Endothelium-Derived Extracellular Vesicles Associate with Poor Prognosis in Metastatic Colorectal Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Isolation of CTCs, tdEVs, CECs, and edEVs
2.3. Enumeration of tdEVs, edEVs, ldEVs, and Nucleated Cells by ACCEPT
2.4. Statistical Analysis
3. Results
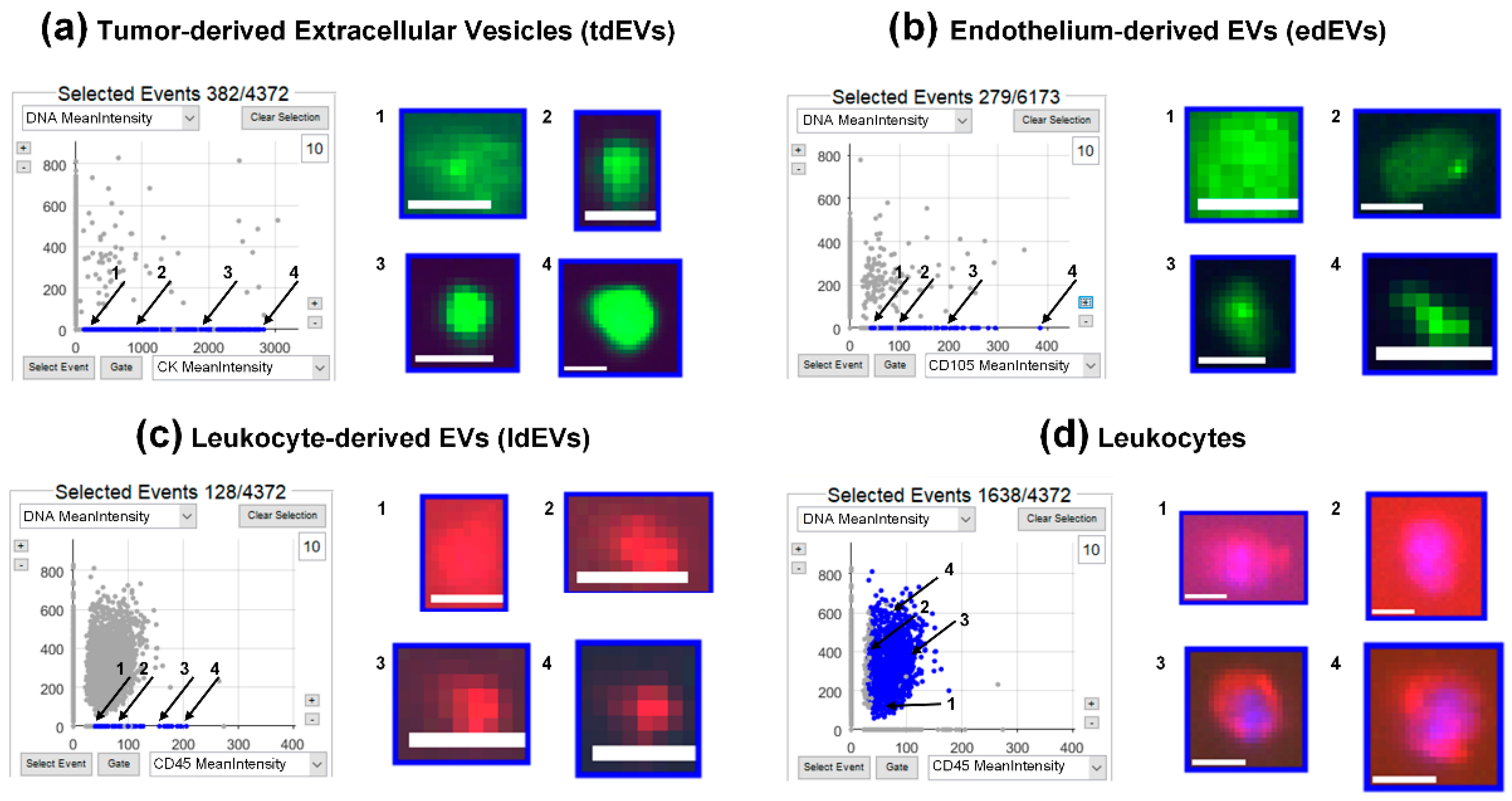
3.1. Identification of tdEVs, edEVs, ldEVs, Leukocytes, and Nucleated Cells by ACCEPT
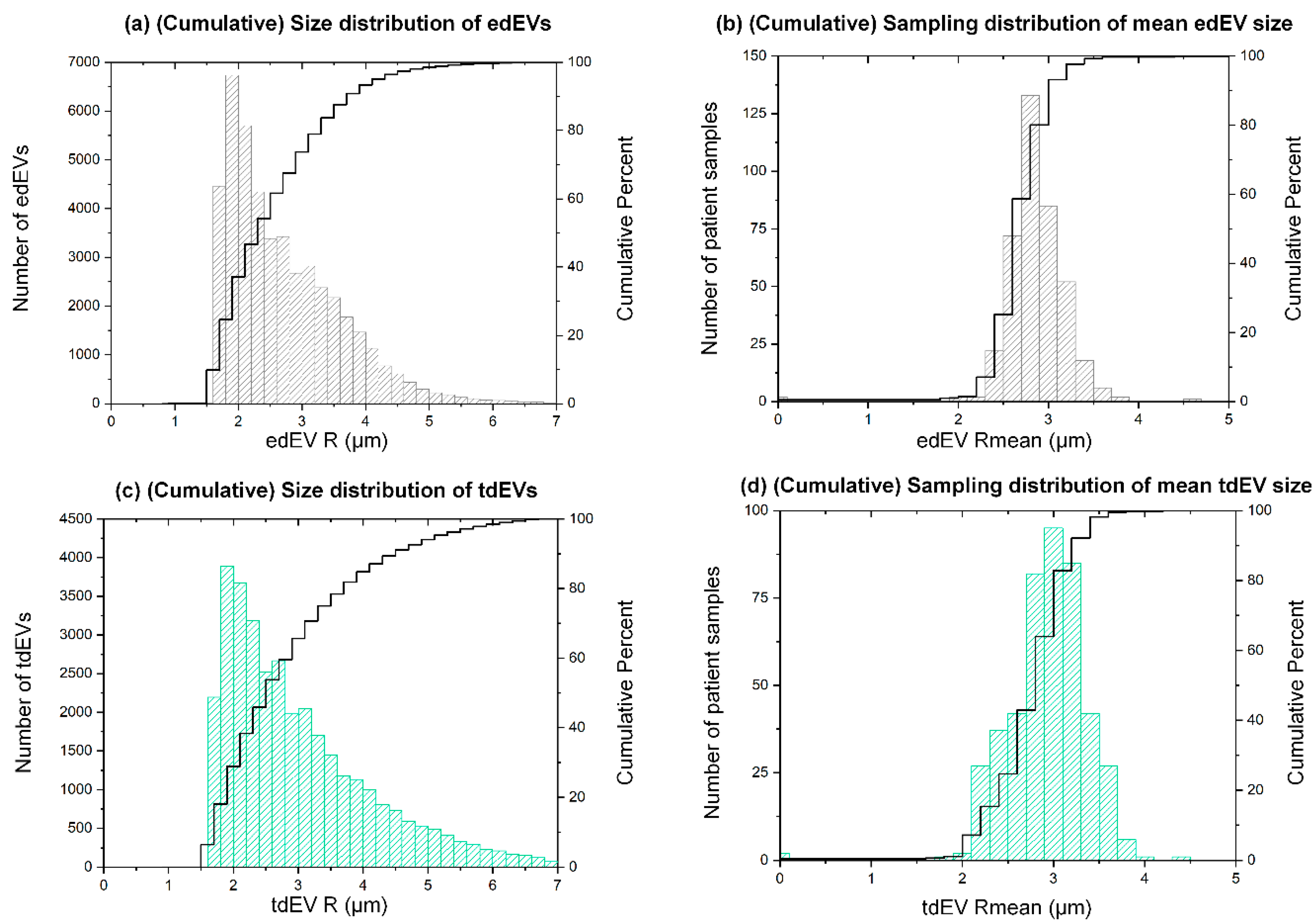
3.2. Estimated Size Distribution of edEVs and tdEVs Derived from Fluorescence Images
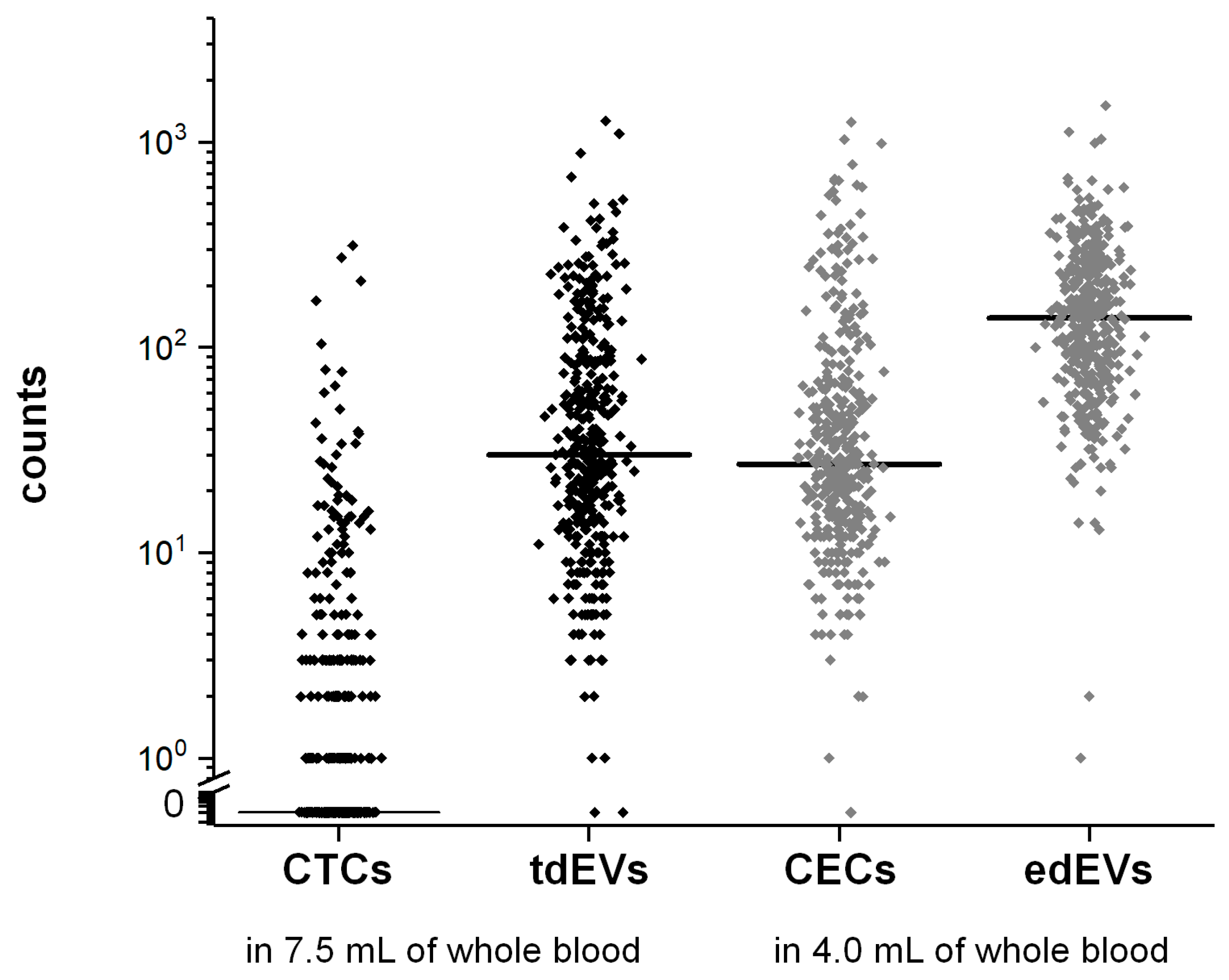
3.3. Frequencies of CTCs, tdEVs, CECs, and edEVs in Colorectal Cancer Patients before the Initiation of a New Therapy
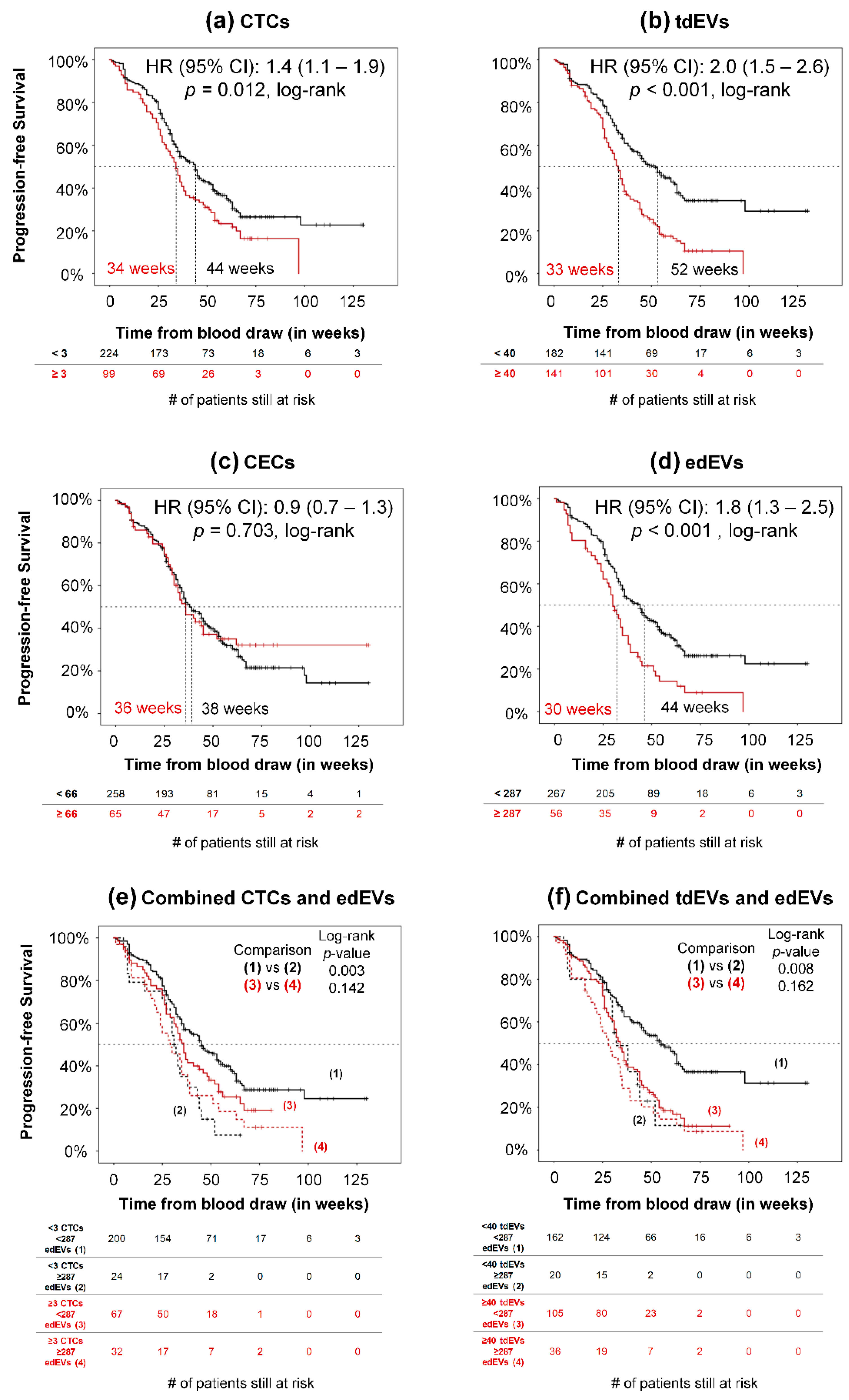
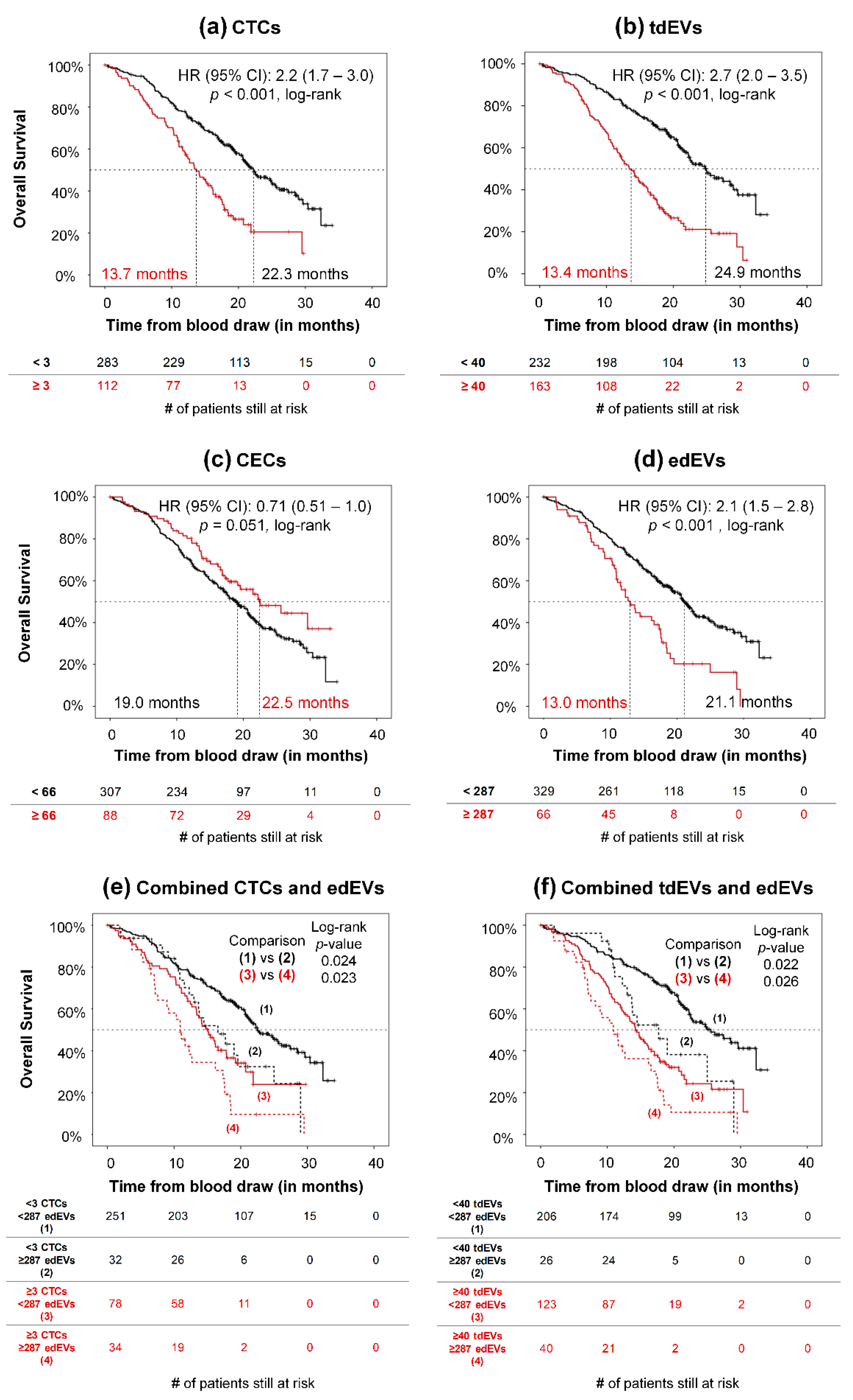
3.4. Association of CTCs, tdEVs, CECs, and edEVs with Progression-Free and Overall Survival of Colorectal Cancer Patients before the Initiation of a New Therapy
3.5. Univariable and Multivariable Associations between Potential Risk Factors and Clinical Outcome (PFS and OS) of mCRC Patients
3.6. Association of Clinical Parameters with tdEVs, CTCs, CECs and edEVs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cohen, S.J.; Punt, C.J.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C.; et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008, 26, 3213–3221. [Google Scholar] [CrossRef] [PubMed]
- Tol, J.; Koopman, M.; Miller, M.C.; Tibbe, A.; Cats, A.; Creemers, G.J.M.; Vos, A.H.; Nagtegaal, I.D.; Terstappen, L.W.M.M.; Punt, C.J.A. Circulating tumour cells early predict progression-free and overall survival in advanced colorectal cancer patients treated with chemotherapy and targeted agents. Ann. Oncol. Off. J. Eur. Soc. Med Oncol. 2010, 21, 1006–1012. [Google Scholar] [CrossRef]
- Nanou, A.; Coumans, F.A.W.; van Dalum, G.; Zeune, L.L.; Dolling, D.; Onstenk, W.; Crespo, M.; Fontes, M.S.; Rescigno, P.; Fowler, G.; et al. Circulating tumor cells, tumor-derived extracellular vesicles and plasma cytokeratins in castration-resistant prostate cancer patients. Oncotarget 2018, 9, 19283–19293. [Google Scholar] [CrossRef] [PubMed]
- De Wit, S.; Rossi, E.; Weber, S.; Tamminga, M.; Manicone, M.; Swennenhuis, J.F.; Groothuis-Oudshoorn, C.G.M.; Vidotto, R.; Facchinetti, A.; Zeune, L.L.; et al. Single tube liquid biopsy for advanced non-small cell lung cancer. Int. J. Cancer 2018. [Google Scholar] [CrossRef]
- Nanou, A.; Miller, M.C.; Zeune, L.L.; de Wit, S.; Punt, C.J.A.; Groen, H.J.M.; Hayes, D.F.; de Bono, J.S.; Terstappen, L. Tumour-derived extracellular vesicles in blood of metastatic cancer patients associate with overall survival. Br. J. Cancer 2020. [Google Scholar] [CrossRef] [PubMed]
