Advances and Prospects in Vaccine Development against Enterococci
Abstract
1. Introduction
2. Enterococcal Infections
3. Translocation and Colonization
4. Host Immune Response against Enterococcal Infections
5. Antibiotic Resistance and Options for Treatment
6. Serotyping of Enterococci
7. Enterococcal Polysaccharides and Proteins as Potential Vaccine Candidates
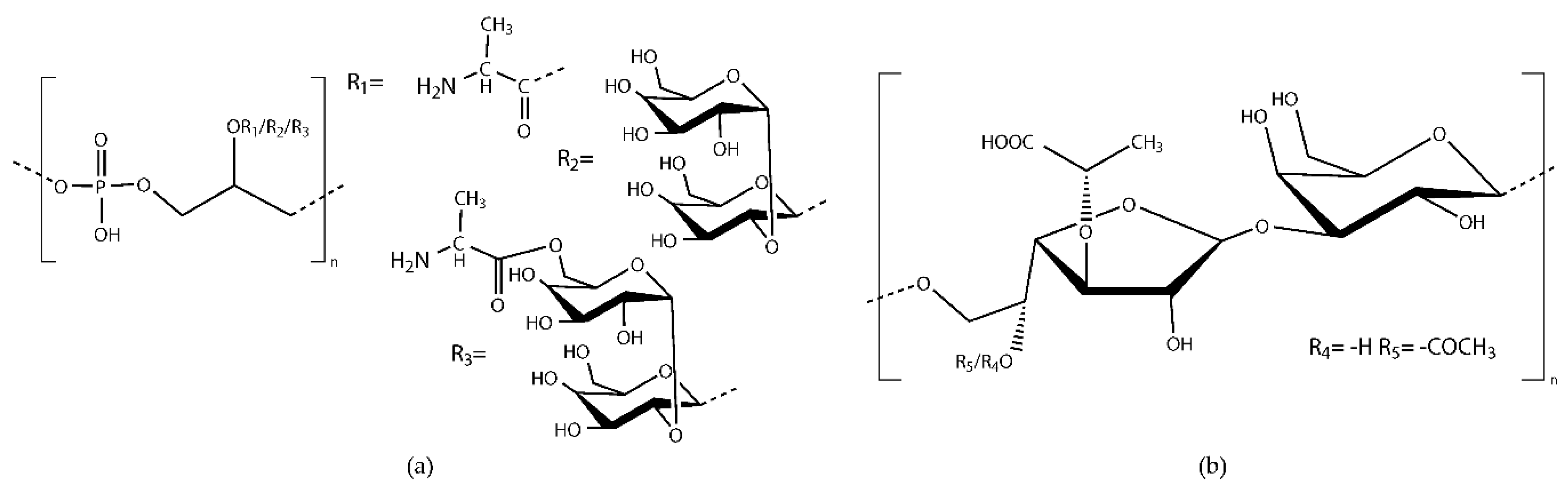
7.1. Enterococcal Polysaccharides
7.2. Enterococcal Proteins
8. Prospects and Pitfalls in the Development of Immunotherapies against Enterococci
9. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Arias, C.A.; Murray, B.E. The rise of the Enterococcus: Beyond vancomycin resistance. Nat. Rev. Microbiol. 2012, 10, 266–278. [Google Scholar] [CrossRef]
- Landete, J.M.; Peirotén, Á.; Medina, M.; Arqués, J.L.; Rodríguez-Mínguez, E. Virulence and Antibiotic Resistance of Enterococci Isolated from Healthy Breastfed Infants. Microb. Drug Resist. 2018, 24, 63–69. [Google Scholar] [CrossRef]
- Foulquié Moreno, M.R.; Sarantinopoulos, P.; Tsakalidou, E.; De Vuyst, L. The role and application of enterococci in food and health. Int. J. Food Microbiol. 2006, 106, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Ben Braïek, O.; Smaoui, S. Enterococci: Between Emerging Pathogens and Potential Probiotics. Biomed Res. Int. 2019, 2019, 5938210. [Google Scholar] [CrossRef] [PubMed]
- Jett, B.D.; Huycke, M.M.; Gilmore, M.S. Virulence of enterococci. Clin. Microbiol. Rev. 1994, 7, 462–478. [Google Scholar] [CrossRef] [PubMed]
- Huycke, M.; Sahm, D.F.; Gilmore, M.S. Multiple-Drug Resistant Enterococci: The Nature of the Problem and an Agenda for the Future. Emerg. Infect. Dis. 1998, 4, 239–249. [Google Scholar] [CrossRef]
- Hollenbeck, B.L.; Rice, L.B. Intrinsic and acquired resistance mechanisms in enterococcus. Virulence 2012, 3, 421–433. [Google Scholar] [CrossRef]
- Ch’ng, J.-H.; Chong, K.K.L.; Lam, L.N.; Wong, J.J.; Kline, K.A. Biofilm-associated infection by enterococci. Nat. Rev. Microbiol. 2019, 17, 82–94. [Google Scholar] [CrossRef]
- Koch, S.; Hufnagel, M.; Theilacker, C.; Huebner, J. Enterococcal infections: Host response, therapeutic, and prophylactic possibilities. Vaccine 2004, 22, 822–830. [Google Scholar] [CrossRef]
- MacCallum, W.G.; Hastings, T.W. A case of acute endocarditis cause by Micrococcous zymogenes (NOV. SPEC.), with a description of the microorganism. J. Exp. Med. 1899, 4, 521–534. [Google Scholar] [CrossRef]
- Goh, H.M.S.; Yong, M.H.A.; Chong, K.K.L.; Kline, K.A. Model systems for the study of Enterococcal colonization and infection. Virulence 2017, 8, 1525–1562. [Google Scholar] [CrossRef] [PubMed]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2018. [Google Scholar] [CrossRef]
- Werner, G.; Coque, T.M.; Hammerum, A.M.; Hope, R.; Hryniewicz, W.; Johnson, A.; Klare, I.; Kristinsson, K.G.; Leclercq, R.; Lester, C.H.; et al. Emergence and spread of vancomycin resistance among enterococci in Europe. Eur. Surveill. 2008, 13, 19046. [Google Scholar]
- Barros, C.H.N.; Fulaz, S.; Stanisic, D.; Tasic, L. Biogenic Nanosilver against Multidrug-Resistant Bacteria (MDRB). Antibiotics 2018, 7, 69. [Google Scholar] [CrossRef] [PubMed]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef]
- Brandl, K.; Plitas, G.; Mihu, C.N.; Ubeda, C.; Jia, T.; Fleisher, M.; Schnabl, B.; DeMatteo, R.P.; Pamer, E.G. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 2008, 455, 804–807. [Google Scholar] [CrossRef]
- Kinnebrew, M.A.; Ubeda, C.; Zenewicz, L.A.; Smith, N.; Flavell, R.A.; Pamer, E.G. Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. J. Infect. Dis. 2010, 201, 534–543. [Google Scholar] [CrossRef]
- Ubeda, C.; Taur, Y.; Jenq, R.R.; Equinda, M.J.; Son, T.; Samstein, M.; Viale, A.; Socci, N.D.; van den Brink, M.R.M.; Kamboj, M.; et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J. Clin. Investig. 2010, 120, 4332–4341. [Google Scholar] [CrossRef]
- Singh, K.V.; Nallapareddy, S.R.; Sillanpää, J.; Murray, B.E. Importance of the Collagen Adhesin Ace in Pathogenesis and Protection against Enterococcus faecalis Experimental Endocarditis. PLoS Pathog. 2010, 6, e1000716. [Google Scholar] [CrossRef]
- Flores-Mireles, A.L.; Walker, J.N.; Bauman, T.M.; Potretzke, A.M.; Schreiber, H.L.; Park, A.M.; Pinkner, J.S.; Caparon, M.G.; Hultgren, S.J.; Desai, A. Fibrinogen Release and Deposition on Urinary Catheters Placed during Urological Procedures. J. Urol. 2016, 196, 416–421. [Google Scholar] [CrossRef]
