RNA Extraction from Endoscopic Ultrasound-Acquired Tissue of Pancreatic Cancer Is Feasible and Allows Investigation of Molecular Features
Abstract
1. Introduction
2. Materials and Methods
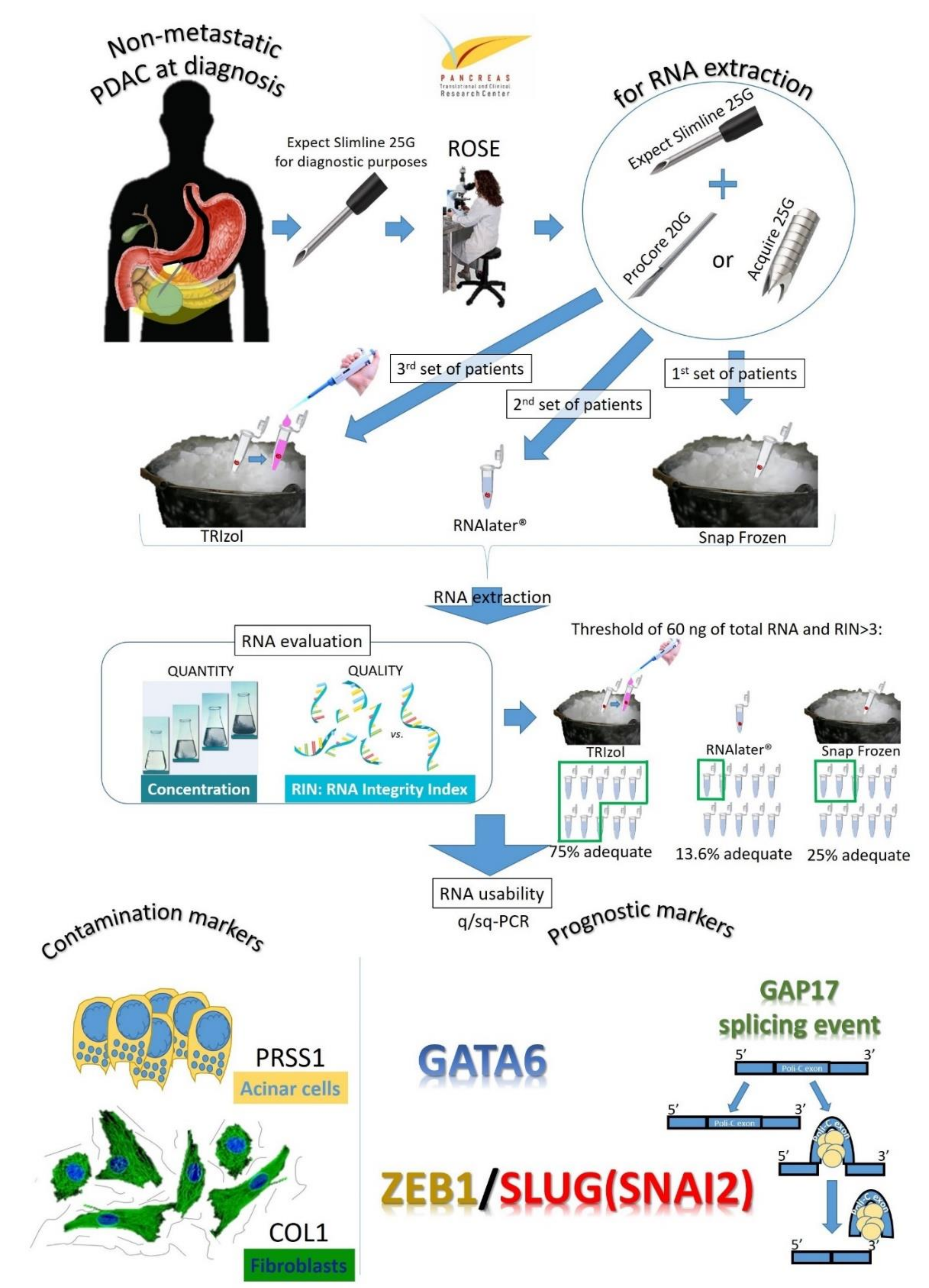
2.1. Study Design and Population
2.2. Endoscopic Ultrasound Procedure Description and Specimen Processing
2.3. RNA Extraction and Analysis
2.4. Gene Expression and Splicing Assay Analysis
2.5. Statistical Analysis
3. Results
3.1. Patients and Tumor Characteristics
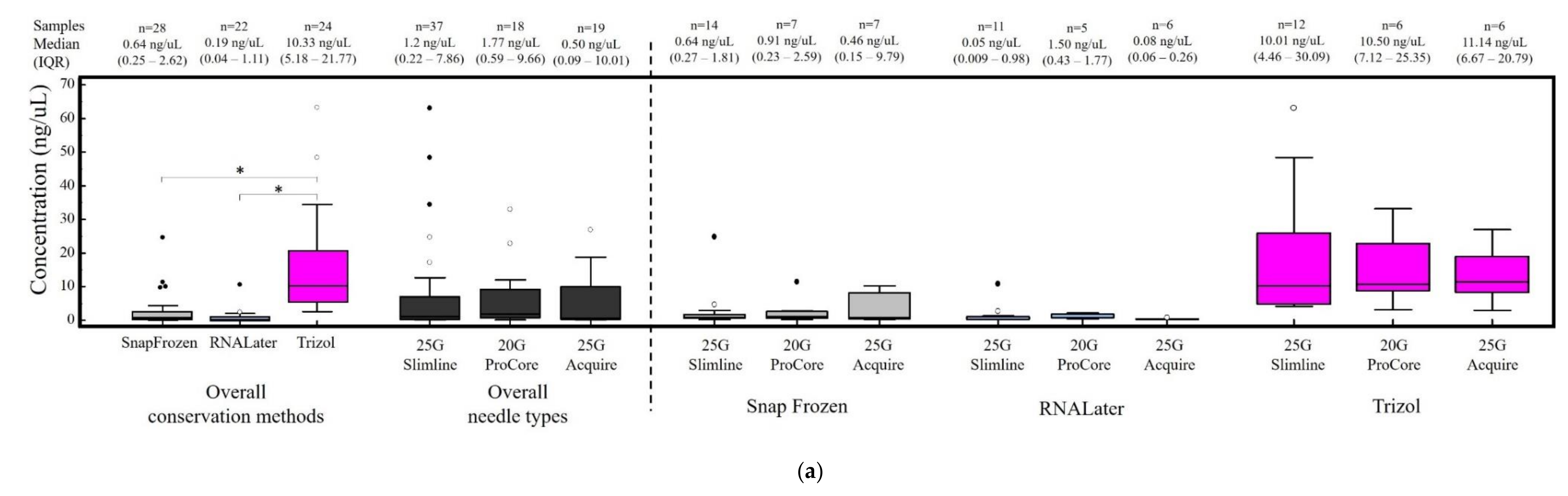
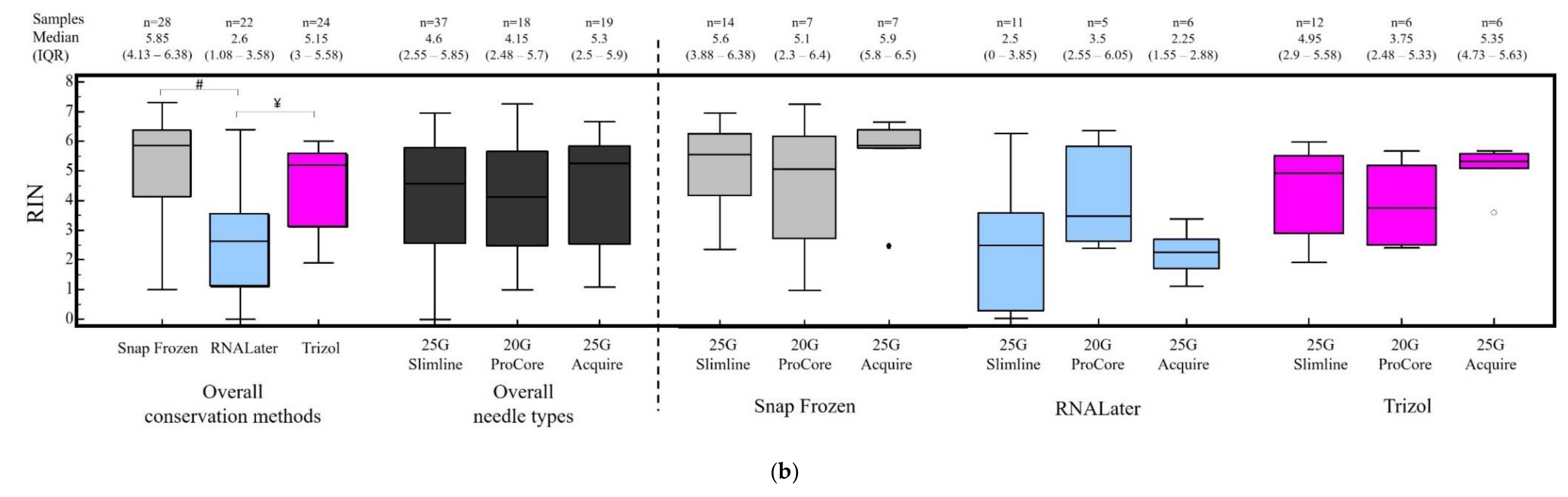
3.2. RNA Quantification and RIN Evaluation
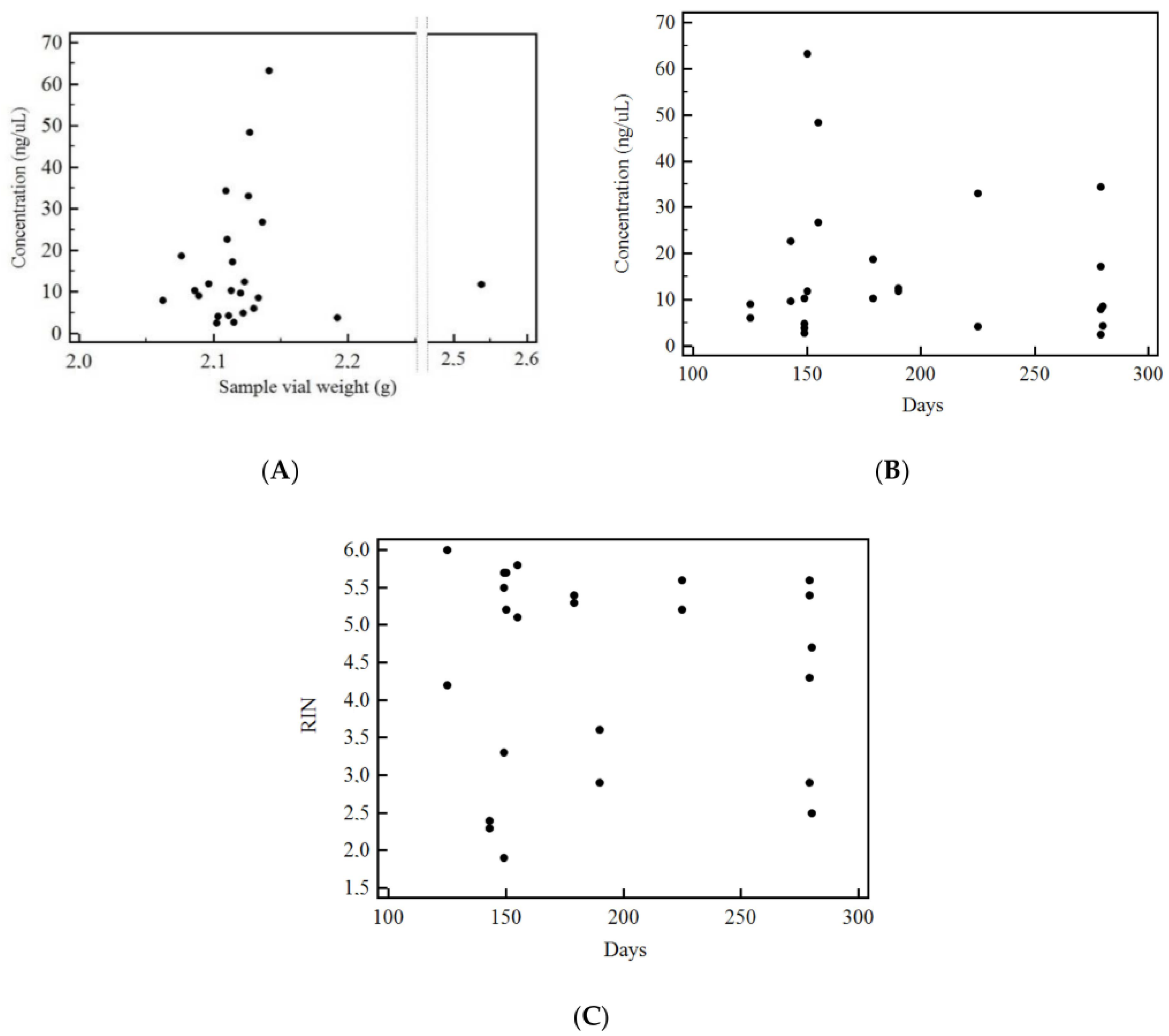
3.3. Factors Correlated with RNA Concentration and Integrity Index
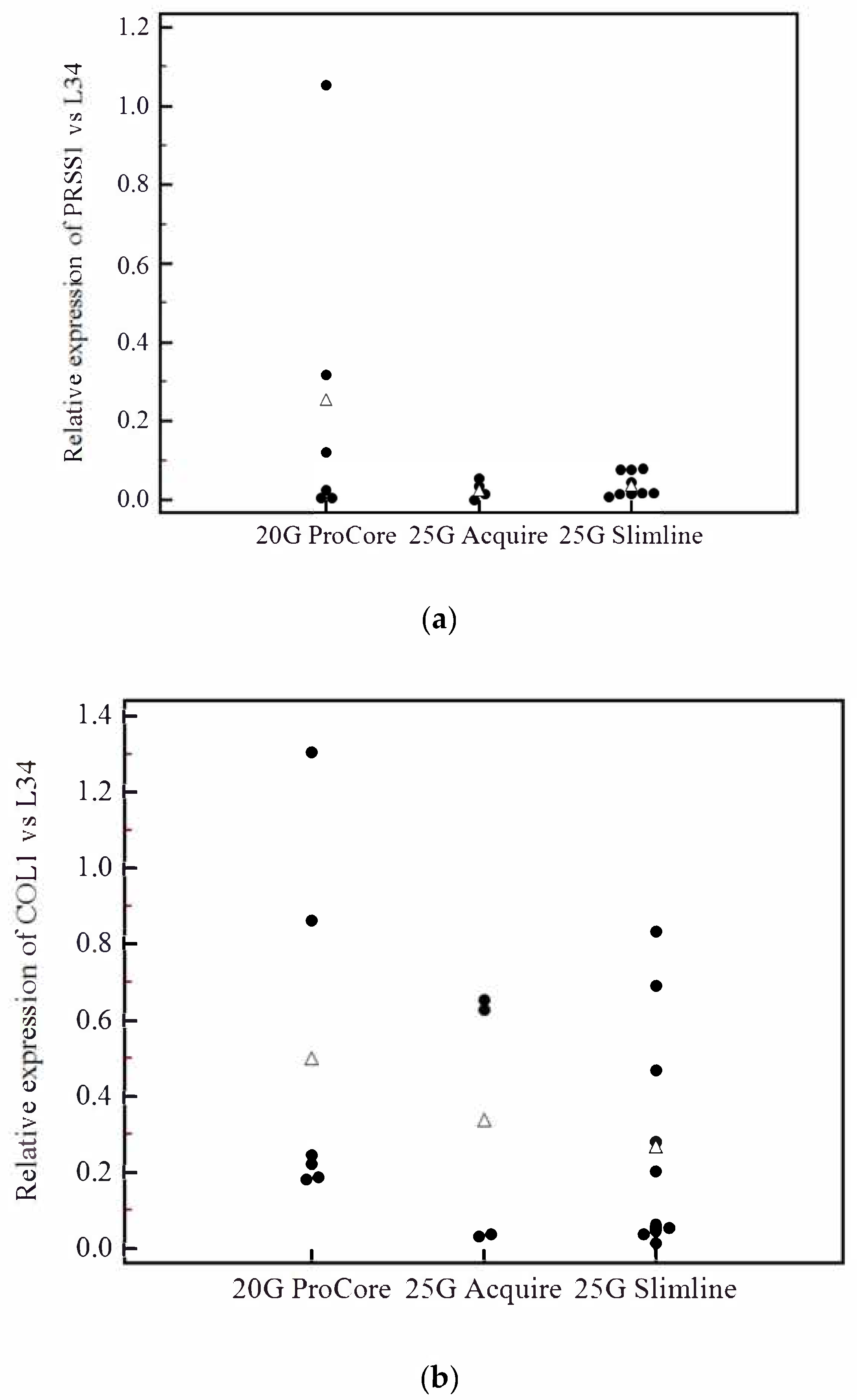
3.4. Analysis of Contamination of Acinar and Stromal on EUS-Derived PDAC Samples
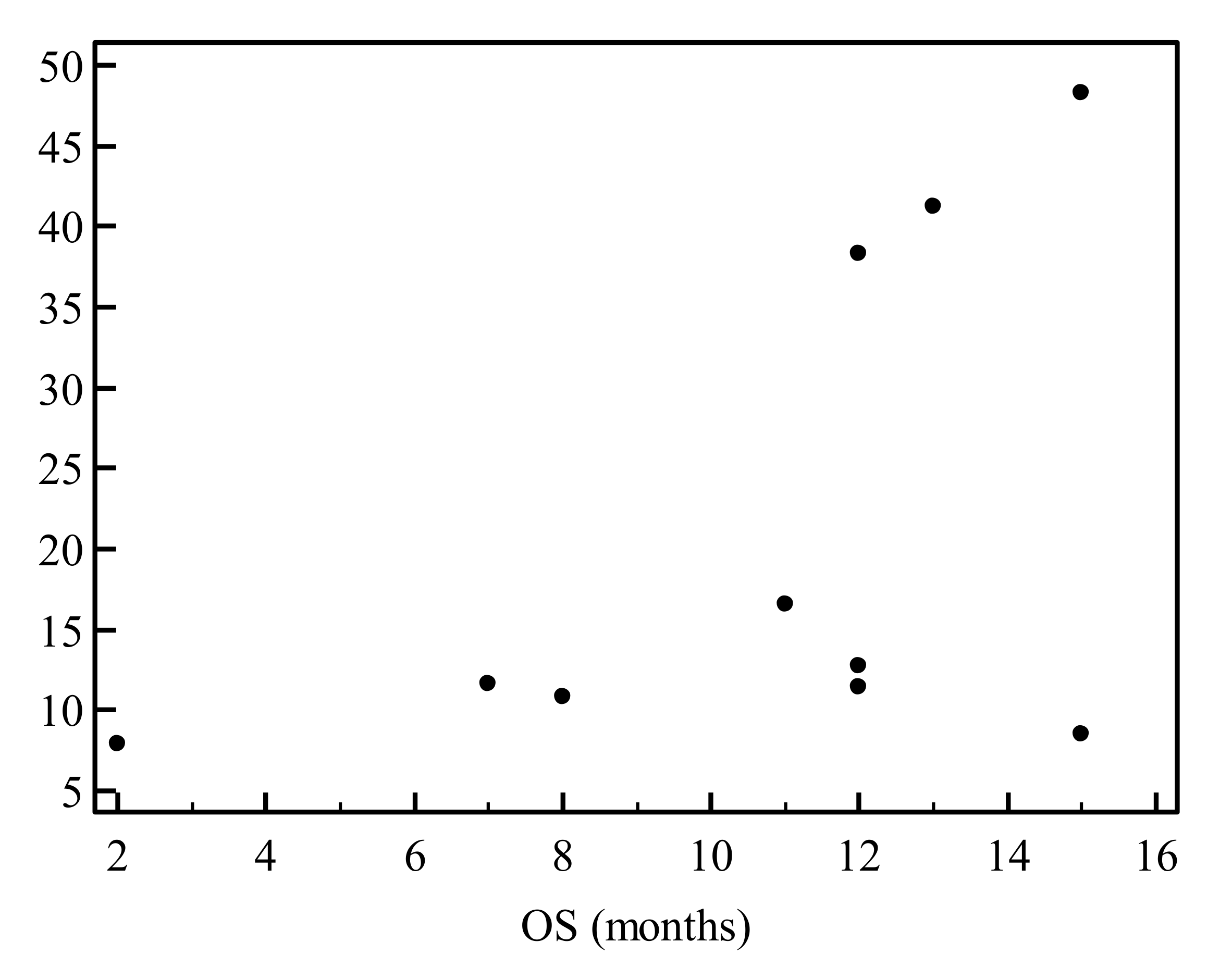
3.5. Evaluation of Prognostic Markers is Feasible on EUS-Derived Samples and May Allow Outcome Prediction
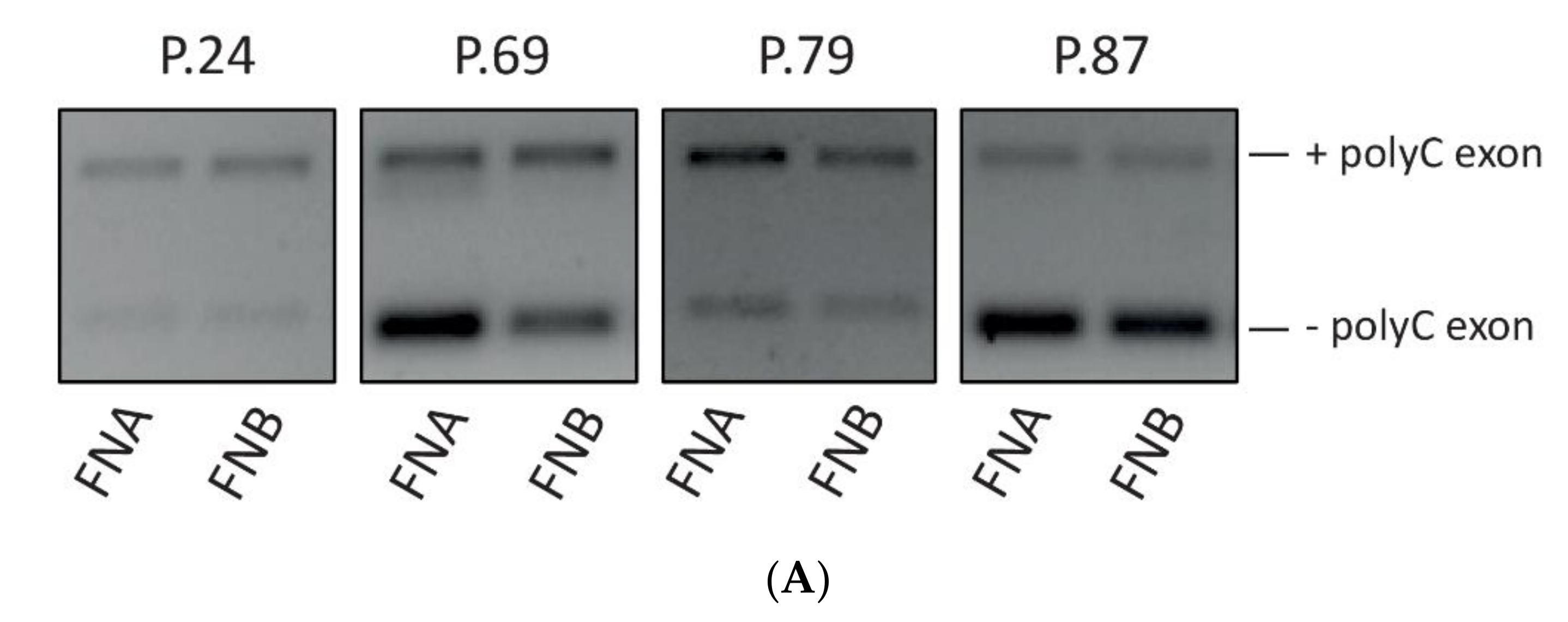
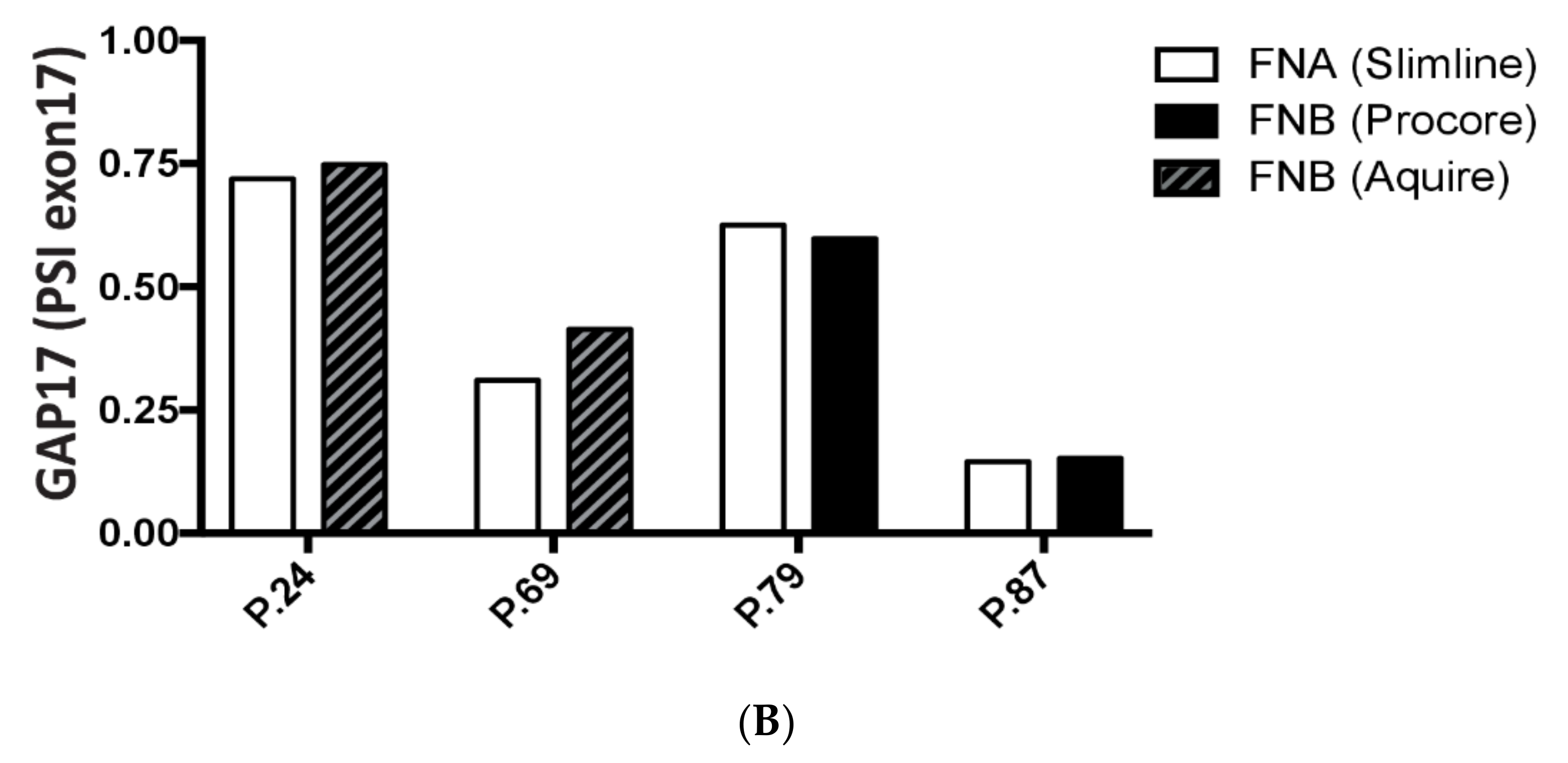
3.6. Analysis of GAP17 Splicing Variants is Feasible on EUS-Derived Samples
4. Discussions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Golan, T.; Hammel, P.; Reni, M.; Van Cutsem, E.; Macarulla, T.; Hall, M.J.; Park, J.O.; Hochhauser, D.; Arnold, D.; Oh, D.Y.; et al. Maintenance Olaparib for Germline BRCA-Mutated Metastatic Pancreatic Cancer. N. Engl. J. Med. 2019, 381, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Tempero, M.A. NCCN Guidelines Updates: Pancreatic Cancer. J. Natl. Compr. Cancer Netw. 2019, 17, 603–605. [Google Scholar]