- Rowand, J.L.; Martin, G.; Doyle, G.V.; Miller, M.C.; Pierce, M.S.; Connelly, M.C.; Rao, C.; Terstappen, L.W. Endothelial cells in peripheral blood of healthy subjects and patients with metastatic carcinomas. Cytometry A 2007, 71, 105–113. [Google Scholar] [CrossRef]
- Simkens, L.H.; Tol, J.; Terstappen, L.W.; Teerenstra, S.; Punt, C.J.; Nagtegaal, I.D. The predictive and prognostic value of circulating endothelial cells in advanced colorectal cancer patients receiving first-line chemotherapy and bevacizumab. Ann. Oncol. 2010, 21, 2447–2448. [Google Scholar] [CrossRef]
- Tol, J.; Koopman, M.; Cats, A.; Rodenburg, C.J.; Creemers, G.J.M.; Schrama, J.G.; Erdkamp, F.L.G.; Vos, A.H.; van Groeningen, C.J.; Sinnige, H.A.M.; et al. Chemotherapy, bevacizumab, and cetuximab in metastatic colorectal cancer. N. Engl. J. Med. 2009, 360, 563–572. [Google Scholar] [CrossRef]
- Therasse, P.; Arbuck, S.G.; Eisenhauer, E.A.; Wanders, J.; Kaplan, R.S.; Rubinstein, L.; Verweij, J.; Van Glabbeke, M.; van Oosterom, A.T.; Christian, M.C.; et al. New guidelines to evaluate the response to treatment in solid tumors. European organization for research and treatment of cancer, national cancer institute of the United States, national cancer institute of Canada. J. Natl. Cancer Inst. 2000, 92, 205–216. [Google Scholar] [CrossRef]
- Tol, J.; Koopman, M.; Rodenburg, C.J.; Cats, A.; Creemers, G.J.; Schrama, J.G.; Erdkamp, F.L.G.; Vos, A.H.; Mol, L.; Antonini, N.F.; et al. A randomised phase III study on capecitabine, oxaliplatin and bevacizumab with or without cetuximab in first-line advanced colorectal cancer, the CAIRO2 study of the Dutch Colorectal Cancer Group (DCCG). An interim analysis of toxicity. Ann. Oncol. J. Eur. Soc. Med Oncol. 2008, 19, 734–738. [Google Scholar] [CrossRef]
- Coumans, F.; Terstappen, L. detection and characterization of circulating tumor cells by the cellsearch approach. Methods Mol Biol. 2015, 1347, 263–278. [Google Scholar] [CrossRef] [PubMed]
- Budczies, J.; Klauschen, F.; Sinn, B.V.; Gyorffy, B.; Schmitt, W.D.; Darb-Esfahani, S.; Denkert, C. Cutoff finder: A comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS ONE. 2012, 7, e51862. [Google Scholar] [CrossRef] [PubMed]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, P.J.; McKenzie, A.; Fisicaro, N.; Rockman, S.P.; Pearse, M.J.; d’Apice, A.J. Endoglin: A 180-kD endothelial cell and macrophage restricted differentiation molecule. Clin Exp. Immunol. 1992, 90, 154–159. [Google Scholar] [CrossRef]