- Flores-Mireles, A.L.; Pinkner, J.S.; Caparon, M.G.; Hultgren, S.J. EbpA vaccine antibodies block binding of Enterococcus faecalis to fibrinogen to prevent catheter-associated bladder infection in mice. Sci. Transl. Med. 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002; ISBN-10: 0-8153-3218-1. [Google Scholar]
- Schwandner, R.; Dziarski, R.; Wesche, H.; Rothe, M.; Kirschning, C.J. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by toll-like receptor 2. J. Biol. Chem. 1999, 274, 17406–17409. [Google Scholar] [CrossRef] [PubMed]
- Leendertse, M.; Willems, R.J.L.; Giebelen, I.A.J.; Roelofs, J.J.T.H.; Bonten, M.J.M.; van der Poll, T. Neutrophils Are Essential for Rapid Clearance of Enterococcus faecium in Mice. Infect. Immun. 2009, 77, 485–491. [Google Scholar] [CrossRef]
- Leendertse, M.; Willems, R.J.L.; Giebelen, I.A.J.; Roelofs, J.J.T.H.; van Rooijen, N.; Bonten, M.J.M.; van der Poll, T. Peritoneal macrophages are important for the early containment of Enterococcus faecium peritonitis in mice. Innate Immun. 2009, 15, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Leendertse, M.; Willems, R.J.L.; Flierman, R.; de Vos, A.F.; Bonten, M.J.M.; van der Poll, T. The Complement System Facilitates Clearance of Enterococcus faecium during Murine Peritonitis. J. Infect. Dis. 2010, 201, 544–552. [Google Scholar] [CrossRef]
- Flannagan, R.S.; Cosío, G.; Grinstein, S. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat. Rev. Microbiol. 2009, 7, 355–366. [Google Scholar] [CrossRef]
- Van Kessel, K.P.M.; Bestebroer, J.; van Strijp, J.A.G. Neutrophil-Mediated Phagocytosis of Staphylococcus aureus. Front. Immunol. 2014, 5, 467. [Google Scholar] [CrossRef]
- Hufnagel, M.; Koch, S.; Kropec, A.; Huebner, J. Opsonophagocytic assay as a potentially useful tool for assessing safety of enterococcal preparations. Int. J. Food Microbiol. 2003, 88, 263–267. [Google Scholar] [CrossRef]
- Arduino, R.C.; Murray, B.E.; Rakita, R.M. Roles of antibodies and complement in phagocytic killing of enterococci. Infect. Immun. 1994, 62, 987–993. [Google Scholar] [CrossRef]
- Huebner, J.; Wang, Y.; Krueger, W.A.; Madoff, L.C.; Martirosian, G.; Boisot, S.; Goldmann, D.A.; Kasper, D.L.; Tzianabos, A.O.; Pier, G.B. Isolation and chemical characterization of a capsular polysaccharide antigen shared by clinical isolates of Enterococcus faecalis and vancomycin-resistant Enterococcus faecium. Infect. Immun. 1999, 67, 1213–1219. [Google Scholar] [CrossRef]
- Kropec, A.; Hufnagel, M.; Zimmermann, K.; Huebner, J. In vitro Assessment of the Host Response against Enterococcus faecalis Used in Probiotic Preparations. Infection 2005, 33, 377–379. [Google Scholar] [CrossRef] [PubMed]
- Romero-Saavedra, F.; Laverde, D.; Wobser, D.; Michaux, C.; Budin-Verneuil, A.; Bernay, B.; Benachour, A.; Hartke, A.; Huebner, J. Identification of Peptidoglycan-Associated Proteins as Vaccine Candidates for Enterococcal Infections. PLoS ONE 2014, 9, e111880. [Google Scholar] [CrossRef]
- Arduino, R.C.; Jacques-Palaz, K.; Murray, B.E.; Rakita, R.M. Resistance of Enterococcus faecium to neutrophil-mediated phagocytosis. Infect. Immun. 1994, 62, 5587–5594. [Google Scholar] [CrossRef] [PubMed]
- Rakita, R.M.; Vanek, N.N.; Jacques-Palaz, K.; Mee, M.; Mariscalco, M.M.; Dunny, G.M.; Snuggs, M.; Van Winkle, W.B.; Simon, S.I. Enterococcus faecalis bearing aggregation substance is resistant to killing by human neutrophils despite phagocytosis and neutrophil activation. Infect. Immun. 1999, 67, 6067–6075. [Google Scholar] [CrossRef]
- Baldassarri, L.; Cecchini, R.; Bertuccini, L.; Ammendolia, M.G.; Iosi, F.; Arciola, C.R.; Montanaro, L.; Di Rosa, R.; Gherardi, G.; Dicuonzo, G.; et al. Enterococcus spp. produces slime and survives in rat peritoneal macrophages. Med. Microbiol. Immunol. 2001, 190, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Gentry-Weeks, C.R.; Karkhoff-Schweizer, R.; Pikis, A.; Estay, M.; Keith, J.M. Survival of Enterococcus faecalis in mouse peritoneal macrophages. Infect. Immun. 1999, 67, 2160–2165. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; Sievert, D.M.; Hageman, J.C.; Boulton, M.L.; Tenover, F.C.; Downes, F.P.; Shah, S.; Rudrik, J.T.; Pupp, G.R.; Brown, W.J.; et al. Infection with Vancomycin-Resistant Staphylococcus aureus Containing the vanA Resistance Gene. N. Engl. J. Med. 2003, 348, 1342–1347. [Google Scholar] [CrossRef]