- Sohal, D.P.S.; Kennedy, E.B.; Cinar, P.; Conroy, T.; Copur, M.S.; Crane, C.H.; Garrido-Laguna, I.; Lau, M.W.; Johnson, T.; Krishnamurthi, S.; et al. Metastatic Pancreatic Cancer: ASCO Guideline Update. J. Clin. Oncol. 2020, 38, 3217–3230. [Google Scholar] [CrossRef]
- Pishvaian, M.J.; Blais, E.M.; Brody, J.R.; Lyons, E.; DeArbeloa, P.; Hendifar, A.; Mikhail, S.; Chung, V.; Sahai, V.; Sohal, D.P.S.; et al. Overall survival in patients with pancreatic cancer receiving matched therapies following molecular profiling: A retrospective analysis of the Know Your Tumor registry trial. Lancet Oncol. 2020, 21, 508–518. [Google Scholar] [CrossRef]
- Singhi, A.D.; George, B.; Greenbowe, J.R.; Chung, J.; Suh, J.; Maitra, A.; Klempner, S.J.; Hendifar, A.; Milind, J.M.; Golan, T.; et al. Real-Time Targeted Genome Profile Analysis of Pancreatic Ductal Adenocarcinomas Identifies Genetic Alterations That Might Be Targeted With Existing Drugs or Used as Biomarkers. Gastroenterology 2019, 156, 2242–2253. [Google Scholar] [CrossRef]
- Puleo, F.; Nicolle, R.; Blum, Y.; Cros, J.; Marisa, L.; Demetter, P.; Quertinmont, E.; Svrcek, M.; Elarouci, N.; Iovanna, J.; et al. Stratification of Pancreatic Ductal Adenocarcinomas Based on Tumor and Microenvironment Features. Gastroenterology 2018, 155, 1999–2013. [Google Scholar] [CrossRef]
- Juiz, N.; Elkaoutari, A.; Bigonnet, M.; Gayet, O.; Roques, J.; Nicolle, R.; Iovanna, J.; Dusetti, N. Basal-like and classical cells coexist in pancreatic cancer revealed by single-cell analysis on biopsy-derived pancreatic cancer organoids from the classical subtype. FASEB J. 2020, 34, 12214–12228. [Google Scholar] [CrossRef]