- Lastres, P.; Bellon, T.; Cabanas, C.; Sanchez-Madrid, F.; Acevedo, A.; Gougos, A.; Letarte, M.; Bernabeu, C. Regulated expression on human macrophages of endoglin, an Arg-Gly-Asp-containing surface antigen. Eur. J. Immunol. 1992, 22, 393–397. [Google Scholar] [CrossRef]
- Dignat-George, F.; Sabatier, F.; Blann, A.; Woywodt, A. Detection of circulating endothelial cells: CD146-based magnetic separation enrichment or flow cytometric assay? J. Clin. Oncol. 2007, 25, e1–e2. [Google Scholar] [CrossRef]
- Elshal, M.F.; Khan, S.S.; Takahashi, Y.; Solomon, M.A.; McCoy, J.P. CD146 (Mel-CAM), an adhesion marker of endothelial cells, is a novel marker of lymphocyte subset activation in normal peripheral blood. Blood 2005, 106, 2923–2924. [Google Scholar] [CrossRef]
- Pickl, W.F.; Majdic, O.; Fischer, G.F.; Petzelbauer, P.; Fae, I.; Waclavicek, M.; Stockl, J.; Scheinecker, C.; Vidicki, T.; Aschauer, H.; et al. MUC18/MCAM (CD146), an activation antigen of human T lymphocytes. J. Immunol. 1997, 158, 2107–2115. [Google Scholar]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef]
- Scher, H.I.; Heller, G.; Molina, A.; Attard, G.; Danila, D.C.; Jia, X.; Peng, W.; Sandhu, S.K.; Olmos, D.; Riisnaes, R.; et al. Circulating tumor cell biomarker panel as an individual-level surrogate for survival in metastatic castration-resistant prostate cancer. J. Clin. Oncol. 2015, 33, 1348–1355. [Google Scholar] [CrossRef]
- Bidard, F.C.; Fehm, T.; Ignatiadis, M.; Smerage, J.B.; Alix-Panabieres, C.; Janni, W.; Messina, C.; Paoletti, C.; Muller, V.; Hayes, D.F.; et al. Clinical application of circulating tumor cells in breast cancer: Overview of the current interventional trials. Cancer Metastasis Rev. 2013, 32, 179–188. [Google Scholar] [CrossRef] [PubMed]
- El-Heliebi, A.; Hille, C.; Laxman, N.; Svedlund, J.; Haudum, C.; Ercan, E.; Kroneis, T.; Chen, S.; Smolle, M.; Rossmann, C. In situ detection and quantification of AR-V7, AR-FL, PSA, and KRAS point mutations in circulating tumor cells. Clin. Chem. 2018, 64, 536–546. [Google Scholar] [CrossRef] [PubMed]
- Swennenhuis, J.F.; Reumers, J.; Thys, K.; Aerssens, J.; Terstappen, L.W. Efficiency of whole genome amplification of single circulating tumor cells enriched by CellSearch and sorted by FACS. Genome Med. 2013, 5, 106. [Google Scholar] [CrossRef] [PubMed]
- Coumans, F.A.; Ligthart, S.T.; Uhr, J.W.; Terstappen, L.W. Challenges in the enumeration and phenotyping of CTC. Clin. Cancer Res. 2012, 18, 5711–5718. [Google Scholar] [CrossRef]
- Nanou, A.; Zeune, L.L.; Bidard, F.-C.; Pierga, J.-Y. Terstappen, L.W.M.M. HER2 expression on tumor-derived extracellular vesicles and circulating tumor cells in metastatic breast cancer. Breast Cancer Res. 2020, 22, 1–11. [Google Scholar] [CrossRef]