- Anderson, A.C.; Jonas, D.; Huber, I.; Karygianni, L.; Wölber, J.; Hellwig, E.; Arweiler, N.; Vach, K.; Wittmer, A.; Al-Ahmad, A. Enterococcus faecalis from food, clinical specimens, and oral sites: Prevalence of virulence factors in association with biofilm formation. Front. Microbiol. 2016, 6, 1534. [Google Scholar] [CrossRef]
- Moon, T.M.; D’Andréa, É.D.; Lee, C.W.; Soares, A.; Jakoncic, J.; Desbonnet, C.; Garcia-Solache, M.; Rice, L.B.; Page, R.; Peti, W. The structures of penicillin-binding protein 4 (PBP4) and PBP5 from Enterococci provide structural insights into β-lactam resistance. J. Biol. Chem. 2018, 293, 18574–18584. [Google Scholar] [CrossRef]
- Moellering, R.C.; Weinberg, A.N.; Weinberg, A.N. Studies on antibiotic syngerism against enterococci. II. Effect of various antibiotics on the uptake of 14 C-labeled streptomycin by enterococci. J. Clin. Investig. 1971, 50, 2580–2584. [Google Scholar] [CrossRef]
- Chow, J.W. Aminoglycoside Resistance in Enterococci. Clin. Infect. Dis. 2000, 31, 586–589. [Google Scholar] [CrossRef] [PubMed]
- Zanella, R.C.; Valdetaro, F.; Lovgren, M.; Tyrrel, G.J.; Bokermann, S.; Almeida, S.C.G.; Vieira, V.S.D.; Brandileone, M.C.C. First Confirmed Case of a Vancomycin-Resistant Enterococcus faecium with vanA Phenotype from Brazil: Isolation from a Meningitis Case in São Paulo. Microb. Drug Resist. 1999, 5, 159–162. [Google Scholar] [CrossRef]
- Kristich, C.J.; Rice, L.B.; Arias, C.A. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Enterococcal Infection—Treatment and Antibiotic Resistance; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- Arthur, M.; Depardieu, F.; Reynolds, P.; Courvalin, P. Quantitative analysis of the metabolism of soluble cytoplasmic peptidoglycan precursors of glycopeptide-resistant enterococci. Mol. Microbiol. 1996, 21, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Iseppi, R.; Di Cerbo, A.; Messi, P.; Sabia, C. Antibiotic resistance and virulence traits in vancomycin-resistant enterococci (Vre) and extended-spectrum β-lactamase/ampc-producing (ESBL/ampc) enterobacteriaceae from humans and pets. Antibiotics 2020, 9, 152. [Google Scholar] [CrossRef]
- Rende-Fournier, R.; Leclercq, R.; Galimand, M.; Duval, J.; Courvalin, P. Identification of the satA gene encoding a streptogramin A acetyltransferase in Enterococcus faecium BM4145. Antimicrob. Agents Chemother. 1993, 37, 2119–2125. [Google Scholar] [CrossRef]
- Werner, G.; Witte, W. Characterization of a new enterococcal gene, satG, encoding a putative acetyltransferase conferring resistance to Streptogramin A compounds. Antimicrob. Agents Chemother. 1999, 43, 1813–1814. [Google Scholar] [CrossRef] [PubMed]
- Portillo, A.; Ruiz-Larrea, F.; Zarazaga, M.; Alonso, A.; Martinez, J.L.; Torres, C. Macrolide resistance genes in Enterococcus spp. Antimicrob. Agents Chemother. 2000, 44, 967–971. [Google Scholar] [CrossRef]
- Singh, K.V.; Weinstock, G.M.; Murray, B.E. An Enterococcus faecalis ABC homologue (Lsa) is required for the resistance of this species to clindamycin and quinupristin-dalfopristin. Antimicrob. Agents Chemother. 2002, 46, 1845–1850. [Google Scholar] [CrossRef]
- Bethea, J.A.; Walko, C.M.; Targos, P.A. Treatment of Vancomycin-Resistant Enterococcus with Quinupristin/Dalfopristin and High-Dose Ampicillin. Ann. Pharmacother. 2004, 38, 989–991. [Google Scholar] [CrossRef]
- Matsumura, S.; Simor, A.E. Treatment of Endocarditis Due to Vancomycin-Resistant Enterococcus faecium with Quinupristin/Dalfopristin, Doxycycline, and Rifampin: A Synergistic Drug Combination. Clin. Infect. Dis. 1998, 27, 1554–1556. [Google Scholar] [CrossRef][Green Version]
- Arias, C.A.; Panesso, D.; McGrath, D.M.; Qin, X.; Mojica, M.F.; Miller, C.; Diaz, L.; Tran, T.T.; Rincon, S.; Barbu, E.M.; et al. Genetic Basis for In Vivo Daptomycin Resistance in Enterococci. N. Engl. J. Med. 2011, 365, 892–900. [Google Scholar] [CrossRef]
- Munita, J.M.; Arias, C.A.; Murray, B.E. Enterococcal endocarditis: Can we win the war? Curr. Infect. Dis. Rep. 2012, 14, 339–349. [Google Scholar] [CrossRef][Green Version]
- Arias, C.A.; Murray, B.E. Emergence and management of drug-resistant enterococcal infections. Expert Rev. Anti. Infect. Ther. 2008, 6, 637–655. [Google Scholar] [CrossRef]
