lujan, L.D.; Robinson, N.J. Poly(γ-glutamylcysteinyl)glycine Synthesis in Datura innoxia and Binding with Cadmium: Role in Cadmium Tolerance. Plant Physiol. 1989, 89, 700–706. [Google Scholar] [CrossRef] [Green Version]
- De Knecht, J.A.; van Dillen, M.; Koevoets, P.; Schat, H.; Verkleij, J.; Ernst, W. Phytochelatins in Cadmium-Sensitive and Cadmium-Tolerant Silene vulgaris. Plant Physiol. 1994, 104, 255–261. [Google Scholar] [CrossRef] [Green Version]
- Dixon, R.A.; Paiva, N.L. Stress-Induced Phenylpropanoid Metabolism. Plant Cell 1995, 7, 1085–1097. [Google Scholar] [CrossRef]
- Kováčik, J.; Klejdus, B. Dynamics of phenolic acids and lignin accumulation in metal-treated Matricaria chamomilla roots. Plant Cell Rep. 2008, 27, 605–615. [Google Scholar] [CrossRef]
- Rahoui, S.; Martinez, Y.; Sakouhi, L.; Ben, C.; Rickauer, M.; El Ferjani, E.; Gentzbittel, L.; Chaoui, A. Cadmium-induced changes in antioxidative systems and differentiation in roots of contrasted Medicago truncatula lines. Protoplasma 2017, 254, 473–489. [Google Scholar] [CrossRef]
- Da Cunha, A. The estimation of l-phenylalanine ammonia-lyase shows phenylpropanoid biosynthesis to be regulated by l-phenylalanine supply and availability. Phytochemistry 1987, 26, 2723–2727. [Google Scholar] [CrossRef]
- Jiang, Y.; Joyce, D.C. ABA effects on ethylene production, PAL activity, anthocyanin and phenolic contents of strawberry fruit. Plant Growth Regul. 2003, 39, 171–174. [Google Scholar] [CrossRef]
- Chalutz, E. Ethylene-induced Phenylalanine Ammonia-Lyase Activity in Carrot Roots. Plant Physiol. 1973, 51, 1033–1036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heredia, J.B.; Cisneros-Zevallos, L. The effect of exogenous ethylene and methyl jasmonate on pal activity, phenolic profiles and antioxidant capacity of carrots (Daucus carota) under different wounding intensities. Postharvest Biol. Technol. 2009, 51, 242–249. [Google Scholar] [CrossRef]
- Pawlak-Sprada, S.; Stobiecki, M.; Deckert, J. Activation of phenylpropanoid pathway in legume plants exposed to heavy metals. Part II. Profiling of isoflavonoids and their glycoconjugates induced in roots of lupine (Lupinus luteus) seedlings treated with cadmium and lead. Acta Biochim. Pol. 2011, 58, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Dixon, R.A. The “ins” and “outs” of flavonoid transport. Trends Plant Sci. 2010, 15, 72–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.-J.; Blount, J.W.; Steele, C.L.; Dixon, R.A. Bottlenecks for metabolic engineering of isoflavone glycoconjugates in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 14578–14583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montesinos, M.C.; Ubeda, A.; Terencio, M.C.; Payá, M.; Alcaraz, M.J. Antioxidant Profile of Mono- and Dihydroxylated Flavone Derivatives in Free Radical Generating Systems. Zeitschrift Naturforsch. Sect. C J. Biosci. 1995, 50, 552–560. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, H. Structure-dependent membrane interaction of flavonoids associated with their bioactivity. Food Chem. 2010, 120, 1089–1096. [Google Scholar] [CrossRef]
