The Regulatory Properties of the Ccr4–Not Complex
Abstract
1. Introduction
2. Structure of the Ccr4–Not (CNOT) Complex
3. mRNA Turnover and Deadenylation
4. The Ccr4–Not Complex and the Regulation of Protein Level: the Role of CNOT4/Not4 as an E3 Ligase
5. The Ccr4–Not Complex and Transcription
6. A Potential Role for the Ccr4–Not Complex in Repair of Transcription-Dependent Replication Stress
7. The Role of the Ccr4–Not Complex in the Export of Nascent RNA through Nuclear Pores
8. The Role of the Ccr4–Not Complex in Cell Cycle Regulation
9. The Role of the Ccr4–Not Complex in Senescence, Apoptosis and Autophagy
9.1. Senescence
9.2. Programmed Cell Death/Apoptosis
9.3. Autophagy
10. The Ccr4–Not Complex and the DNA Damage Response
11. The CNOT Complex and Human Disease
12. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Denis:, C.L.; Chen, J. The CCR4-NOT complex plays diverse roles in mRNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 2003, 73, 221–250. [Google Scholar]
- Chen, J.; Rappsilber, J.; Chiang, Y.-C.; Russell, P.; Mann, M.; Denis, C.L. Purification and characterization of the 1.0 MDa CCR4-NOT complex identifies two novel components of the complex. J. Mol. Biol. 2001, 314, 683–694. [Google Scholar] [CrossRef] [PubMed]
- Kruk, J.A.; Dutta, A.; Fu, J.; Gilmour, D.S.; Reese, J.C. The multifunctional Ccr4–Not complex directly promotes transcription elongation. Genes Dev. 2011, 25, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Collart, M.A.; Panasenko, O.O. The Ccr4–not complex. Gene 2012, 492, 42–53. [Google Scholar] [CrossRef]
- Chapat, C.; Corbo, L. Novel roles of the CCR4–NOT complex. Wiley Interdiscip. Rev. RNA 2014, 5, 883–901. [Google Scholar] [CrossRef] [PubMed]
- Collart, M.A. Global control of gene expression in yeast by the Ccr4-Not complex. Gene 2003, 313, 1–16. [Google Scholar] [CrossRef]
- Collart, M.A. The Ccr4-Not complex is a key regulator of eukaryotic gene expression. Wiley Interdiscip. Rev. RNA 2016, 7, 438–454. [Google Scholar] [CrossRef]
- Stowell, J.A.; Webster, M.W.; Kögel, A.; Wolf, J.; Shelley, K.L.; Passmore, L.A. Reconstitution of targeted deadenylation by the Ccr4-Not complex and the YTH domain protein Mmi1. Cell Rep. 2016, 17, 1978–1989. [Google Scholar] [CrossRef]
- Bhaskar, V.; Roudko, V.; Basquin, J.; Sharma, K.; Urlaub, H.; Séraphin, B.; Conti, E. Structure and RNA-binding properties of the Not1–Not2–Not5 module of the yeast Ccr4–Not complex. Nat. Struct. Mol. Biol. 2013, 20, 1281–1288. [Google Scholar] [CrossRef]
- Buschauer, R.; Matsuo, Y.; Sugiyama, T.; Chen, Y.-H.; Alhusaini, N.; Sweet, T.; Ikeuchi, K.; Cheng, J.; Matsuki, Y.; Nobuta, R.; et al. The Ccr4-Not complex monitors the translating ribosome for codon optimality. Science 2020, 368. [Google Scholar] [CrossRef]
- Collart, M.A.; Struhl, K. NOT1 (CDC39), NOT2 (CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 1994, 8, 525–537. [Google Scholar] [CrossRef]
- Tucker, M.; Valencia-Sanchez, M.A.; Staples, R.R.; Chen, J.; Denis, C.L.; Parker, R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 2001, 104, 377–386. [Google Scholar] [CrossRef]
- Tucker, M.; Staples, R.R.; Valencia-Sanchez, M.A.; Muhlrad, D.; Parker, R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002, 21, 1427–1436. [Google Scholar] [CrossRef] [PubMed]
- Temme, C.; Zhang, L.; Kremmer, E.; Ihling, C.; Chartier, A.; Sinz, A.; Simonelig, M.; Wahle, E. Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation. RNA 2010, 16, 1356–1370. [Google Scholar] [CrossRef]
- Petit, A.-P.; Wohlbold, L.; Bawankar, P.; Huntzinger, E.; Schmidt, S.; Izaurralde, E.; Weichenrieder, O. The structural basis for the interaction between the CAF1 nuclease and the NOT1 scaffold of the human CCR4–NOT deadenylase complex. Nucleic Acids Res. 2012, 40, 11058–11072. [Google Scholar] [CrossRef]
- Basquin, J.; Roudko, V.V.; Rode, M.; Basquin, C.; Séraphin, B.; Conti, E. Architecture of the nuclease module of the yeast Ccr4-not complex: The Not1-Caf1-Ccr4 interaction. Mol. Cell 2012, 48, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Collart, M.A. The NOT4 RING E3 ligase: A relevant player in cotranslational quality control. ISRN Mol. Biol. 2013. [Google Scholar] [CrossRef] [PubMed]