- Moffitt, R.A.; Marayati, R.; Flate, E.L.; Volmar, K.E.; Loeza, S.G.; Hoadley, K.A.; Rashid, N.U.; Williams, L.A.; Eaton, S.C.; Chung, A.H.; et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 2015, 47, 1168–1178. [Google Scholar] [CrossRef]
- Chan-Seng-Yue, M.; Kim, J.C.; Wilson, G.W.; Ng, K.; Figueroa, E.F.; O’Kane, G.M.; Connor, A.A.; Denroche, R.E.; Grant, R.C.; McLeod, J.; et al. Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution. Nat. Genet. 2020, 52, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Jansen, L.; Balavarca, Y.; Molina-Montes, E.; Babaei, M.; van der Geest, L.; Lemmens, V.; Van Eycken, L.; De Schutter, H.; Johannesen, T.B.; et al. Resection of pancreatic cancer in Europe and USA: An international large-scale study highlighting large variations. Gut 2019, 68, 130–139. [Google Scholar] [CrossRef]
- Martinelli, P.; Carrillo-de Santa Pau, E.; Cox, T.; Sainz, B., Jr.; Dusetti, N.; Greenhalf, W.; Rinaldi, L.; Costello, E.; Ghaneh, P.; Malats, N.; et al. GATA6 regulates EMT and tumour dissemination, and is a marker of response to adjuvant chemotherapy in pancreatic cancer. Gut 2016, 66, 1665–1676. [Google Scholar] [CrossRef] [PubMed]
- O’Kane, G.M.; Grünwald, B.T.; Jang, G.H.; Masoomian, M.; Picardo, S.; Grant, R.C.; Denroche, R.E.; Zhang, A.; Wang, Y.; Lam, B.; et al. GATA6 Expression Distinguishes Classical and Basal-like Subtypes in Advanced Pancreatic Cancer. Clin. Cancer Res. 2020, 26, 4901–4910. [Google Scholar] [CrossRef]