- Samsonowicz, M.; Regulska, E. Spectroscopic study of molecular structure, antioxidant activity and biological effects of metal hydroxyflavonol complexes. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 173, 757–771. [Google Scholar] [CrossRef]
- Samsonowicz, M.; Regulska, E.; Kalinowska, M. Hydroxyflavone metal complexes—Molecular structure, antioxidant activity and biological effects. Chem. Biol. Interact. 2017, 273, 245–256. [Google Scholar] [CrossRef]
- Martinoia, E.; Maeshima, M.; Neuhaus, H.E. Vacuolar transporters and their essential role in plant metabolism. J. Exp. Bot. 2007, 58, 83–102. [Google Scholar] [CrossRef]
- Zhu, G.; Xiao, H.; Guo, Q.; Zhang, Z.; Zhao, J.; Yang, D. Effects of cadmium stress on growth and amino acid metabolism in two Compositae plants. Ecotoxicol. Environ. Saf. 2018, 158, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Zemanová, V.; Pavlík, M.; Pavlíková, D.; Tlustoš, P. The significance of methionine, histidine and tryptophan in plant responses and adaptation to cadmium stress. Plant Soil Environ. 2014, 60, 426–432. [Google Scholar] [CrossRef]
- Verbruggen, N.; Hermans, C. Proline accumulation in plants: A review. Amino Acids 2008, 35, 753–759. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.S.; Schat, H.; Vooijs, R. In vitro alleviation of heavy metal-induced enzyme inhibition by proline. Phytochemistry 1998, 49, 1531–1535. [Google Scholar] [CrossRef]
- Bottari, E.; Festa, M.R. Asparagine as a ligand for cadmium (II), lead (II) and zinc (II). Chem. Speciat. Bioavailab. 1996, 8, 75–83. [Google Scholar] [CrossRef] [Green Version]
- Krämer, U.; Cotter-Howells, J.D.; Charnock, J.M.; Baker, A.J.M.; Smith, J.A.C. Free histidine as a metal chelator in plants that accumulate nickel. Nature 1996, 379, 635–638. [Google Scholar] [CrossRef]
- Ali, Q.; Ashraf, M.; Shahbaz, M.; Humera, H. Ameliorating effect of foliar applied proline on nutrient uptake in water stressed maize (Zea mays L.) plants. Pakistan J. Bot. 2008, 40, 211–219. [Google Scholar]
- Liang, X.; Zhang, L.; Natarajan, S.K.; Becker, D.F. Proline Mechanisms of Stress Survival. Antioxid. Redox Signal. 2013, 19, 998–1011. [Google Scholar] [CrossRef] [Green Version]
- Rejeb, K.B.; Abdelly, C.; Savouré, A. How reactive oxygen species and proline face stress together. Plant Physiol. Biochem. 2014, 80, 278–284. [Google Scholar] [CrossRef]
- Kim, T.H.; Bigot, J.; Ourry, A.; Boucaud, J. Amino acid content in xylem sap of regrowing alfalfa (Medicago sativa L.): Relations with N uptake, N2 fixation and N remobilization. Plant Soil 1993, 149, 167–174. [Google Scholar] [CrossRef]