- Lau, N.; Kolkman, A.; van Schaik, F.; Mulder, K.; Pijnappel, W.; Heck, A.; Timmers, H.T.M. Human Ccr4-Not complexes contain variable deadenylase subunits. Biochem. J. 2009, 422, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, D.; Takahashi, A.; Yanagiya, A.; Yamaguchi, T.; Abe, T.; Kureha, T.; Kuba, K.; Kanegae, Y.; Furuta, Y.; Yamamoto, T.; et al. Essential functions of the CNOT7/8 catalytic subunits of the CCR4-NOT complex in mRNA regulation and cell viability. RNA Biol. 2020, 17. [Google Scholar] [CrossRef]
- Mauxion, F.; Prève, B.; Séraphin, B. C2ORF29/CNOT11 and CNOT10 form a new module of the CCR4-NOT complex. RNA Biol. 2013, 10, 267–276. [Google Scholar] [CrossRef]
- Azzouz, N.; Panasenko, O.O.; Deluen, C.; Hsieh, J.; Theiler, G.; Collart, M.A. Specific roles for the Ccr4-Not complex subunits in expression of the genome. RNA 2009, 15, 377–383. [Google Scholar] [CrossRef]
- Nasertorabi, F.; Batisse, C.; Diepholz, M.; Suck, D.; Böttcher, B. Insights into the structure of the CCR4-NOT complex by electron microscopy. FEBS Lett. 2011, 585, 2182–2186. [Google Scholar] [CrossRef] [PubMed]
- Boland, A.; Chen, Y.; Raisch, T.; Jonas, S.; Kuzuoğlu-Öztürk, D.; Wohlbold, L.; Weichenrieder, O.; Izaurralde, E. Structure and assembly of the NOT module of the human CCR4–NOT complex. Nat. Struct. Mol. Biol. 2013, 20, 1289–1297. [Google Scholar] [CrossRef]
- Ukleja, M.; Cuellar, J.; Siwaszek, A.; Kasprzak, J.M.; Czarnocki-Cieciura, M.; Bujnicki, J.M.; Janusz, M.; Dziembowski, A.; Valpuesta, J.M. The architecture of the Schizosaccharomyces pombe CCR4-NOT complex. Nat. Commun. 2016, 7, 10433. [Google Scholar] [CrossRef]
- Raisch, T.; Chang, C.-T.; Levdansky, Y.; Muthukumar, S.; Raunser, S.; Valkov, E. Reconstitution of recombinant human CCR4-NOT reveals molecular insights into regulated deadenylation. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Raisch, T.; Sandmeir, F.; Weichenrieder, O.; Valkov, E.; Izaurralde, E. Structural and biochemical analysis of a NOT1 MIF4G-like domain of the CCR4-NOT complex. J. Struct. Biol. 2018, 204, 388–395. [Google Scholar] [CrossRef]
- Collart, M.A.; Panasenko, O.O. The Ccr4-Not. Complex: Architecture and Structural Insights. Macromolecular Protein Complexes; Springer: Cham, Switzerland, 2017; pp. 349–379. [Google Scholar]
- Bawankar, P.; Loh, B.; Wohlbold, L.; Schmidt, S.; Izaurralde, E. NOT10 and C2orf29/NOT11 form a conserved module of the CCR4-NOT complex that docks onto the NOT1 N-terminal domain. RNA Biol. 2013, 10, 228–244. [Google Scholar] [CrossRef] [PubMed]
- Albert, T.K.; Lemaire, M.; van Berkum, N.L.; Gentz, R.; Collart, M.A.; Timmers, H.T.M. Isolation and characterization of human orthologs of yeast CCR4–NOT complex subunits. Nucleic Acids Res. 2000, 28, 809–817. [Google Scholar] [CrossRef][Green Version]
- Hagkarim, N.C.; Ryan, E.L.; Byrd, P.J.; Hollingworth, R.; Shimwell, N.J.; Agathanggelou, A.; Vavasseur, M.; Kolbe, V.; Speiseder, T.; Dobner, T.; et al. Degradation of a novel DNA damage response protein, tankyrase 1 binding protein 1, following adenovirus infection. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed]
- Keskeny, C.; Raisch, T.; Sgromo, A.; Igreja, C.; Bhandari, D.; Weichenrieder, O.; Izaurralde, E. A conserved CAF40-binding motif in metazoan NOT4 mediates association with the CCR4–NOT complex. Genes Dev. 2019, 33, 236–252. [Google Scholar] [CrossRef]
- Garces, R.G.; Gillon, W.; Pai, E.F. Atomic model of human Rcd-1 reveals an armadillo-like-repeat protein with in vitro nucleic acid binding properties. Protein Sci. 2007, 16, 176–188. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Bai, Y.; Zhang, A.; Zhang, Q.; Bartlam, M.G. Insights into the structure and architecture of the CCR4–NOT complex. Front. Genet. 2014, 5, 137. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Boland, A.; Kuzuoğlu-Öztürk, D.; Bawankar, P.; Loh, B.; Chang, C.-T.; Weichenrieder, O.; Izaurralde, E. A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol. Cell 2014, 54, 737–750. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Takahashi, A.; Morita, M.; Suzuki, T.; Yamamoto, T. The role of the CNOT1 subunit of the CCR4-NOT complex in mRNA deadenylation and cell viability. Protein Cell 2011, 2, 755–763. [Google Scholar] [CrossRef]