- Marshall, S.H.; Donskey, C.J.; Hutton-Thomas, R.; Salata, R.A.; Rice, L.B. Gene dosage and linezolid resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob. Agents Chemother. 2002, 46, 3334–3336. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Wu, C.; Shen, Z.; Schwarz, S.; Du, X.-D.; Dai, L.; Zhang, W.; Zhang, Q.; Shen, J. First Report of the Multidrug Resistance Gene cfr in Enterococcus faecalis of Animal Origin. Antimicrob. Agents Chemother. 2012, 56, 1650–1654. [Google Scholar] [CrossRef] [PubMed]
- Lancefield, R.C. A serological differentiation of human and other groups of hemolytic streptococci. J. Exp. Med. 1933, 57, 571–594. [Google Scholar] [CrossRef]
- Maekawa, S.; Yoshioka, M.; Kumamoto, Y. Proposal of a new scheme for the serological typing of Enterococcus faecalis strains. Microbiol. Immunol. 1992, 36, 671–681. [Google Scholar] [CrossRef]
- Hufnagel, M.; Hancock, L.E.; Koch, S.; Theilacker, C.; Gilmore, M.S.; Huebner, J. Serological and Genetic Diversity of Capsular Polysaccharides in Enterococcus faecalis. J. Clin. Microbiol. 2004, 42, 2548–2557. [Google Scholar] [CrossRef]
- Hufnagel, M.; Carey, V.J.; Baldassarri, L.; Reinert, R.R.; Huebner, J.; Baldasarri, L.; Reinert, R.R.; Huebner, J. Distribution of Four Capsular Serotypes of Enterococcus faecalis among Clinical Isolates from Different Geographical Origins and Infection Sites. Infection 2006, 34, 22–25. [Google Scholar] [CrossRef]
- Hancock, L.E.; Gilmore, M.S. The capsular polysaccharide of Enterococcus faecalis and its relationship to other polysaccharides in the cell wall. Proc. Natl. Acad. Sci. USA 2002, 99, 1574–1579. [Google Scholar] [CrossRef]
- Thurlow, L.R.; Thomas, V.C.; Hancock, L.E. Capsular polysaccharide production in Enterococcus faecalis and contribution of CpsF to capsule serospecificity. J. Bacteriol. 2009, 191, 6203–6210. [Google Scholar] [CrossRef] [PubMed]
- Thurlow, L.R.; Thomas, V.C.; Fleming, S.D.; Hancock, L.E. Enterococcus faecalis capsular polysaccharide serotypes C and D and their contributions to host innate immune evasion. Infect. Immun. 2009, 77, 5551–5557. [Google Scholar] [CrossRef] [PubMed]
- Theilacker, C.; Kaczyński, Z.; Kropec, A.; Sava, I.; Ye, L.; Bychowska, A.; Holst, O.; Huebner, J. Serodiversity of Opsonic Antibodies against Enterococcus faecalis—Glycans of the Cell Wall Revisited. PLoS ONE 2011, 6, e17839. [Google Scholar] [CrossRef] [PubMed][Green Version]
- McBride, S.M.; Fischetti, V.A.; LeBlanc, D.J.; Moellering, R.C.; Gilmore, M.S. Genetic diversity among Enterococcus faecalis. PLoS ONE 2007, 2, e582. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.A.; Contreras, G.A.; Murray, B.E. Management of multidrug-resistant enterococcal infections. Clin. Microbiol. Infect. 2010, 16, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Navarre, W.W.; Schneewind, O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 1999, 63, 174–229. [Google Scholar] [CrossRef] [PubMed]
- Hancock, L.E.; Murray, B.E.; Sillanpää, J. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Enterococcal Cell Wall Components and Structures; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- Rakita, R.M.; Quan, V.C.; Jacques-Palaz, K.; Singh, K.V.; Arduino, R.C.; Mee, M.; Murray, B.E. Specific antibody promotes opsonization and PMN-mediated killing of phagocytosis-resistant Enterococcus faecium. FEMS Immunol. Med. Microbiol. 2000, 28, 291–299. [Google Scholar] [CrossRef]
- Huebner, J.; Quaas, A.; Krueger, W.A.; Goldmann, D.A.; Pier, G.B. Prophylactic and therapeutic efficacy of antibodies to a capsular polysaccharide shared among vancomycin-sensitive and -resistant enterococci. Infect. Immun. 2000, 68, 4631–4636. [Google Scholar] [CrossRef]
- Wang, Y.; Huebner, J.; Tzianabos, A.O.; Martirosian, G.; Kasper, D.L.; Pier, G.B. Structure of an antigenic teichoic acid shared by clinical isolates of Enterococcus faecalis and vancomycin-resistant Enterococcus faecium. Carbohydr. Res. 1999, 316, 155–160. [Google Scholar] [CrossRef]
- Theilacker, C.; Kaczynski, Z.; Kropec, A.; Fabretti, F.; Sange, T.; Holst, O.; Huebner, J. Opsonic antibodies to Enterococcus faecalis strain 12030 are directed against lipoteichoic acid. Infect. Immun. 2006, 74, 5703–5712. [Google Scholar] [CrossRef]