- Passacantilli, I.; Panzeri, V.; Bielli, P.; Farini, D.; Pilozzi, E.; Fave, G.D.; Capurso, G.; Sette, C. Alternative polyadenylation of ZEB1 promotes its translation during genotoxic stress in pancreatic cancer cells. Cell Death Dis. 2017, 8, e3168. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Sheng, W.; Jia, C.; Shi, X.; Cao, R.; Wang, G.; Lin, Y.; Zhu, F.; Dong, Q.; Dong, M. Musashi2 promotes the progression of pancreatic cancer through a novel ISYNA1-p21/ZEB-1 pathway. J. Cell. Mol. Med. 2020, 24, 10560–10572. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhang, Y.; Yang, J.; Cui, X.; Zhou, Z.; Zhan, H.; Ding, K.; Tian, X.; Yang, Z.; Fung, K.A.; et al. ZIP4 Increases Expression of Transcription Factor ZEB1 to Promote Integrin α3β1 Signaling and Inhibit Expression of the Gemcitabine Transporter ENT1 in Pancreatic Cancer Cells. Gastroenterology 2020, 158, 679–692. [Google Scholar] [CrossRef]
- Escobar-Hoyos, L.F.; Penson, A.; Kannan, R.; Cho, H.; Pan, C.H.; Singh, R.K.; Apken, L.H.; Hobbs, G.A.; Luo, R.; Lecomte, N.; et al. Altered RNA Splicing by Mutant p53 Activates Oncogenic RAS Signaling in Pancreatic Cancer. Cancer Cell 2020, 38, 198–211. [Google Scholar] [CrossRef]
- Marchetti, G.; Ricardo, V.D.; Ardengh, A.O.; de Almeida, A.F.; Taglieri, E.; Micelli-Neto, O.; Kemp, R.; Dos Santos, J.S.; Ardengh, J.C. Adverse events and mortality: Comparative analysis between diagnostic and interventional endoscopic ultrasound. Scand. J. Gastroenterol. 2020, 55, 995–1001. [Google Scholar] [CrossRef]
- Carrara, S.; Soldà, G.; Di Leo, M.; Rahal, D.; Peano, C.; Giunta, M.; Lamonaca, L.; Auriemma, F.; Anderloni, A.; Fugazza, A.; et al. Side-by-side comparison of next-generation sequencing, cytology, and histology in diagnosing locally advanced pancreatic adenocarcinoma. Gastrointest. Endosc. 2020. [Google Scholar] [CrossRef]