- Shirai, Y.-T.; Suzuki, T.; Morita, M.; Takahashi, A.; Yamamoto, T. Multifunctional roles of the mammalian CCR4–NOT complex in physiological phenomena. Front. Genet. 2014, 5, 286. [Google Scholar] [CrossRef]
- Winkler, G.S. The mammalian anti-proliferative BTG/Tob protein family. J. Cell. Phys. 2010, 222, 66–72. [Google Scholar] [CrossRef]
- Mauxion, F.; Faux, C.; Séraphin, B. The BTG2 protein is a general activator of mRNA deadenylation. EMBO J. 2008, 27, 1039–1048. [Google Scholar] [CrossRef]
- Yang, X.; Morita, M.; Wang, H.; Suzuki, T.; Yang, W.; Luo, Y.; Zhao, C.; Yu, Y.; Bartlam, M.; Yamamoto, T.; et al. Crystal structures of human BTG2 and mouse TIS21 involved in suppression of CAF1 deadenylase activity. Nucleic Acids Res. 2008, 36, 6872–6881. [Google Scholar] [CrossRef]
- Miyasaka, T.; Morita, M.; Ito, K.; Suzuki, T.; Fukuda, H.; Takeda, S.; Inoue, J.; Semba, K.; Yamamoto, T. Interaction of antiproliferative protein Tob with the CCR4-NOT deadenylase complex. Cancer Sci. 2008, 99, 755–761. [Google Scholar] [CrossRef]
- Webster, M.W.; Chen, Y.-H.; Stowell, J.A.; Alhusaini, N.; Sweet, T.; Graveley, B.R.; Coller, J.; Passmore, L.A. mRNA deadenylation is coupled to translation rates by the differential activities of Ccr4-Not nucleases. Mol. Cell 2018, 70, 1089–1100. [Google Scholar] [CrossRef]
- Braun, J.E.; Huntzinger, E.; Fauser, M.; Izaurralde, E. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol. Cell 2011, 44, 120–133. [Google Scholar] [CrossRef]
- Chekulaeva, M.; Mathys, H.; Zipprich, J.T.; Attig, J.; Colic, M.; Parker, R.; Filipowicz, W. miRNA repression involves GW182-mediated recruitment of CCR4–NOT through conserved W-containing motifs. Nat. Struct. Mol. Biol. 2011, 18, 1218–1226. [Google Scholar] [CrossRef]
- Fabian, M.R.; Cieplak, M.K.; Frank, F.; Morita, M.; Green, J.; Srikumar, T.; Nagar, B.; Yamamoto, T.; Raught, B.; Duchaine, T.F.; et al. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4–NOT. Nat. Struct. Mol. Biol. 2011, 18, 1211–1217. [Google Scholar] [CrossRef] [PubMed]
- Mathys, H.; Basquin, J.; Ozgur, S.; Czarnocki-Cieciura, M.; Bonneau, F.; Aartse, A.; Dziembowski, A.; Nowotny, M.; Conti, E.; Filipowicz, W. Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Mol. Cell 2014, 54, 751–765. [Google Scholar] [CrossRef] [PubMed]
- Rouya, C.; Siddiqui, N.; Morita, M.; Duchaine, T.F.; Fabian, M.R.; Sonenberg, N. Human DDX6 effects miRNA-mediated gene silencing via direct binding to CNOT1. RNA 2014, 20, 1398–1409. [Google Scholar] [CrossRef] [PubMed]
- Ozgur, S.; Basquin, J.; Kamenska, A.; Filipowicz, W.; Standart, N.; Conti, E. Structure of a human 4E-T/DDX6/CNOT1 complex reveals the different interplay of DDX6-binding proteins with the CCR4-NOT complex. Cell Rep. 2015, 13, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Sgromo, A.; Raisch, T.; Backhaus, C.; Keskeny, C.; Alva, V.; Weichenrieder, O.; Izaurralde, E. Drosophila Bag-of-marbles directly interacts with the CAF40 subunit of the CCR4–NOT complex to elicit repression of mRNA targets. RNA 2018, 24, 381–395. [Google Scholar] [CrossRef]
- Rodriguez-Gil, A.; Ritter, O.; Hornung, J.; Stekman, H.; Krüger, M.; Braun, T.; Kremmer, E.; Kracht, M.; Schmitz, M.L. HIPK family kinases bind and regulate the function of the CCR4-NOT complex. Mol. Biol. Cell 2016, 27, 1969–1980. [Google Scholar] [CrossRef]
- Yang, C.-Y.; Ramamoorthy, S.; Boller, S.; Rosenbaum, M.; Gil, A.R.; Mittler, G.; Imai, Y.; Kuba, K.; Grosschedl, R. Interaction of CCR4–NOT with EBF1 regulates gene-specific transcription and mRNA stability in B lymphopoiesis. Genes Dev. 2016, 30, 2310–2324. [Google Scholar] [CrossRef]
- Deluen, C.; James, N.; Maillet, L.; Molinete, M.; Theiler, G.; Lemaire, M.; Paquet, N.; Collart, M.A. The Ccr4-Not complex and yTAF1 (yTafII130p/yTafII145p) show physical and functional interactions. Mol. Cell. Biol. 2002, 22, 6735–6749. [Google Scholar] [CrossRef]