- Swoboda, J.G.; Campbell, J.; Meredith, T.C.; Walker, S. Wall teichoic acid function, biosynthesis, and inhibition. Chembiochem 2010, 11, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Theilacker, C.; Kropec, A.; Hammer, F.; Sava, I.; Wobser, D.; Sakinc, T.; Codée, J.D.C.; Hogendorf, W.F.J.; van der Marel, G.A.; Huebner, J. Protection Against Staphylococcus aureus by Antibody to the Polyglycerolphosphate Backbone of Heterologous Lipoteichoic Acid. J. Infect. Dis. 2012, 205, 1076–1085. [Google Scholar] [CrossRef] [PubMed]
- Teng, F.; Singh, K.V.; Bourgogne, A.; Zeng, J.; Murray, B.E. Further characterization of the epa gene cluster and epa polysaccharides of Enterococcus faecalis. Infect. Immun. 2009, 77, 3759–3767. [Google Scholar] [CrossRef]
- Geiss-Liebisch, S.; Rooijakkers, S.H.M.; Beczala, A.; Sanchez-Carballo, P.; Kruszynska, K.; Repp, C.; Sakinc, T.; Vinogradov, E.; Holst, O.; Huebner, J.; et al. Secondary cell wall polymers of Enterococcus faecalis are critical for resistance to complement activation via mannose-binding lectin. J. Biol. Chem. 2012, 287, 37769–37777. [Google Scholar] [CrossRef]
- Guerardel, Y.; Sadovskaya, I.; Maes, E.; Furlan, S.; Chapot-Chartier, M.P.; Mesnage, S.; Rigottier-Gois, L.; Serror, P. Complete structure of the enterococcal polysaccharide antigen (EPA) of vancomycin-resistant enterococcus faecalis v583 reveals that EPA decorations are teichoic acids covalently linked to a rhamnopolysaccharide backbone. MBio 2020, 11. [Google Scholar] [CrossRef]
- Teng, F.; Jacques-Palaz, K.D.; Weinstock, G.M.; Murray, B.E. Evidence that the Enterococcal Polysaccharide Antigen Gene (epa) Cluster Is Widespread in Enterococcus faecalis and Influences Resistance to Phagocytic Killing of E. faecalis. Infect. Immun. 2002, 70. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xu, Y.; Singh, K.V.; Qin, X.; Murray, B.E.; Weinstock, G.M. Analysis of a gene cluster of Enterococcus faecalis involved in polysaccharide biosynthesis. Infect. Immun. 2000, 68, 815–823. [Google Scholar] [CrossRef]
- Chatterjee, A.; Johnson, C.N.; Luong, P.; Hullahalli, K.; McBride, S.W.; Schubert, A.M.; Palmer, K.L.; Carlson, P.E.; Duerkop, B.A. Bacteriophage resistance alters antibiotic-mediated intestinal expansion of enterococci. Infect. Immun. 2019, 87. [Google Scholar] [CrossRef]
- Pazur, J.H.; Dropkin, D.J.; Scott Forsberg, L. Glycans from streptococcal cell-walls: The molecular structure of an antigenic diheteroglycan of D-glucose and L-rhamnose from Streptococcus bovis. Carbohydr. Res. 1978, 66, 155–166. [Google Scholar] [CrossRef]
- Krylov, V.B.; Gerbst, A.G.; Argunov, D.A.; Dmitrenok, A.S.; Shashkov, A.S.; Kaczynski, Z.; Huebner, J.; Holst, O.; Nifantiev, N.E. Definitive Structural Assessment of Enterococcal Diheteroglycan. Chem. Eur. J. 2015, 21, 1749–1754. [Google Scholar] [CrossRef]
- Micoli, F.; Del Bino, L.; Alfini, R.; Carboni, F.; Romano, M.R.; Adamo, R. Glycoconjugate vaccines: Current approaches towards faster vaccine design. Expert Rev. Vaccines 2019, 18, 881–895. [Google Scholar] [CrossRef]
- Wang, L.; Berni, F.; Enotarpi, J.; Overkleeft, H.S.; Van Der Marel, G.; Codée, J.D.C. Reagent controlled stereoselective synthesis of teichoic acid α-(1,2)-glucans. Org. Biomol. Chem. 2020, 18, 2038–2050. [Google Scholar] [CrossRef] [PubMed]
- Van der Es, D.; Berni, F.; Hogendorf, W.F.J.; Meeuwenoord, N.; Laverde, D.; van Diepen, A.; Overkleeft, H.S.; Filippov, D.V.; Hokke, C.H.; Huebner, J.; et al. Streamlined Synthesis and Evaluation of Teichoic Acid Fragments. Chem. Eur. J. 2018, 24, 4014–4018. [Google Scholar] [CrossRef] [PubMed]
- Laverde, D.; Wobser, D.; Romero-Saavedra, F.; Hogendorf, W.; van der Marel, G.; Berthold, M.; Kropec, A.; Codee, J.; Huebner, J. Synthetic Teichoic Acid Conjugate Vaccine against Nosocomial Gram-Positive Bacteria. PLoS ONE 2014, 9, e110953. [Google Scholar] [CrossRef]
- Hogendorf, W.F.J.; Meeuwenoord, N.; Overkleeft, H.S.; Filippov, D.V.; Laverde, D.; Kropec, A.; Huebner, J.; Van der Marel, G.A.; Codée, J.D.C. Automated solid phase synthesis of teichoic acids. Chem. Commun. 2011, 47, 8961. [Google Scholar] [CrossRef]
- Laverde, D.; Romero-Saavedra, F.; Argunov, D.A.; Enotarpi, J.; Krylov, V.B.; Kalfopoulou, E.; Martini, C.; Torelli, R.; Van Der Marel, G.A.; Sanguinetti, M.; et al. Synthetic Oligomers Mimicking Capsular Polysaccharide Diheteroglycan are Potential Vaccine Candidates against Encapsulated Enterococcal Infections. ACS Infect. Dis. 2020, 6, 1816–1826. [Google Scholar] [CrossRef]