- Marino, D.; Damiani, I.; Gucciardo, S.; Mijangos, I.; Pauly, N.; Puppo, A. Inhibition of nitrogen fixation in symbiotic Medicago truncatula upon Cd exposure is a local process involving leghaemoglobin. J. Exp. Bot. 2013, 64, 5651–5660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schat, H.; Sharma, S.S.; Vooijs, R. Heavy metal-induced accumulation of free proline in a metal-tolerant and a nontolerant ecotype of Silene vulgaris. Physiol. Plant. 1997, 101, 477–482. [Google Scholar] [CrossRef]
- Costa, G.; Spitz, E. Influence of cadmium on soluble carbohydrates, free amino acids, protein content of in vitro cultured Lupinus albus. Plant Sci. 1997, 128, 131–140. [Google Scholar] [CrossRef]
- Dobra, J.; Motyka, V.; Dobrev, P.; Malbeck, J.; Prasil, I.T.; Haisel, D.; Gaudinova, A.; Havlova, M.; Gubis, J.; Vankova, R. Comparison of hormonal responses to heat, drought and combined stress in tobacco plants with elevated proline content. J. Plant Physiol. 2010, 167, 1360–1370. [Google Scholar] [CrossRef]
- Zemanová, V.; Pavlík, M.; Pavlíková, D.; Tlustoš, P. The changes of contents of selected free amino acids associated with cadmium stress in Noccaea caerulescens and Arabidopsis halleri. Plant Soil Environ. 2013, 59, 417–422. [Google Scholar] [CrossRef] [Green Version]
- Aziz, A.; Martin-Tanguy, J.; Larher, F. Stress-induced changes in polyamine and tyramine levels can regulate proline accumulation in tomato leaf discs treated with sodium chloride. Physiol. Plant. 1998, 104, 195–202. [Google Scholar] [CrossRef]
- Ghabriche, R.; Ghnaya, T.; Mnasri, M.; Zaier, H.; Baioui, R.; Vromman, D.; Ghabriche, R.; Lutts, S. Polyamine and tyramine involvement in NaCl-induced improvement of Cd resistance in the halophyte Inula chrithmoides L. J. Plant Physiol. 2017, 216, 136–144. [Google Scholar] [CrossRef]
- Gupta, K.; Dey, A.; Gupta, B. Plant polyamines in abiotic stress responses. Acta Physiol. Plant. 2013, 35, 2015–2036. [Google Scholar] [CrossRef]
- Nahar, K.; Rahman, M.; Hasanuzzaman, M.; Alam, M.M.; Rahman, A.; Suzuki, T.; Fujita, M. Physiological and biochemical mechanisms of spermine-induced cadmium stress tolerance in mung bean (Vigna radiata L.) seedlings. Environ. Sci. Pollut. Res. 2016, 23, 21206–21218. [Google Scholar] [CrossRef]
- Groppa, M.D.; Ianuzzo, M.P.; Tomaro, M.L.; Benavides, M.P. Polyamine metabolism in sunflower plants under long-term cadmium or copper stress. Amino Acids 2007, 32, 265–275. [Google Scholar] [CrossRef]
- Zhao, H.; Yang, H. Exogenous polyamines alleviate the lipid peroxidation induced by cadmium chloride stress in Malus hupehensis Rehd. Sci. Hortic. 2008, 116, 442–447. [Google Scholar] [CrossRef]