- Funakoshi, Y.; Doi, Y.; Hosoda, N.; Uchida, N.; Osawa, M.; Shimada, I.; Tsujimoto, M.; Suzuki, T.; Katada, T.; Hoshino, S. Mechanism of mRNA deadenylation: Evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev. 2007, 21, 3135–3148. [Google Scholar] [CrossRef] [PubMed]
- Beilharz, T.H.; Preiss, T. Widespread use of poly (A) tail length control to accentuate expression of the yeast transcriptome. RNA 2007, 13, 982–997. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Sweet, T.J.; Chamnongpol, S.; Baker, K.E.; Coller, J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature 2009, 461, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Traven, A.; Hammet, A.; Tenis, N.; Denis, C.L.; Heierhorst, J. Ccr4-not complex mRNA deadenylase activity contributes to DNA damage responses in Saccharomyces cerevisiae. Genetics 2005, 169, 65–75. [Google Scholar] [CrossRef][Green Version]
- Morel, A.-P.; Sentis, S.; Bianchin, C.; Le Romancer, M.; Jonard, L.; Rostan, M.-C.; Rimokh, R.; Corbo, L. BTG2 antiproliferative protein interacts with the human CCR4 complex existing in vivo in three cell-cycle-regulated forms. J. Cell Sci. 2003, 116, 2929–2936. [Google Scholar] [CrossRef]
- Winkler, G.S.; Balacco, D.L. Heterogeneity and complexity within the nuclease module of the Ccr4-Not complex. Front. Genet. 2013, 4, 296. [Google Scholar] [CrossRef][Green Version]
- Bianchin, C.; mauxion, F.; Sentis, S.; Séraphin, B.; Corbo, L. Conservation of the deadenylase activity of proteins of the Caf1 family in human. RNA 2005, 11, 487–494. [Google Scholar] [CrossRef][Green Version]
- Aslam, A.; Mittal, S.; Koch, F.; Andrau, J.-C.; Winkler, G.S. The Ccr4–NOT deadenylase subunits CNOT7 and CNOT8 have overlapping roles and modulate cell proliferation. Mol. Biol. Cell 2009, 20, 3840–3850. [Google Scholar] [CrossRef]
- Berthet, C.; Morera, A.-M.; Asensio, M.-J.; Chauvin, M.-A.; Morel, A.-P.; Dijoud, F.; Magaud, J.; Durand, P.; Rouault, J. CCR4-associated factor CAF1 is an essential factor for spermatogenesis. Mol. Cell. Biol. 2004, 24, 5808–5820. [Google Scholar] [CrossRef]
- Nakamura, T.; Yao, R.; Ogawa, T.; Suzuki, T.; Ito, C.; Tsunekawa, N.; Inoue, K.; Ajima, R.; Miyasaka, T.; Yoshida, Y.; et al. Oligo-astheno-teratozoospermia in mice lacking Cnot7, a regulator of retinoid X receptor beta. Nat. Genet. 2004, 36, 528–533. [Google Scholar] [CrossRef]
- Malvar, T.; Biron, R.W.; Kaback, D.B.; Denis, C.L. The CCR4 protein from Saccharomyces cerevisiae contains a leucine-rich repeat region which is required for its control of ADH2 gene expression. Genetics 1992, 132, 951–962. [Google Scholar]
- Dupressoir, A.; Morel, A.-P.; Barbot, W.; Loireau, M.-P.; Corbo, L.; Heidmann, T. Identification of four families of yCCR4-and Mg 2+-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genom. 2001, 2, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Clark, L.B.; Viswanathan, P.; Quigley, G.; Chiang, Y.-C.; McMahon, J.S.; Yao, G.; Chen, J.; Nelsbach, A.; Denis, C.L. Systematic mutagenesis of the leucine-rich repeat (LRR) domain of CCR4 reveals specific sites for binding to CAF1 and a separate critical role for the LRR in CCR4 deadenylase activity. J. Biol. Chem. 2004, 279, 13616–13623. [Google Scholar] [CrossRef] [PubMed]
- Morita, M.; Suzuki, T.; Nakamura, T.; Yokoyama, K.; Miyasaka, T.; Yamamoto, T. Depletion of mammalian CCR4b deadenylase triggers elevation of the p27Kip1 mRNA level and impairs cell growth. Mol. Cell. Biol. 2007, 27, 4980–4990. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, A.; Chang, T.-C.; Yamashita, Y.; Zhu, W.; Zhong, Z.; Chen, C.-Y.A.; Shyu, A.-B. Concerted action of poly (A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 2005, 12, 1054–1063. [Google Scholar] [CrossRef]
- Wahle, E.; Winkler, G.S. RNA decay machines: Deadenylation by the Ccr4–Not and Pan2–Pan3 complexes. Biochim. Biophys. Acta Gene Regul. Mech. 2013, 1829, 561–570. [Google Scholar] [CrossRef]