- Ciardiello, C.; Cavallini, L.; Spinelli, C.; Yang, J.; Reis-Sobreiro, M.; de Candia, P.; Minciacchi, V.R.; Di Vizio, D. Focus on extracellular vesicles: New frontiers of cell-to-cell communication in cancer. Int. J. Mol. Sci. 2016, 17, 175. [Google Scholar] [CrossRef]
- Koukourakis, M.I.; Giatromanolaki, A.; Simopoulos, C.; Polychronidis, A.; Sivridis, E. Lactate dehydrogenase 5 (LDH5) relates to up-regulated hypoxia inducible factor pathway and metastasis in colorectal cancer. Clinical Exp. Metastasis 2005, 22, 25–30. [Google Scholar] [CrossRef]
- Jurisic, V.; Radenkovic, S.; Konjevic, G. The actual role of LDH as tumor marker, biochemical and clinical aspects. Adv. Exp. Med. Biol. 2015, 867, 115–124. [Google Scholar]
- Koukourakis, M.I.; Giatromanolaki, A.; Sivridis, E.; Bougioukas, G.; Didilis, V.; Gatter, K.; Harris, A. Lactate dehydrogenase-5 (LDH-5) overexpression in non-small-cell lung cancer tissues is linked to tumour hypoxia, angiogenic factor production and poor prognosis. Br. J. Cancer 2003, 89, 877–885. [Google Scholar] [CrossRef]
- Kolev, Y.; Uetake, H.; Takagi, Y.; Sugihara, K. Lactate dehydrogenase-5 (LDH-5) expression in human gastric cancer: Association with hypoxia-inducible factor (HIF-1α) pathway, angiogenic factors production and poor prognosis. Ann. Surg. Oncol. 2008, 15, 2336–2344. [Google Scholar] [CrossRef]
- Zweitzig, D.R.; Smirnov, D.A.; Connelly, M.C.; Terstappen, L.W.; O’Hara, S.M.; Moran, E. Physiological stress induces the metastasis marker AGR2 in breast cancer cells. Mol. Cell. Biochem. 2007, 306, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Kallergi, G.; Markomanolaki, H.; Giannoukaraki, V.; Papadaki, M.A.; Strati, A.; Lianidou, E.S.; Georgoulias, V.; Mavroudis, D.; Agelaki, S. Hypoxia-inducible factor-1α and vascular endothelial growth factor expression in circulating tumor cells of breast cancer patients. Breast Cancer Res. 2009, 11, R84. [Google Scholar] [CrossRef] [PubMed]
- Strijbos, M.H.; Gratama, J.W.; Kraan, J.; Lamers, C.H.; den Bakker, M.A.; Sleijfer, S. Circulating endothelial cells in oncology: Pitfalls and promises. Br. J. Cancer 2008, 98, 1731–1735. [Google Scholar] [CrossRef] [PubMed]
- Strijbos, M.H.; Gratama, J.W.; Schmitz, P.I.; Rao, C.; Onstenk, W.; Doyle, G.V.; Miller, M.C.; de Wit, R.; Terstappen, L.W.; Sleijfer, S. Circulating endothelial cells, circulating tumour cells, tissue factor, endothelin-1 and overall survival in prostate cancer patients treated with docetaxel. Eur. J. Cancer 2010, 46, 2027–2035. [Google Scholar] [CrossRef]
- Smirnov, D.A.; Foulk, B.W.; Doyle, G.V.; Connelly, M.C.; Terstappen, L.W.M.M.; O’Hara, S.M. Global gene expression profiling of circulating endothelial cells in patients with metastatic carcinomas. Cancer Res. 2006, 66, 2918–2922. [Google Scholar] [CrossRef]