- Yang, H.Y.; Shi, G.X.; Li, W.L.; Wu, W.L. Exogenous spermidine enhances Hydrocharis dubia cadmium tolerance. Russ. J. Plant Physiol. 2013, 60, 770–775. [Google Scholar] [CrossRef]
- Geuns, J.M.C.; Cuypers, A.J.F.; Michiels, T.; Colpaert, J.V.; Van Laere, A.; Van Den Broeck, K.A.O.; Vandecasteele, C.H.A. Mung bean seedlings as bio-indicators for soil and water contamination by cadmium. Sci. Total Environ. 1997, 203, 183–197. [Google Scholar] [CrossRef]
- Vishwakarma, K.; Upadhyay, N.; Kumar, N.; Yadav, G.; Singh, J.; Mishra, R.K.; Kumar, V.; Verma, R.; Upadhyay, R.G.; Pandey, M.; et al. Abscisic Acid Signaling and Abiotic Stress Tolerance in Plants: A Review on Current Knowledge and Future Prospects. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Amjad, M.; Akhtar, J.; Anwar-ul-Haq, M.; Yang, A.; Akhtar, S.S.; Jacobsen, S.E. Integrating role of ethylene and ABA in tomato plants adaptation to salt stress. Sci. Hortic. 2014, 172, 109–116. [Google Scholar] [CrossRef]
- Hsu, Y.T.; Kao, C.H. Role of abscisic acid in cadmium tolerance of rice (Oryza sativa L.) seedlings. Plant. Cell Environ. 2003, 26, 867–874. [Google Scholar] [CrossRef] [Green Version]
- Salt, D.E.; Prince, R.C.; Pickering, I.J.; Raskin, I. Mechanisms of Cadmium Mobility and Accumulation in Indian Mustard. Plant Physiol. 1995, 109, 1427–1433. [Google Scholar] [CrossRef] [Green Version]
- Watkins, J.; Chapman, J.M.; Muday, G.K. Abscisic acid-induced reactive oxygen species are modulated by flavonols to control stomata aperture. Plant Physiol. 2017, 175. [Google Scholar] [CrossRef] [Green Version]

| Gene Annotation | Contig ID M. sativa | Rel. Norm. Gene Expression | p-Value |
|---|---|---|---|
| Polyamine synthesis | |||
| arginine decarboxylase | 46,745 | 1.123 | 0.548 |
| diamine oxidase | 14,769 | 0.800 | 0.042 |
| glutamate decarboxylase | 103,359 | 1.685 | 0.019 |
| SAM decarboxylase | 9152 | 1.141 | 0.459 |
| spermidine synthase | 5341 | 1.213 | 0.042 |
| spermine synthase | 92,526 | 1.341 | 0.032 |
| Flavone synthesis | |||
| chalcone isomerase | 60,809 | 1.809 | 0.019 |
| chalcone reductase | 59,796 | 2.753 | 0.019 |
| chalcone synthase | 99,463 | 1.247 | 0.371 |
| isoflavone synthase | 10,252 | 2.699 | 0.019 |
| Glutathione synthesis and redox cycle | |||
| homoglutathione synthetase | [63] | 1.355 | 0.01 |
| γ-glutamylcysteine synthetase | [63] | 0.959 | 0.361 |
| glutathione reductase 1 | [63] | 1.105 | 0.071 |
| glutathione reductase 2 | [63] | 1.043 | 0.681 |
| monodehydroascorbate reductase | [63] | 1.177 | 0.048 |
| glutathione synthetase 2 | 10,444 | 1.384 | 0.004 |
| Rt (min) | Relative Abundance | Fold | [M − H] | Error (ppm) | Molecular Formula | MSMS [M − H] | Putative Identity | Reference | |
|---|---|---|---|---|---|---|---|---|---|
| Cadmium-Exposed | Control | ||||||||