- Yi, H.; Park, J.; Ha, M.; Lim, J.; Chang, H.; Kim, V.N. PABP cooperates with the CCR4-NOT complex to promote mRNA deadenylation and block precocious decay. Mol. Cell 2018, 70, 1081–1088. [Google Scholar] [CrossRef]
- Tang, T.T.; Stowell, J.A.; Hill, C.H.; Passmore, L.A. The intrinsic structure of poly (A) RNA determines the specificity of Pan2 and Caf1 deadenylases. Nat. Struct. Mol. Biol. 2019, 26, 433–442. [Google Scholar] [CrossRef]
- Parker, R.; Song, H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004, 11, 121–127. [Google Scholar] [CrossRef]
- Goldstrohm, A.C.; Wickens, M. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 2008, 9, 337–344. [Google Scholar] [CrossRef]
- Garneau, N.L.; Wilusz, J.; Wilusz, C.J. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007, 8, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Parker, R.; Sheth, U. P bodies and the control of mRNA translation and degradation. Mol. Cell 2007, 25, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Houseley, J.; Tollervey, D. The many pathways of RNA degradation. Cell 2009, 136, 763–776. [Google Scholar] [CrossRef]
- Chahar, H.S.; Chen, S.; Manjunath, N. P-body components LSM1, GW182, DDX3, DDX6 and XRN1 are recruited to WNV replication sites and positively regulate viral replication. Virology 2013, 436, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Charenton, C.; Taverniti, V.; Gaudon-Plesse, C.; Back, R.; Séraphin, B.; Graille, M. Structure of the active form of Dcp1–Dcp2 decapping enzyme bound to m 7 GDP and its Edc3 activator. Nat. Struct. Mol. Biol. 2016, 23, 982–986. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, D.; Parker, R. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol. Biol. Cell 2007, 18, 2274–2287. [Google Scholar] [CrossRef]
- Lin, M.-D.; Jiao, X.; Grima, D.; Newbury, S.F.; Kiledjian, M.; Chou, T.-B. Drosophila processing bodies in oogenesis. Dev. Biol. 2008, 322, 276–288. [Google Scholar] [CrossRef]
- Eulalio, A.; Behm-Ansmant, I.; Schweizer, D.; Izaurralde, E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 2007, 27, 3970–3981. [Google Scholar] [CrossRef]
- Miller, J.E.; Reese, J.C. Ccr4-Not complex: The control freak of eukaryotic cells. Crit. Rev. Biochem. Mol. Biol. 2012, 47, 315–333. [Google Scholar] [CrossRef]
- Muhlrad, D.; Parker, R. The yeast EDC1 mRNA undergoes deadenylation-independent decapping stimulated by Not2p, Not4p, and Not5p. EMBO J. 2005, 24, 1033–1045. [Google Scholar] [CrossRef]
- Andrei, M.A.; Ingelfinger, D.; Heintzmann, R.; Achsel, T.; Rivera-Pomar, R.; Lührmann, R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 2005, 11, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Inoue, T.; Yokoyama, K.; Morita, M.; Suzuki, T.; Yamamoto, T. CNOT2 depletion disrupts and inhibits the CCR4–NOT deadenylase complex and induces apoptotic cell death. Genes Cell. 2011, 16, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Igarashi, K.; Aisaki, K.-. i Kanno, J.; Saga, Y. NANOS2 interacts with the CCR4-NOT deadenylation complex and leads to suppression of specific RNAs. Proc. Natl. Acad. Sci. USA 2010, 107, 3594–3599. [Google Scholar] [CrossRef] [PubMed]
- Mugler, C.F.; Hondele, M.; Heinrich, S.; Sachdev, R.; Vallotton, P.; Koek, A.Y.; Chan, L.Y.; Weis, K. ATPase activity of the DEAD-box protein Dhh1 controls processing body formation. eLife 2016, 5, e18746. [Google Scholar] [CrossRef] [PubMed]
- Maillet, L.; Collart, M.A. Interaction between Not1p, a component of the Ccr4-not complex, a global regulator of transcription, and Dhh1p, a putative RNA helicase. J. Biol. Chem. 2002, 277, 2835–2842. [Google Scholar] [CrossRef]
- Hwang, J.; Sato, H.; Tang, Y.; Matsuda, D.; Maquat, L.E. UPF1 association with the cap-binding protein, CBP80, promotes nonsense-mediated mRNA decay at two distinct steps. Mol. Cell 2010, 39, 396–409. [Google Scholar] [CrossRef] [PubMed]
- Loh, B.; Jonas, S.; Izaurralde, E. The SMG5–SMG7 heterodimer directly recruits the CCR4–NOT deadenylase complex to mRNAs containing nonsense codons via interaction with POP2. Genes Dev. 2013, 27, 2125–2138. [Google Scholar] [CrossRef]
- Kurosaki, T.; Popp, M.W.; Maquat, L.E. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat. Rev. Mol. Cell Biol. 2019, 20, 406–420. [Google Scholar] [CrossRef]