- Larghi, A.; Lawlor, R.T.; Crinò, S.F.; Luchini, C.; Rizzatti, G.; Curatolo, M.; Gabbrielli, A.; Inzani, F.; Scarpa, A. Endoscopic ultrasound guided fine needle biopsy samples to drive personalized medicine: A proof of concept study. Pancreatology 2020, 20, 778–780. [Google Scholar] [CrossRef] [PubMed]
- Park, J.K.; Lee, J.H.; Noh, D.H.; Park, J.K.; Lee, K.T.; Lee, J.K.; Lee, K.H.; Jang, K.T.; Cho, J. Factors of Endoscopic Ultrasound-Guided Tissue Acquisition for Successful Next-Generation Sequencing in Pancreatic Ductal Adenocarcinoma. Gut Liver 2020, 14, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Laurell, H.; Bouisson, M.; Berthelemy, P.; Rochaix, P.; Dejean, S.; Besse, P.; Susini, C.; Pradayrol, L.; Vaysse, N.; Buscail, L. Identification of biomarkers of human pancreatic adenocarcinomas by expression profiling and validation with gene expression analysis in endoscopic ultrasound-guided fine needle aspiration samples. World J. Gastroenterol. 2006, 12, 3344–3351. [Google Scholar] [CrossRef] [PubMed]
- Berry, W.; Algar, E.; Kumar, B.; Desmond, C.; Swan, M.; Jenkins, B.J.; Croagh, D. Endoscopic ultrasound-guided fine-needle aspirate-derived preclinical pancreatic cancer models reveal panitumumab sensitivity in KRAS wild-type tumors. Int. J. Cancer 2017, 140, 2331–2343. [Google Scholar] [CrossRef] [PubMed]
- Gleeson, F.C.; Levy, M.J.; Jackson, R.A.; Murphy, S.J.; Halling, K.C.; Kipp, B.R.; Graham, R.P.; Zhang, L. Endoscopic ultrasound may be used to deliver gene expression signatures using digital mRNA detection methods to immunophenotype pancreatic ductal adenocarcinoma to facilitate personalized