- Gootjes, E.C.; Kraan, J.; Buffart, T.; Verhoef, C.; Verheul, H.M.; Sleijfer, S.; Group, O.S. CD276-positive circulating endothelial cells in advanced colorectal cancer. Am. Soc. Clin. Oncol. 2019, 37, 572. [Google Scholar] [CrossRef]
- Duval, A.; Helley, D.; Capron, L.; Youinou, P.; Renaudineau, Y.; Dubucquoi, S.; Fischer, A.M.; Hachulla, E. Endothelial dysfunction in systemic lupus patients with low disease activity: Evaluation by quantification and characterization of circulating endothelial microparticles, role of anti-endothelial cell antibodies. Rheumatology 2010, 49, 1049–1055. [Google Scholar] [CrossRef]
- Dignat-George, F.; Boulanger, C.M. The many faces of endothelial microparticles. Arter. Thromb. Vasc. Biol. 2011, 31, 27–33. [Google Scholar] [CrossRef]
- Hromada, C.; Muhleder, S.; Grillari, J.; Redl, H.; Holnthoner, W. Endothelial extracellular vesicles-promises and challenges. Front Physiol. 2017, 8, 275. [Google Scholar] [CrossRef]
- Jy, W.; Horstman, L.L.; Jimenez, J.J.; Ahn, Y.S.; Biro, E.; Nieuwland, R.; Sturk, A.; Dignat-George, F.; Sabatier, F.; Camoin-Jau, L.; et al. Measuring circulating cell-derived microparticles. J. Thromb Haemost. 2004, 2, 1842–1851. [Google Scholar] [CrossRef]
- Lacroix, R.; Robert, S.; Poncelet, P.; Dignat-George, F. Overcoming limitations of microparticle measurement by flow cytometry. Semin. Thromb Hemost. 2010, 36, 807–818. [Google Scholar] [CrossRef] [PubMed]
- Caruso, S.; Poon, I.K. Apoptotic cell-derived extracellular vesicles: More than just debris. Front. Immunol. 2018, 9, 1486. [Google Scholar] [CrossRef]
- De Wit, S.; Zeune, L.L.; Hiltermann, T.J.N.; Groen, H.J.M.; Dalum, G.V.; Terstappen, L. Classification of cells in CTC-enriched samples by advanced image analysis. Cancers 2018, 10, 377. [Google Scholar] [CrossRef] [PubMed]
- Bhindi, B.; Hermanns, T.; Wei, Y.; Yu, J.; Richard, P.O.; Wettstein, M.S.; Templeton, A.; Li, K.; Sridhar, S.S.; Jewett, M.A.; et al. Identification of the best complete blood count-based predictors for bladder cancer outcomes in patients undergoing radical cystectomy. Br. J. Cancer. 2016, 114, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Taussky, D.; Soulieres, D.; Azoulay, L.; Yin, H.; Bahig, H.; Bahary, J.P.; Delouya, G. A combination of testosterone and white blood cell count as a predictive factor of overall survival in localized prostate cancer. Target Oncol. 2017, 12, 695–701. [Google Scholar] [CrossRef] [PubMed]