| 2.23 | 446 | 281 | 1.59 | 164.0719 | 1.22 | C9H11NO2 | 147.0434 103.0539- 164.0693 72.0108 | L-Phe * | |
| 2.50 | 1 | 89 | 89.00 | 274.0932 [M − H2O − H] | −0.11 | C11H19NO8 | 89.0280 274.0929 100.0413 159.8914 232.0821 | N-acetyl-hexosamine derivative | |
| 15.37 | 21 | 1 | 39.40 | 669.1687 [M + FA − H] | 2.18 | C28H32O16 | 299.0537 284.0294 461.1077 | Trihydroxy-methoxy (iso)flavone di-hexoside | [64,65,66] |
| 18.18 | 284 | 30 | 9.36 | 299.0768 | −1.47 | C13H16O8 | 137.0227 109.0266 135.0047 153.0146 91.0181 | Hydroxybenzoyl-hexoside | [67] |
| 19.07 | 464 | 69 | 6.69 | 605.1163 | 2.50 | C27H26O16 | 113.0234 253.0478 351.0579 85.0294 193.0316 | Dihydroxy(iso)flavone di-hexuronide | [64,65] |
| 19.41 | 21 | 1 | 24.86 | 299.0773 | 0.20 | C13H16O8 | 137.0198 135.0049 100.9291 | Hydroxybenzoyl-hexoside | [65,67] |
| 19.49 | 171 | 15 | 11.37 | 605.1164 | 2.63 | C27H26O16 | 113.0211 253.0500 85.0285 351.0572 193.0314 | Dihydroxy(iso)flavone di-hexuronide | [64,65] |
| 19.64 | 63 | 6 | 10.26 | 461.1093 | 0.78 | C22H22O11 | 284.0273 299.0545 255.0303 135.0046 | Trihydroxy methoxy-(iso)flavone hexoside | [64,65,66] |
| 20.06 | 228 | 50 | 4.58 | 429.0824 | −0.75 | C21H18O10 | 253.0489 85.0297 113.0224 135.0058 117.0322 | Dihydroxy(iso)flavone-hexuronide | [64,65] |
| 20.41 | 50 | 7 | 6.88 | 429.0823 | −0.98 | C21H18O10 | 253.0481 117.0344 135.0079 85.0281 280.7946 | Dihydroxy(iso)flavone-hexuronide | [64,65] |
| 21.83 | 52 | 123 | 2.35 | 447.0931 | −0.42 | C21H20O11 | 284.0327 447.1017 | Tetrahydroxy(iso)flavone-hexoside | [64,68] |
| 23.22 | 26 | nd | treated only | 283.0610 | −0.71 | C16H12O5 | 268.0387 147.9081 211.0346 283.0632 | Dihydroxy- methoxy(iso)flavone | [64,66,69] |
| 23.80 | 59 | 2 | 33.23 | 283.0608 | −1.41 | C16H12O5 | 268.0340 211.0382 239.0319 195.0406 | Dihydroxy-methoxy(iso)flavone | [65,70] |
| 23.96 | 68 | 10 | 6.59 | 253.0504 | −0.91 | C15H10O4 | 253.0470 117.0329 135.0080 91.0203 133.0290 | Dihydroxy(iso)flavone | [65] |
| 24.03 | 28 | 1 | 19.01 | 313.0712 | −1.79 | C17H14O6 | 255.0286 298.0491 283.0201 227.0312 171.0404 | Dihydroxy- dimethoxy-(iso)flavone | [65] |
| 25.11 | 151 | 68 | 2.22 | 263.1285 | −1.44 | C15H20O4 | 153.0898 219.1360 204.1122 136.0514 | Abscisic acid* | |
| 26.22 | 29 | 2 | 18.56 | 267.0663 | 0.07 | C16H12O4 | 252.0402 223.0395 195.0422 267.0628 251.0305 | Hydroxy-methoxy(iso)flavone | [64,70] |
| 26.8 | 588 | 88 | 6.69 | 267.0664 | 0.45 | C16H12O4 | 252.0406 223.0371 195.0419 | Hydroxy-methoxy(iso)flavone | |
| 27.73 | 39 | nd | treated only | 299.0920 | −1.62 | C17H16O5 | 135.0072 91.0170 299.0911 284.0303 269.0444 256.0339 | Unknown compound | |
| 28.34 | 129 | 17 | 7.65 | 473.1464 [M + FA − H] | 2.28 | C24H26O10 | 254.0571 209.0582 211.0394 225.0548 269.0838 | Hydroxy-methoxy-pterocarpan malonate hexoside derivative | [71] |