immunotherapy. Pancreatology 2020, 20, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Costache, M.I.; Iordache, S.; Costache, C.A.; Dragos, E.; Dragos, A.; Saftoiu, A. Molecular Analysis of Vascular Endothelial Growth Factor (VEGF) Receptors in EUS-guided Samples Obtained from Patients with Pancreatic Adenocarcinoma. J. Gastrointest. Liver Dis. 2017, 26, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Archibugi, L.; Testoni, S.G.G.; Redegalli, M.; Petrone, M.C.; Reni, M.; Falconi, M.; Doglioni, C.; Capurso, G.; Arcidiacono, P.G. New era for pancreatic endoscopic ultrasound: From imaging to molecular pathology of pancreatic cancer. World J. Gastrointest. Oncol. 2019, 11, 933–945. [Google Scholar] [CrossRef]
- Recouvreux, M.V.; Moldenhauer, M.R.; Galenkamp, K.M.O.; Jung, M.; James, B.; Zhang, Y.; Lowy, A.; Bagchi, A.; Commisso, C. Glutamine depletion regulates Slug to promote EMT and metastasis in pancreatic cancer. J. Exp. Med. 2020, 217, e20200388. [Google Scholar] [CrossRef]
- Wang, H.; Li, Q.F.; Chow, H.Y.; Choi, S.C.; Leung, Y.C. Arginine deprivation inhibits pancreatic cancer cell migration, invasion and EMT via the down regulation of Snail, Slug, Twist, and MMP1/9. J. Physiol. Biochem. 2020, 76, 73–83. [Google Scholar] [CrossRef]
- Tsukasa, K.; Ding, Q.; Yoshimitsu, M.; Miyazaki, Y.; Matsubara, S.; Takao, S. Slug contributes to gemcitabine resistance through epithelial-mesenchymal transition in CD133(+) pancreatic cancer cells. Hum. Cell 2015, 28, 167–174. [Google Scholar] [CrossRef]