- Tibaldi, C.; Vasile, E.; Bernardini, I.; Orlandini, C.; Andreuccetti, M.; Falcone, A. Baseline elevated leukocyte count in peripheral blood is associated with poor survival in patients with advanced non-small cell lung cancer: A prognostic model. J. Cancer Res. Clin. Oncol. 2008, 134, 1143–1149. [Google Scholar] [CrossRef]
- Mandrekar, S.J.; Schild, S.E.; Hillman, S.L.; Allen, K.L.; Marks, R.S.; Mailliard, J.A.; Krook, J.E.; Maksymiuk, A.W.; Chansky, K.; Kelly, K.; et al. A prognostic model for advanced stage nonsmall cell lung cancer. Pooled analysis of north central cancer treatment group trials. Cancer 2006, 107, 781–792. [Google Scholar] [CrossRef]
- Bournazos, S.; Wang, T.T.; Ravetch, J.V. The role and function of fcgamma receptors on myeloid cells. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Pincetic, A.; Bournazos, S.; DiLillo, D.J.; Maamary, J.; Wang, T.T.; Dahan, R.; Fiebiger, B.M.; Ravetch, J.V. Type I and type II Fc receptors regulate innate and adaptive immunity. Nat. Immunol. 2014, 15, 707–716. [Google Scholar] [CrossRef]
- Uracz, W.; Pituch-Noworolska, A.; Popiela, T.; Zembala, M. The Fc receptors of normal and cancer patients monocytes. Immunobiology 1982, 163, 450–459. [Google Scholar] [CrossRef]
- Trzpis, M.; McLaughlin, P.M.; de Leij, L.M.; Harmsen, M.C. Epithelial cell adhesion molecule: More than a carcinoma marker and adhesion molecule. Am. J. Pathol. 2007, 171, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Breuhahn, K.; Baeuerle, P.A.; Peters, M.; Prang, N.; Tox, U.; Kohne-Volland, R.; Dries, V.; Schirmacher, P.; Leo, E. Expression of epithelial cellular adhesion molecule (Ep-CAM) in chronic (necro-)inflammatory liver diseases and hepatocellular carcinoma. Hepatol Res. 2006, 34, 50–56. [Google Scholar] [CrossRef] [PubMed]
- De Boer, C.J.; van Krieken, J.H.; Janssen-van Rhijn, C.M.; Litvinov, S.V. Expression of Ep-CAM in normal, regenerating, metaplastic, and neoplastic liver. J. Pathol. 1999, 188, 201–206. [Google Scholar] [CrossRef]
- Amann, M.; Friedrich, M.; Lutterbuese, P.; Vieser, E.; Lorenczewski, G.; Petersen, L.; Brischwein, K.; Kufer, P.; Kischel, R.; Baeuerle, P.A.; et al. Therapeutic window of an EpCAM/CD3-specific BiTE antibody in mice is determined by a subpopulation of EpCAM-expressing lymphocytes that is absent in humans. Cancer Immunol. Immunother 2009, 58, 95–109. [Google Scholar] [CrossRef]
- Nelson, A.J.; Dunn, R.J.; Peach, R.; Aruffo, A.; Farr, A.G. The murine homolog of human Ep-CAM, a homotypic adhesion molecule, is expressed by thymocytes and thymic epithelial cells. Eur. J. Immunol. 1996, 26, 401–408. [Google Scholar] [CrossRef]
- Dailey, M.O. Expression of T iymphocyte adhesion molecules: Regulation during antigen-induced T cell activation and differentiation. Crit. Rev. Immunol. 1998, 18, 153–184. [Google Scholar] [CrossRef]