- Inada, T.; Makino, S. Novel roles of the multi-functional CCR4-NOT complex in post-transcriptional regulation. Front. Genet. 2014, 5, 135. [Google Scholar] [CrossRef]
- Sandler, H.; Kreth, J.; Timmers, H.T.M.; Stoecklin, G. Not1 mediates recruitment of the deadenylase Caf1 to mRNAs targeted for degradation by tristetraprolin. Nucleic Acids Res. 2011, 39, 4373–4386. [Google Scholar] [CrossRef]
- Sanduja, S.; Blanco, F.F.; Dixon, D.A. The roles of TTP and BRF proteins in regulated mRNA decay. Wiley interdisciplinary reviews: RNA 2011, 2, 42–57. [Google Scholar] [CrossRef]
- Chamboredon, S.; Ciais, D.; Desroches-Castan, A.; Savi, P.; Bono, F.; Feige, J.-J.; Cherradi, N. Hypoxia-inducible factor-1α mRNA: A new target for destabilization by tristetraprolin in endothelial cells. Mol. Biol. Cell 2011, 22, 3366–3378. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.; Seong, H.; Yoon, N.; Seo, S.W.; Park, J.W.; Kang, S.S.; Park, J.M.; Han, Y.S. Tristetraprolin regulates the decay of the hypoxia-induced vascular endothelial growth factor mRNA in ARPE-19 cells. Mol. Med. Rep. 2016, 14, 5395–5400. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Carballo, E.; Lai, W.S.; Blackshear, P.J. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science 1998, 281, 1001–1005. [Google Scholar] [CrossRef]
- Ogilvie, R.L.; Abelson, M.; Hau, H.H.; Vlasova, I.; Blackshear, P.J.; Bohjanen, P.R. Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. J. Immunol. 2005, 174, 953–961. [Google Scholar] [CrossRef] [PubMed]
- Stoecklin, G.; Ming, X.-F.; Looser, R.; Moroni, C. Somatic mRNA turnover mutants implicate tristetraprolin in the interleukin-3 mRNA degradation pathway. Mol. Cell. Biol. 2000, 20, 3753–3763. [Google Scholar] [CrossRef] [PubMed]
- Patino, W.D.; Kang, J.-G.; Matoba, S.; Mian, O.Y.; Gochuico, B.R.; Hwang, P.M. Atherosclerotic plaque macrophage transcriptional regulators are expressed in blood and modulated by tristetraprolin. Circ. Res. 2006, 98, 1282–1289. [Google Scholar] [CrossRef]
- Marderosian, M.; Sharma, A.; Funk, A.; Vartanian, R.; Masri, J.; Jo, O.; Gera, F.J. Tristetraprolin regulates Cyclin D1 and c-Myc mRNA stability in response to rapamycin in an Akt-dependent manner via p38 MAPK signaling. Oncogene 2006, 25, 6277–6290. [Google Scholar] [CrossRef]
- Polesskaya, A.; Pinna, G.; Sassi, Y.; Vandamme, M.; Bigot, A.; Mouly, V.; Nadya, M.; Annick, H.-B.; Cindy, D. Post-transcriptional modulation of interleukin 8 by CNOT6L regulates skeletal muscle differentiation. Biochimi. Biophys. Acta Mol. Cell Res. 2016, 1863, 263–270. [Google Scholar] [CrossRef]
- Polesskaya, A.; Degerny, C.; Pinna, G.; Maury, Y.; Kratassiouk, G.; Mouly, V.; Nadya, M.; Jeremie, K.; Niels, F.; Annick, H.-B. Genome-wide exploration of miRNA function in mammalian muscle cell differentiation. PLoS ONE 2013, 8, e71927. [Google Scholar] [CrossRef]
- Milewska, M.; Domoradzki, T.; Majewska, A.; Błaszczyk, M.; Gajewska, M.; Hulanicka, M.; Ciecierska, A.; Grzelkowska-Kowalczyk, K. Interleukin-8 enhances myocilin expression, Akt-FoxO3 signaling and myogenic differentiation in rat skeletal muscle cells. J. Cell. Phys. 2019, 234, 19675–19690. [Google Scholar] [CrossRef] [PubMed]
- Ezzeddine, N.; Chang, T.-C.; Zhu, W.; Yamashita, A.; Chen, C.-Y.A.; Zhong, Z.; Yamashita, Y.; Zheng, D.; Shyu, A. Human TOB, an antiproliferative transcription factor, is a poly (A)-binding protein-dependent positive regulator of cytoplasmic mRNA deadenylation. Mol. Cell. Biol. 2007, 27, 7791–7801. [Google Scholar] [CrossRef] [PubMed]
- Prévôt, D.; Morel, A.-P.; Voeltzel, T.; Rostan, M.-C.; Rimokh, R.; Magaud, J.-P.; Corbo, L. Relationships of the Antiproliferative Proteins BTG1 and BTG2 with CAF1, the Human Homolog of a Component of the Yeast CCR4 Transcriptional Complex INVOLVEMENT IN ESTROGEN RECEPTOR α SIGNALING PATHWAY. J. Biol. Chem. 2001, 276, 9640–9648. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, M.; Sasaoka, Y.; Kiso, M.; Abe, K.; Haraguchi, S.; Kobayashi, S.; Saga, Y. Conserved role of nanos proteins in germ cell development. Science 2003, 301, 1239–1241. [Google Scholar] [CrossRef] [PubMed]