| 30.19 | 12 | 1 | 10.12 | 297.0763 | −1.85 | C17H14O5 | 239.0363 282.0544 183.0049 195.0418 254.0552 211.0394 | Dimethoxy-hydroxy(iso)flavone | [64] |
| 34.76 | 38 | 3 | 14.57 | 313.0715 | −0.83 | C17H14O6 | 255.0277 298.0473 227.0329 270.0526 183.0420 | Dihydroxy- dimethoxy(iso)flavone | [64,65] |
| 36.49 | 71 | 3 | 22.64 | 509.1244 | 0.41 | C30H22O8 | 237.0909 263.0704 135.0058 373.1054 399.0872 | Hydroxylated chalcone dimer | [65] |
| Rt (min) | Relative Abundance | Fold | [M + H] | Error (ppm) | Molecular Formula | MSMS [M + H] | Putative Identity | Reference | |
|---|---|---|---|---|---|---|---|---|---|
| Cadmium-Exposed | Control | ||||||||
| 2.5 | 1 | 114 | 113.29 | 276.107 | −2.82 | C11H19NO8 | 138.0521 96.0424 144.0625 126.0524 84.0413 | N-acetyl-hexosamine derivative | |
| 16.62 | 82 | 312 | 3.92 | 271.0593 | −2.95 | C15H10O5 | 215.0675 149.0229 57.0700 43.0562 | Trihydroxy(iso)flavone conjugated | [64,65] |
| 18.17 | 288 | 16 | 17.84 | 241.0853 | −2.57 | C15H12O3 | 107.0438 131.0451 147.0454 123.0414 77.0363 | Unknown compound | |
| 19.04 | 380 | 65 | 5.83 | 607.1284 | −1.58 | C27H26O16 | 255.0602 | Dihydroxy(iso)flavone-di-hexuronide | [64,65] |
| 19.46 | 139 | 15 | 9.17 | 607.01278 | −2.57 | C27H26O16 | 255.0560 | Dihydroxy(iso)flavone di-hexuronide | [64,65] |
| 19.65 | 36 | 3 | 12.82 | 463.1224 | −2.35 | C22H22O11 | 301.0698 241.0524 269.0445 213.0523 145.0240 | Trihydroxy-methoxy-(iso)flavone hexoside | [64,65,66] |
| 20.07 | 281 | 70 | 3.99 | 431.0957 | −3.64 | C21H18O10 | 255.0646 137.0214 145.0236 119.0462 | Dihydroxy(iso)flavone hexuronide | [64,65] |
| 20.37 | 58 | 10 | 6.03 | 431.0959 | −3.18 | C21H18O10 | 255.0629 137.0224 145.0267 119.0485 | Dihydroxy(iso)flavone hexuronide | [64,65] |
| 21.13 | 76 | nd | treated only | 489.1382 | −1.9 | C24H26O12 | 241.0828 131.0497 147.0428 213.0866 107.0483 | Unknown compound | [65] |
| 21.82 | 24 | 53 | 2.22 | 449.1063 | −3.43 | C21H20O11 | 287.0529 153.0133 269.0445 259.0586 | Tetrahydroxy(iso)flavone-hexoside | [64,68] |
| 23.07 | 104 | 4 | 23.42 | 563.1382 | −2.36 | C26H26O14 | 315.0839 299.0534 254.0531 | Dihydroxy- dimethoxy-(iso)flavone malonate-hexoside derivative | [64,65] |
| 23.22 | 70 | 1 | 61.76 | 533.1279 | −2.01 | C25H24O13 | 285.0756 270.0515 253.0510 225.0545 242.0553 | Dihydroxy methoxy-(iso)flavone malonate-hexoside derivative | [64,65] |
| 23.94 | 61 | 17 | 3.51 | 255.0640 | −4.67 | C15H10O4 | 255.0638 137.0209 145.0263 119.0462 93.0307 | Dihydroxy(iso)flavone | [65] |
| 24.02 | 86 | 2 | 37.18 | 563.1376 | −3.43 | C26H26O14 | 315.0834 300.0593 283.0562 255.0645 | Dihydroxy- dimethoxy-(iso)flavone malonate-hexoside derivative | [64,65] |
| 24.78 | 70 | 1 | 90.39 | 563.1376 | −3.43 | C26H26O14 | 315.0868 207.0628 175.0371 300.0625 147.0419 | Dihydroxy- dimethoxy-(iso)flavone malonate-hexoside derivative | [64,65] |