- McKelvey, K.J.; Powell, K.L.; Ashton, A.W.; Morris, J.M.; McCracken, S.A. Exosomes: mechanisms of uptake. J. Circ. Biomark. 2015, 4, 7. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654. [Google Scholar] [CrossRef]
- Chennakrishnaiah, S.; Meehan, B.; D’Asti, E.; Montermini, L.; Lee, T.H.; Karatzas, N.; Buchanan, M.; Tawil, N.; Choi, D.; Divangahi, M. Leukocytes as a reservoir of circulating oncogenic DNA and regulatory targets of tumor-derived extracellular vesicles. J. Thromb. Haemostasis. 2018, 16, 1800–1813. [Google Scholar] [CrossRef]
- Zech, D.; Rana, S.; Buchler, M.W.; Zoller, M. Tumor-exosomes and leukocyte activation: An ambivalent crosstalk. Cell Commun. Signal. 2012, 10, 37. [Google Scholar] [CrossRef]
| Variable | Cut-Off | PFS Risk | OS Risk | |||
|---|---|---|---|---|---|---|
| HR (95% CI) | p-Value | HR (95% CI) | p-Value | |||
| Prior adjuvant therapy | 0.7 (0.4–1.0) | 0.043 | 0.7 (0.4–1.0) | 0.046 | ||
| Number of metastatic sites | 1.3 (1.2–1.5) | <0.001 | 1.3 (1.2–1.5) | < 0.001 | ||
| Primary tumor in situ | 1.5 (1.1–2.0) | 0.013 | 1.8 (1.3–2.4) | < 0.001 | ||
| Presence of KRAS mutation | 1.0 (0.7–1.3) | 0.839 | 1.1 (0.8–1.5) | 0.666 | ||
| Presence of BRAF mutation | 3.0 (2.0–4.6) | <0.001 | 2.7 (1.8–4.0) | < 0.001 | ||
| Presence of NRAS mutation | 1.19 (0.60–2.40) | 0.629 | 1.0 (0.5–2.0) | 1.000 | ||
| Right-sidedness of primary tumor (transverse colon, hepatic flexure, ascending colon, cecum) | 1.3 (1.0–1.8) | 0.042 | 1.6 (1.2–2.1) | 0.001 | ||
| Treatment arm | 1.1 (0.9–1.4) | 0.465 | 1.2 (0.9–1.5) | 0.313 | ||
| ECOG performance status | 1.4 (1.1–1.8) | 0.012 | 1.4 (1.1–1.9) | 0.006 | ||
| age | 1.0 (1.0–1.0) | 0.078 | 1.0 (1.0–1.0) | 0.290 | ||
| gender | 1.0 (0.7–1.3) | 0.958 | 1.0 (0.7–1.3) | 0.773 | ||
| CellSearch Populations | ||||||
| CTCs (log transformed) | - | 1.2 (1.1–1.4) | 0.004 | 1.6 (1.4–1.9) | <0.001 | |
| ≥ 3 CTCs | 3 | 1.4 (1.1–1.9) | 0.012 | 2.2 (1.7–3.0) | <0.001 | |
| tdEVs (log-transformed) | - | 1.8 (1.4–2.3) | <0.001 | 2.5 (2.0–3.2) | <0.001 | |
| ≥ 40 tdEVs | 40 | 2.0 (1.5–2.6) | <0.001 | 2.7 (2.0–3.5) | <0.001 | |
| CECs (log-transformed) | - | 1.1 (0.9–1.5) | 0.338 | 1.0 (0.8–1.2) | 0.767 | |
| ≥ 66 CECs | 66 | 0.9 (0.7–1.3) | 0.703 | 0.7 (0.5–1.0) | 0.051 | |
| edEVs (log-transformed) | - | 1.7 (1.2–2.5) | 0.004 | 1.9 (1.3–2.8) | 0.002 | |
| ≥ 287 edEVs | 287 | 1.8 (1.3–2.5) | <0.001 | 2.1 (1.5–2.8) | <0.001 | |
| Other CellSearch Populations | ||||||
| Leukocytes a (log-transformed) | - | 1.3 (1.1–1.5) | 0.009 | 1.3 (1.1–1.6) | 0.001 | |
| ldEVs a (log-transformed) | - | 1.1 (0.9–1.4) | 0.437 | 1.2 (0.9–1.5) | 0.219 | |