- Saga, Y. Mouse germ cell development during embryogenesis. Curr. Opin. Genet. Dev. 2008, 18, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Saba, R.; Miyoshi, K.; Morita, Y.; Saga, Y. Interaction between NANOS2 and the CCR4-NOT deadenylation complex is essential for male germ cell development in mouse. PLoS ONE 2012, 7, e33558. [Google Scholar] [CrossRef]
- Zheng, X.; Dumitru, R.; Lackford, B.L.; Freudenberg, J.M.; Singh, A.P.; Archer, T.K.; Jothi, R.; Hu, G. Cnot1, Cnot2, and Cnot3 maintain mouse and human ESC identity and inhibit extraembryonic differentiation. Stem Cell 2012, 30, 910–922. [Google Scholar] [CrossRef]
- Hu, G.; Kim, J.; Xu, Q.; Leng, Y.; Orkin, S.H.; Elledge, S.J. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009, 23, 837–848. [Google Scholar] [CrossRef]
- Suzuki, T.; Kikuguchi, C.; Sharma, S.; Sasaki, T.; Tokumasu, M.; Adachi, S.; Natsume, T.; Kanegae, Y.; Yamamoto, T. CNOT3 suppression promotes necroptosis by stabilizing mRNAs for cell death-inducing proteins. Sci. Rep. 2015, 5, 14779. [Google Scholar] [CrossRef]
- Semotok, J.L.; Cooperstock, R.L.; Pinder, B.D.; Vari, H.K.; Lipshitz, H.D.; Smibert, C.A. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr. Biol. 2005, 15, 284–294. [Google Scholar] [CrossRef]
- Chicoine, J.; Benoit, P.; Gamberi, C.; Paliouras, M.; Simonelig, M.; Lasko, P. Bicaudal-C recruits CCR4-NOT deadenylase to target mRNAs and regulates oogenesis, cytoskeletal organization, and its own expression. Dev. Cell 2007, 13, 691–704. [Google Scholar] [CrossRef] [PubMed]
- Joly, W.; Chartier, A.; Rojas-Rios, P.; Busseau, I.; Simonelig, M. The CCR4 deadenylase acts with Nanos and Pumilio in the fine-tuning of Mei-P26 expression to promote germline stem cell self-renewal. Stem Cell Rep. 2013, 1, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.Z.; Hong, A.; Lilly, M.A.; Lehmann, R. Twin, a CCR4 homolog, regulates cyclin poly (A) tail length to permit Drosophila oogenesis. Development 2005, 132, 1165–1174. [Google Scholar] [CrossRef]
- Zaessinger, S.; Busseau, I.; Simonelig, M. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development 2006, 133, 4573–4583. [Google Scholar] [CrossRef]
- Behm-Ansmant, I.; Rehwinkel, J.; Doerks, T.; Stark, A.; Bork, P.; Izaurralde, E. mRNA degradation by miRNAs and GW182 requires both CCR4: NOT deadenylase and DCP1: DCP2 decapping complexes. Genes Dev. 2006, 20, 1885–1898. [Google Scholar] [CrossRef] [PubMed]
- Fabian, M.R.; Sonenberg, N. The mechanics of miRNA-mediated gene silencing: A look under the hood of miRISC. Nat. Struct. Mol. Biol. 2012, 19, 586–593. [Google Scholar] [CrossRef] [PubMed]
- Park, O.H.; Ha, H.; Lee, Y.; Boo, S.H.; Kwon, D.H.; Song, H.K.; Do, H.; Song, H.K.; Kim, Y.K. Endoribonucleolytic cleavage of m6A-containing RNAs by RNase P/MRP complex. Mol. Cell 2019, 74, 494–507. [Google Scholar] [CrossRef]
- Zaccara, S.; Jaffrey, S.R. A Unified Model for the Function of YTHDF Proteins in Regulating m6A-Modified. mRNA Cell 2020, 181, 1582–1595. [Google Scholar] [CrossRef]
- Du, H.; Zhao, Y.; He, J.; Zhang, Y.; Xi, H.; Liu, M.; Ma, J.; Wu, L. YTHDF2 destabilizes m 6 A-containing RNA through direct recruitment of the CCR4–NOT deadenylase complex. Nat. Commun. 2016, 7, 1–11. [Google Scholar] [CrossRef]
- Hu, G.; McQuiston, T.; Bernard, A.; Park, Y.-D.; Qiu, J.; Vural, A.; Zhang, N.; Waterman, S.R.; Blewett, N.H.; Myers, T.G.; et al. A conserved mechanism of TOR-dependent RCK-mediated mRNA degradation regulates autophagy. Nat. Cell Biol. 2015, 17, 930–942. [Google Scholar] [CrossRef]
- Radhakrishnan, A.; Chen, Y.-H.; Martin, S.; Alhusaini, N.; Green, R.; Coller, J. The DEAD-box protein Dhh1p couples mRNA decay and translation by monitoring codon optimality. Cell 2016, 167, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Laribee, R.N.; Hosni-Ahmed, A.; Workman, J.J.; Chen, H. Ccr4-not regulates RNA polymerase I transcription and couples nutrient signaling to the control of ribosomal RNA biogenesis. PLoS Genet. 2015, 11, 1005113. [Google Scholar] [CrossRef] [PubMed]