| 25.19 | 32 | 9 | 3.75 | 299.0907 | −2.34 | C17H14O5 | 299.0955 284.0663 93.0674 256.0754 166.0265 | Dimethoxy-hydroxy(iso)flavone | [64] |
| 26.18 | 74 | 7 | 9.93 | 517.1324 | −3.19 | C25H24O12 | 269.0809 254.0608 237.0551 213.0890 107.0488 | Hydroxy-methoxy(iso)flavone malonyl hexose | [64,69,71] |
| 26.79 | 1999 | 287 | 6.97 | 517.1324 | −3.19 | C25H24O12 | 269.0804 254.0569 213.0902 237.0544 226.0615 | Hydroxy-methoxy-(iso)flavone malonate-hexoside derivative | [64,71] |
| 26.90 | 593 | 50 | 11.85 | 547.1433 | −2.41 | C26H26O13 | 299.0914 284.0680 256.0706 239.0703 | Dimethoxy-hydroxy(iso)flavone malonate-hexoside derivative | [64] |
| 27.55 | 46 | nd | treated only | 533.1272 | −3.26 | C25H24O13 | 151.0376 123.0401 285.0806 | Unknown compound | |
| 28.35 | 611 | 100 | 6.10 | 519.1478 | −3.70 | C25H26O12 | 137.0575 271.0961 161.0581 123.0421 201.1603 | Hydroxy methoxy-pterocarpan malonate-hexoside derivative | [64,69,71] |
| 28.50 | 32 | nd | treated only | 329.1011 | −2.70 | C18H16O6 | 329.1008 313.0722 285.0746 295.0591 267.0649 | Unknown compound | |
| 30.18 | 387 | 105 | 3.69 | 299.0904 | −3.34 | C17H14O5 | 299.0915 284.0663 256.0721 132.0539 227.0699 | Dimethoxy-hydroxy(iso)flavone | [64,65] |
| 31.13 | 73 | 19 | 3.86 | 271.0954 | −4.02 | C16H14O4 | 137.0573 109.0621 79.0524 123.0413 161.0578 | Pterocarpan | [64,69] |
| 36.48 | 23 | nd | treated-only | 511.1372 | −3.01 | C30H22O8 | 137.0214 375.1088 439.2446 265.0972 | Hydroxylated chalcone dimer | [65] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gutsch, A.; Hendrix, S.; Guerriero, G.; Renaut, J.; Lutts, S.; Alseekh, S.; Fernie, A.R.; Hausman, J.-F.; Vangronsveld, J.; Cuypers, A.; et al. Long-Term Cd Exposure Alters the Metabolite Profile in Stem Tissue of Medicago sativa. Cells 2020, 9, 2707. https://doi.org/10.3390/cells9122707
Gutsch A, Hendrix S, Guerriero G, Renaut J, Lutts S, Alseekh S, Fernie AR, Hausman J-F, Vangronsveld J, Cuypers A, et al. Long-Term Cd Exposure Alters the Metabolite Profile in Stem Tissue of Medicago sativa. Cells. 2020; 9(12):2707. https://doi.org/10.3390/cells9122707
Chicago/Turabian StyleGutsch, Annelie, Sophie Hendrix, Gea Guerriero, Jenny Renaut, Stanley Lutts, Saleh Alseekh, Alisdair R. Fernie, Jean-Francois Hausman, Jaco Vangronsveld, Ann Cuypers, and et al. 2020. "Long-Term Cd Exposure Alters the Metabolite Profile in Stem Tissue of Medicago sativa" Cells 9, no. 12: 2707. https://doi.org/10.3390/cells9122707
APA StyleGutsch, A., Hendrix, S., Guerriero, G., Renaut, J., Lutts, S., Alseekh, S., Fernie, A. R., Hausman, J.-F., Vangronsveld, J., Cuypers, A., & Sergeant, K. (2020). Long-Term Cd Exposure Alters the Metabolite Profile in Stem Tissue of Medicago sativa. Cells, 9(12), 2707. https://doi.org/10.3390/cells9122707