| Nucleated a (log-transformed) | - | 1.1 (0.9–1.4) | 0.239 | 1.3 (1.0–1.6) | 0.048 | |
| Leukocytes b (log-transformed) | - | 1.1 (0.8–1.5) | 0.737 | 1.1 (0.8–1.5) | 0.751 | |
| ldEVs b (log-transformed) | - | 1.0 (0.7–1.4) | 0.976 | 1.1 (0.8–1.6) | 0.608 | |
| Nucleated b (log-transformed) | - | 1.1 (0.9–1.5) | 0.436 | 1.3 (1.0–1.7) | 0.091 | |
| CD105+ leukocytes b (log-transformed) | - | 1.0 (0.9–1.2) | 0.789 | 1.0 (0.8–1.2) | 0.927 | |
| CD105+ ldEVs b (log-transformed) | - | 1.0 (0.8–1.2) | 0.842 | 1.1 (0.9–1.4) | 0.203 | |
| CBC-Based Parameters | ||||||
| Leukocytes (log-transformed) | - | 6.0 (2.1–16.9) | 0.001 | 9.5 (3.5–25.9) | <0.001 | |
| Platelets (log-transformed) | - | 4.0 (1.6–10.3) | 0.004 | 5.3 (2.1–13.4) | <0.001 | |
| Hemoglobin, per 1 g/dL | - | 0.8 (0.7–0.9) | <0.001 | 0.7 (0.6–0.8) | <0.001 | |
| Alkaline phosphatase (ALP) (log-transformed) | 2.7 (1.6–4.4) | <0.001 | 3.8 (2.4–6.2) | <0.001 | ||
| Baseline serum lactate Dehydrogenase (LDH) (log-transformed) | 1.9 (1.3–2.8) | 0.002 | 2.4 (1.6–3.5) | <0.001 | ||
| Variable in Equation | PFS Risk | OS Risk | ||
|---|---|---|---|---|
| HR (95% CI) | p-Value | HR (95% CI) | p-Value | |
| Presence of BRAF mutation | 2.8 (1.8–4.4) | < 0.001 | 2.3 (1.4–3.6) | <0.001 |
| tdEVs (log-transformed) | 1.8 (1.3–2.4) | 0.001 | ||
| Number of metastatic sites | 1.4 (1.2–1.7) | < 0.001 | 1.3 (1.1–1.5) | 0.003 |
| Right-sidedness of the primary tumor | 1.6 (1.1–2.3) | 0.010 | ||
| Platelets (log transformed) | 4.6 (1.4–15.1) | 0.013 | ||
| Hemoglobin, per 1 g/dL | 0.8 (0.7–1.0) | 0.048 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nanou, A.; Mol, L.; Coumans, F.A.W.; Koopman, M.; Punt, C.J.A.; Terstappen, L.W.M.M. Endothelium-Derived Extracellular Vesicles Associate with Poor Prognosis in Metastatic Colorectal Cancer. Cells 2020, 9, 2688. https://doi.org/10.3390/cells9122688
Nanou A, Mol L, Coumans FAW, Koopman M, Punt CJA, Terstappen LWMM. Endothelium-Derived Extracellular Vesicles Associate with Poor Prognosis in Metastatic Colorectal Cancer. Cells. 2020; 9(12):2688. https://doi.org/10.3390/cells9122688
Chicago/Turabian StyleNanou, Afroditi, Linda Mol, Frank A. W. Coumans, Miriam Koopman, Cornelis J. A. Punt, and Leon W. M. M. Terstappen. 2020. "Endothelium-Derived Extracellular Vesicles Associate with Poor Prognosis in Metastatic Colorectal Cancer" Cells 9, no. 12: 2688. https://doi.org/10.3390/cells9122688
APA StyleNanou, A., Mol, L., Coumans, F. A. W., Koopman, M., Punt, C. J. A., & Terstappen, L. W. M. M. (2020). Endothelium-Derived Extracellular Vesicles Associate with Poor Prognosis in Metastatic Colorectal Cancer. Cells, 9(12), 2688. https://doi.org/10.3390/cells9122688