- Young Bang, J.; Krall, K.; Jhala, N.; Singh, C.; Tejani, M.; Arnoletti, J.P.; Navaneethan, U.; Hawes, R.; Varadarajulu, S. Comparing Needles and Methods of Endoscopic Ultrasound-Guided Fine-Needle Biopsy to Optimize Specimen Quality and Diagnostic Accuracy for Patients With Pancreatic Masses in a Randomized Trial. Clin. Gastroenterol. Hepatol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Ishigaki, K.; Nakai, Y.; Oyama, H.; Kanai, S.; Suzuki, T.; Nakamura, T.; Sato, T.; Hakuta, R.; Saito, K.; Saito, T.; et al. Endoscopic Ultrasound-Guided Tissue Acquisition by 22-Gauge Franseen and Standard Needles for Solid Pancreatic Lesions. Gut Liver 2020. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.; Steuber, B.; Kopp, W.; Kari, V.; Urbach, L.; Wang, X.; Küffer, S.; Bohnenberger, H.; Spyropoulou, D.; Zhang, Z.; et al. EZH2 regulates pancreatic cancer subtype identity and tumor progression via transcriptional repression of GATA6. Cancer Res. 2020, 80, 4620–4632. [Google Scholar] [CrossRef] [PubMed]
- Paronetto, M.P.; Passacantilli, I.; Sette, C. Alternative splicing and cell survival: From tissue homeostasis to disease. Cell Death Differ. 2016, 23, 1919–1929. [Google Scholar] [CrossRef]
- Panzeri, V.; Manni, I.; Capone, A.; Sacconi, A.; Di Agostino, S.; Pilozzi, E.; Piaggio, G.; Capurso, G.; Sette, C. The RNA binding protein MEX3A is a prognostic factor and refulator of resistance to gemcitabine in Pancreatic Ductal Adenocarcinoma. Mol. Oncol. 2020, in press. [Google Scholar] [CrossRef]
| Total Patients Enrolled (n = 37) | |
|---|---|
| Age, years (mean ± SD) | 68.3 (± 10.9) |
| Sex, male, n (%) | 18 (48.6%) |
| Location of the lesion | |
| Head | 24 (64.9%) |
| Body-tail | 13 (35.1%) |
| Resectable | 4 (10.8%) |
| Borderline resectable | 19 (51.4%) |