- Teng, F.; Kawalec, M.; Weinstock, G.M.; Hryniewicz, W.; Murray, B.E. An Enterococcus faecium Secreted Antigen, SagA, Exhibits Broad-Spectrum Binding to Extracellular Matrix Proteins and Appears Essential for E. faecium Growth. Infect. Immun. 2003, 71, 5033–5041. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Howden, B.P.; Stinear, T.P. Evolution of virulence in Enterococcus faecium, a hospital-adapted opportunistic pathogen. Curr. Opin. Microbiol. 2018, 41, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Paganelli, F.L.; de Been, M.; Braat, J.C.; Hoogenboezem, T.; Vink, C.; Bayjanov, J.; Rogers, M.R.C.; Huebner, J.; Bonten, M.J.M.; Willems, R.J.L.; et al. Distinct SagA from Hospital-Associated Clade A1 Enterococcus faecium Strains Contributes to Biofilm Formation. Appl. Environ. Microbiol. 2015, 81, 6873–6882. [Google Scholar] [CrossRef]
- Kropec, A.; Sava, I.G.; Vonend, C.; Sakinc, T.; Grohmann, E.; Huebner, J. Identification of SagA as a novel vaccine target for the prevention of Enterococcus faecium infections. Microbiology 2011, 157, 3429–3434. [Google Scholar] [CrossRef]
- Romero-Saavedra, F.; Laverde, D.; Budin-Verneuil, A.; Muller, C.; Bernay, B.; Benachour, A.; Hartke, A.; Huebner, J. Characterization of two metal binding lipoproteins as vaccine candidates for enterococcal infections. PLoS ONE 2015, 10, e0136625. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kazemian, H.; Pourmand, M.R.; Siadat, S.D.; Mahdavi, M.; Yazdi, M.H.; Majelan, P.A.; Afshar, D.; Yaseri, M.; Davari, M.; Getso, M.I. Molecular cloning and immunogenicity evaluation of PpiC, GeLE, and VS87_01105 proteins of enterococcus faecalis as Vaccine Candidates. Iran. Biomed. J. 2019, 23, 343–353. [Google Scholar] [CrossRef]
- Nallapareddy, S.R.; Singh, K.V.; Murray, B.E. Contribution of the Collagen Adhesin Acm to Pathogenesis of Enterococcus faecium in Experimental Endocarditis. Infect. Immun. 2008, 76, 4120–4128. [Google Scholar] [CrossRef] [PubMed]
- Nallapareddy, S.R.; Singh, K.V.; Sillanpää, J.; Zhao, M.; Murray, B.E. Relative contributions of Ebp Pili and the collagen adhesin ace to host extracellular matrix protein adherence and experimental urinary tract infection by Enterococcus faecalis OG1RF. Infect. Immun. 2011, 79, 2901–2910. [Google Scholar] [CrossRef] [PubMed]
- Sillanpää, J.; Nallapareddy, S.R.; Singh, K.V.; Prakash, V.P.; Fothergill, T.; Ton-That, H.; Murray, B.E. Characterization of the ebpfm pilus-encoding operon of Enterococcus faecium and its role in biofilm formation and virulence in a murine model of urinary tract infection. Virulence 2010, 1, 236. [Google Scholar] [CrossRef] [PubMed]
- Nallapareddy, S.R.; Sillanpää, J.; Ganesh, V.K.; Höök, M.; Murray, B.E. Inhibition of Enterococcus faecium adherence to collagen by antibodies against high-affinity binding subdomains of Acm. Infect. Immun. 2007, 75, 3192–3196. [Google Scholar] [CrossRef][Green Version]
- Sillanpää, J.; Chang, C.; Singh, K.V.; Montealegre, M.C.; Nallapareddy, S.R.; Harvey, B.R.; Ton-That, H.; Murray, B.E. Contribution of Individual Ebp Pilus Subunits of Enterococcus faecalis OG1RF to Pilus Biogenesis, Biofilm Formation and Urinary Tract Infection. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Nielsen, H.V.; Flores-Mireles, A.L.; Kau, A.L.; Kline, K.A.; Pinkner, J.S.; Neiers, F.; Normark, S.; Henriques-Normark, B.; Caparon, M.G.; Hultgren, S.J. Pilin and sortase residues critical for endocarditis- and biofilm-associated pilus biogenesis in Enterococcus faecalis. J. Bacteriol. 2013, 195, 4484–4495. [Google Scholar] [CrossRef]
- Flores-Mireles, A.L.; Walker, J.N.; Potretzke, A.; Schreiber, H.L.; Pinkner, J.S.; Bauman, T.M.; Park, A.M.; Desai, A.; Hultgren, S.J.; Caparon, M.G. Antibody-based therapy for enterococcal catheter-associated urinary tract infections. MBio 2016, 7. [Google Scholar] [CrossRef]
- Grandi, G. Antibacterial vaccine design using genomics and proteomics. Trends Biotechnol. 2001, 19, 181–188. [Google Scholar] [CrossRef]
- Reffuveille, F.; Leneveu, C.; Chevalier, S.; Auffray, Y.; Rince, A. Lipoproteins of Enterococcus faecalis: Bioinformatic identification, expression analysis and relation to virulence. Microbiology 2011, 157, 3001–3013. [Google Scholar] [CrossRef][Green Version]
- Prasanna, M.; Soulard, D.; Camberlein, E.; Ruffier, N.; Lambert, A.; Trottein, F.; Csaba, N.; Grandjean, C. Semisynthetic glycoconjugate based on dual role protein/PsaA as a pneumococcal vaccine. Eur. J. Pharm. Sci. 2019, 129, 31–41. [Google Scholar] [CrossRef]
- Burnie, J.; Carter, T.; Rigg, G.; Hodgetts, S.; Donohoe, M.; Matthews, R. Identification of ABC transporters in vancomycin-resistant Enterococcus faecium as potential targets for antibody therapy. FEMS Immunol. Med. Microbiol. 2002, 33, 179–189. [Google Scholar] [CrossRef]
- Wagner, T.; Joshi, B.; Janice, J.; Askarian, F.; Škalko-Basnet, N.; Hagestad, O.C.; Mekhlif, A.; Wai, S.N.; Hegstad, K.; Johannessen, M. Enterococcus faecium produces membrane vesicles containing virulence factors and antimicrobial resistance related proteins. J. Proteomics 2018, 187, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Paganelli, F.L.; Bierschenk, D.; Kuipers, A.; Bonten, M.J.M.; Willems, R.J.L.; van Schaik, W. Genome-wide identification of ampicillin resistance determinants in Enterococcus faecium. PLoS Genet. 2012, 8, e1002804. [Google Scholar] [CrossRef] [PubMed]
- Cacaci, M.; Giraud, C.; Leger, L.; Torelli, R.; Martini, C.; Posteraro, B.; Palmieri, V.; Sanguinetti, M.; Bugli, F.; Hartke, A. Expression profiling in a mammalian host reveals the strong induction of genes encoding LysM domain-containing proteins in Enterococcus faecium. Sci. Rep. 2018, 8, 12412. [Google Scholar] [CrossRef]
- Reffuveille, F.; Connil, N.; Sanguinetti, M.; Posteraro, B.; Chevalier, S.; Auffray, Y.; Rince, A. Involvement of peptidylprolyl cis/trans isomerases in Enterococcus faecalis virulence. Infect. Immun. 2012, 80, 1728–1735. [Google Scholar] [CrossRef]
- Williamson, R.; Le Bouguenec, C.; Gutmann, L.; Horaud, T. One or Two Low Affinity Penicillin-binding Proteins May Be Responsible for the Range of Susceptibility of Enterococcus faecium to Benzylpenicillin. Microbiology 1985, 131, 1933–1940. [Google Scholar] [CrossRef]
- Koch, S.; Hufnagel, M.; Huebner, J. Treatment and prevention of enterococcal infections-alternative and experimental approaches. Expert Opin. Biol. Ther. 2004, 4, 1519–1531. [Google Scholar] [CrossRef]
- Elizaga, M.L.; Weinstein, R.A.; Hayden, M.K. Patients in Long-Term Care Facilities: A Reservoir for Vancomycin-Resistant Enterococci. Clin. Infect. Dis. 2002, 34, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Husni, R.; Hachem, R.; Hanna, H.; Raad, I. Risk Factors for Vancomycin-Resistant Enterococcus (VRE) Infection in Colonized Patients With Cancer. Infect. Control Hosp. Epidemiol. 2002, 23, 102–103. [Google Scholar] [CrossRef]
- Ali, L.; Blum, H.E.; Sakιnç, T. Detection and characterization of bacterial polysaccharides in drug-resistant enterococci. Glycoconj. J. 2019, 36, 429–438. [Google Scholar] [CrossRef]
- Pollard, A.J.; Perrett, K.P.; Beverley, P.C. Maintaining protection against invasive bacteria with protein–polysaccharide conjugate vaccines. Nat. Rev. Immunol. 2009, 9, 213–220. [Google Scholar] [CrossRef]
- Rappuoli, R. Glycoconjugate vaccines: Principles and mechanisms. Sci. Transl. Med. 2018, 10, 1–7. [Google Scholar] [CrossRef]
- Nielsen, T.B.; Pantapalangkoor, P.; Luna, B.M.; Bruhn, K.W.; Yan, J.; Dekitani, K.; Hsieh, S.; Yeshoua, B.; Pascual, B.; Vinogradov, E.; et al. Monoclonal Antibody Protects Against Acinetobacter baumannii Infection by Enhancing Bacterial Clearance and Evading Sepsis. J. Infect. Dis. 2017, 216, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Diago-Navarro, E.; Motley, M.P.; Ruiz-Peréz, G.; Yu, W.; Austin, J.; Seco, B.M.S.; Xiao, G.; Chikhalya, A.; Seeberger, P.H.; Fries, B.C. Novel, Broadly Reactive Anticapsular Antibodies against Carbapenem-Resistant Klebsiella pneumoniae Protect from Infection. MBio 2018, 9. [Google Scholar] [CrossRef]
- Avery, O.T.; Goebel, W.F. Chemo-immunological studies on conjugated carbohydrate-proteins: II. Immunological specificity of synthetic sugar-protein antigens. J. Exp. Med. 1929, 50, 533–550. [Google Scholar] [CrossRef]
- Micoli, F.; Adamo, R.; Costantino, P. Protein Carriers for Glycoconjugate Vaccines: History, Selection Criteria, Characterization and New Trends. Molecules 2018, 23, 1451. [Google Scholar] [CrossRef]
- Dagan, R.; Poolman, J.; Siegrist, C.-A. Glycoconjugate vaccines and immune interference: A review. Vaccine 2010, 28, 5513–5523. [Google Scholar] [CrossRef]
- Tontini, M.; Romano, M.R.; Proietti, D.; Balducci, E.; Micoli, F.; Balocchi, C.; Santini, L.; Masignani, V.; Berti, F.; Costantino, P. Preclinical studies on new proteins as carrier for glycoconjugate vaccines. Vaccine 2016, 34, 4235–4242. [Google Scholar] [CrossRef] [PubMed]