| Locally advanced | 13 (35.1%) |
| Metastatic | 1 (2.7%) |
| Dimension of lesion at EUS, mm (mean ± SD) | 32.8 (± 10.6) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Archibugi, L.; Ruta, V.; Panzeri, V.; Redegalli, M.; Testoni, S.G.G.; Petrone, M.C.; Rossi, G.; Falconi, M.; Reni, M.; Doglioni, C.; et al. RNA Extraction from Endoscopic Ultrasound-Acquired Tissue of Pancreatic Cancer Is Feasible and Allows Investigation of Molecular Features. Cells 2020, 9, 2561. https://doi.org/10.3390/cells9122561
Archibugi L, Ruta V, Panzeri V, Redegalli M, Testoni SGG, Petrone MC, Rossi G, Falconi M, Reni M, Doglioni C, et al. RNA Extraction from Endoscopic Ultrasound-Acquired Tissue of Pancreatic Cancer Is Feasible and Allows Investigation of Molecular Features. Cells. 2020; 9(12):2561. https://doi.org/10.3390/cells9122561
Chicago/Turabian StyleArchibugi, Livia, Veronica Ruta, Valentina Panzeri, Miriam Redegalli, Sabrina Gloria Giulia Testoni, Maria Chiara Petrone, Gemma Rossi, Massimo Falconi, Michele Reni, Claudio Doglioni, and et al. 2020. "RNA Extraction from Endoscopic Ultrasound-Acquired Tissue of Pancreatic Cancer Is Feasible and Allows Investigation of Molecular Features" Cells 9, no. 12: 2561. https://doi.org/10.3390/cells9122561
APA StyleArchibugi, L., Ruta, V., Panzeri, V., Redegalli, M., Testoni, S. G. G., Petrone, M. C., Rossi, G., Falconi, M., Reni, M., Doglioni, C., Sette, C., Arcidiacono, P. G., & Capurso, G. (2020). RNA Extraction from Endoscopic Ultrasound-Acquired Tissue of Pancreatic Cancer Is Feasible and Allows Investigation of Molecular Features. Cells, 9(12), 2561. https://doi.org/10.3390/cells9122561