- Romero-Saavedra, F.; Laverde, D.; Kalfopoulou, E.; Martini, C.; Torelli, R.; Martinez-Matamoros, D.; Sanguinetti, M.; Huebner, J. Conjugation of different immunogenic enterococcal vaccine target antigens leads to extended strain coverage. J. Infect. Dis. 2019. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A.; Dadachova, E.; Pirofski, L. Passive antibody therapy for infectious diseases. Nat. Rev. Microbiol. 2004, 2, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Kalfopoulou, E.; Laverde, D.; Miklic, K.; Romero-Saavedra, F.; Malic, S.; Carboni, F.; Adamo, R.; Lenac Rovis, T.; Jonjic, S.; Huebner, J. Development of Opsonic Mouse Monoclonal Antibodies against Multidrug-Resistant Enterococci. Infect. Immun. 2019, 87. [Google Scholar] [CrossRef] [PubMed]
- Anish, C.; Schumann, B.; Pereira, C.L.; Seeberger, P.H. Chemical Biology Approaches to Designing Defined Carbohydrate Vaccines. Chem. Biol. 2014, 21, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Sparrow, E.; Friede, M.; Sheikh, M.; Torvaldsen, S. Therapeutic antibodies for infectious diseases. Bull. World Health Organ. 2017, 95, 235–237. [Google Scholar] [CrossRef] [PubMed]
- Rossmann, F.S.; Laverde, D.; Kropec, A.; Romero-Saavedra, F.; Meyer-Buehn, M.; Huebner, J. Isolation of Highly Active Monoclonal Antibodies against Multiresistant Gram-Positive Bacteria. PLoS ONE 2015, 10, e0118405. [Google Scholar] [CrossRef]
- Singh, K.V.; Pinkston, K.L.; Gao, P.; Harvey, B.R.; Murray, B.E. Anti-Ace monoclonal antibody reduces Enterococcus faecalis aortic valve infection in a rat infective endocarditis model. Pathog. Dis. 2018, 76. [Google Scholar] [CrossRef]
- Pinkston, K.L.; Singh, K.V.; Gao, P.; Wilganowski, N.; Robinson, H.; Ghosh, S.; Azhdarinia, A.; Sevick-Muraca, E.M.; Murray, B.E.; Harvey, B.R. Targeting Pili in Enterococcal Pathogenesis. Infect. Immun. 2014, 82, 1540–1547. [Google Scholar] [CrossRef]
- Motley, M.P.; Fries, B.C. A New Take on an Old Remedy: Generating Antibodies against Multidrug-Resistant Gram-Negative Bacteria in a Postantibiotic World. mSphere 2017, 2, e00397-17. [Google Scholar] [CrossRef]
- Oleksiewicz, M.B.; Nagy, G.; Nagy, E. Anti-bacterial monoclonal antibodies: Back to the future? Arch. Biochem. Biophys. 2012, 526, 124–131. [Google Scholar] [CrossRef]
- Koefoed, K.; Steinaa, L.; Søderberg, J.N.; Kjær, I.; Jacobsen, H.J.; Meijer, P.J.; Haurum, J.S.; Jensen, A.; Kragh, M.; Andersen, P.S.; et al. Rational identification of an optimal antibody mixture for targeting the epidermal growth factor receptor. MAbs 2011, 3, 584. [Google Scholar] [CrossRef] [PubMed]
- Rossmann, F.S.; Kropec, A.; Laverde, D.; Saaverda, F.R.; Wobser, D.; Huebner, J. In vitro and in vivo activity of hyperimmune globulin preparations against multiresistant nosocomial pathogens. Infection 2015, 43, 169–175. [Google Scholar] [CrossRef] [PubMed][Green Version]
| Name | Functional Description | Reference |
|---|---|---|
| Ace | collagen adhesin | [19] |
| Acm | collagen adhesin | [96] |
| SagA | secreted antigen a, bacterial growth and biofilm formation | [93,94] |
| AdcAfm | zinc ABC transporter substrate-binding lipoprotein | [94] |
| PsaAfm | manganese ABC transporter substrate-binding lipoprotein | [94] |
| LysM | peptidoglycan-binding protein | [33] |
| DdcP | D-alanyl-D-alanine carboxypeptidase | [33] |
| PpiC | peptidyl-prolyl cis-trans isomerase | [33,95] |
| PBP5 | penicillin-binding protein 5 | [33] |
| EbpA | endocarditis- and biofilm-associated pili A | [21] |
| GelE | gelatinase | [95] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalfopoulou, E.; Huebner, J. Advances and Prospects in Vaccine Development against Enterococci. Cells 2020, 9, 2397. https://doi.org/10.3390/cells9112397
Kalfopoulou E, Huebner J. Advances and Prospects in Vaccine Development against Enterococci. Cells. 2020; 9(11):2397. https://doi.org/10.3390/cells9112397
Chicago/Turabian StyleKalfopoulou, Ermioni, and Johannes Huebner. 2020. "Advances and Prospects in Vaccine Development against Enterococci" Cells 9, no. 11: 2397. https://doi.org/10.3390/cells9112397
APA StyleKalfopoulou, E., & Huebner, J. (2020). Advances and Prospects in Vaccine Development against Enterococci. Cells, 9(11), 2397. https://doi.org/10.3390/cells9112397