- Panasenko, O.O.; Somasekharan, S.P.; Villanyi, Z.; Zagatti, M.; Bezrukov, F.; Rashpa, R.; Cornut, J.; Iqbal, J.; Longis, M.; Carl, S.H.; et al. Co-translational assembly of proteasome subunits in NOT1-containing assemblysomes. Nat. Struct. Mol. Biol. 2019, 26, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Harel-Sharvit, L.; Eldad, N.; Haimovich, G.; Barkai, O.; Duek, L.; Choder, M. RNA polymerase II subunits link transcription and mRNA decay to translation. Cell 2010, 143, 552–563. [Google Scholar] [CrossRef]
- Choder, M. mRNA imprinting: Additional level in the regulation of gene expression. Cell. Logist. 2011, 1, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Gupta, I.; Villanyi, Z.; Kassem, S.; Hughes, C.; Panasenko, O.O.; Steinmetz, L.M.; Collart, M.A. Translational capacity of a cell is determined during transcription elongation via the Ccr4-Not complex. Cell Rep. 2016, 15, 1782–1794. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A. The unravelling of the ubiquitin system. Nat. Rev. Mol. Cell Biol. 2015, 16, 322–324. [Google Scholar] [CrossRef] [PubMed]
- Collins, G.A.; Goldberg, A.L. The logic of the 26S proteasome. Cell 2017, 169, 792–806. [Google Scholar] [CrossRef]
- Albert, T.K.; Hanzawa, H.; Legtenberg, Y.I.; de Ruwe, M.J.; van den Heuvel, F.A.; Collart, M.A.; Boelens, R.; Timmers, H.T. Marc Identification of a ubiquitin–protein ligase subunit within the CCR4–NOT transcription repressor complex. EMBO J. 2002, 21, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Mulder, K.W.; Inagaki, A.; Cameroni, E.; Mousson, F.; Winkler, G.S.; De Virgilio, C.; Collart, M.A.; Timmers, H.T.M. Modulation of Ubc4p/Ubc5p-mediated stress responses by the RING-finger-dependent ubiquitin-protein ligase Not4p in Saccharomyces cerevisiae. Genetics 2007, 176, 181–192. [Google Scholar] [CrossRef]
- Hanzawa, H.; de Ruwe, M.J.; Albert, T.K.; van der Vliet, P.C.; Timmers, H.M.; Boelens, R. The Structure of the C4C4RING finger of human NOT4 reveals features distinct from those of C3HC4 RING fingers. J. Biol. Chem. 2001, 276, 10185–10190. [Google Scholar] [CrossRef] [PubMed]
- Panasenko, O.O.; Collart, M.A. Not4 E3 ligase contributes to proteasome assembly and functional integrity in part through Ecm29. Mol. Cell. Biol. 2011, 31, 1610–1623. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Panasenko, O.O.; Collart, M.A. Presence of Not5 and ubiquitinated Rps7A in polysome fractions depends upon the Not4 E3 ligase. Mol. Microbiol. 2012, 83, 640–653. [Google Scholar] [CrossRef] [PubMed]
- Panasenko, O.; Landrieux, E.; Feuermann, M.; Finka, A.; Paquet, N.; Collart, M.A. The yeast Ccr4-Not complex controls ubiquitination of the nascent-associated polypeptide (NAC-EGD) complex. J. Biol. Chem. 2006, 281, 31389–31398. [Google Scholar] [CrossRef] [PubMed]
- Panasenko, O.O.; David, F.P.; Collart, M.A. Ribosome association and stability of the nascent polypeptide-associated complex is dependent upon its own ubiquitination. Genetics 2009, 181, 447–460. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Wolgast, M.; Beebe, L.M.; Reese, J.C. Ccr4–Not maintains genomic integrity by controlling the ubiquitylation and degradation of arrested RNAPII. Genes Dev. 2019, 33, 705–717. [Google Scholar] [CrossRef] [PubMed]
- Mersman, D.P.; Du, H.-N.; Fingerman, I.M.; South, P.F.; Briggs, S.D. Polyubiquitination of the demethylase Jhd2 controls histone methylation and gene expression. Genes Dev. 2009, 23, 951–962. [Google Scholar] [CrossRef][Green Version]
- Cooper, K.F.; Scarnati, M.S.; Krasley, E.; Mallory, M.J.; Jin, C.; Law, M.J.; Strich, R. Oxidative-stress-induced nuclear to cytoplasmic relocalization is required for Not4-dependent cyclin C destruction. J. Cell Sci. 2012, 125, 1015–1026. [Google Scholar] [CrossRef]
- Laribee, R.N.; Shibata, Y.; Mersman, D.P.; Collins, S.R.; Kemmeren, P.; Roguev, A.; Weissman, J.S.; Briggs, S.D.; Krogan, N.J.; Strahl, B.D. CCR4/NOT complex associates with the proteasome and regulates histone methylation. Proc. Natl. Acad. Sci. USA 2007, 104, 5836–5841. [Google Scholar] [CrossRef]
- Halter, D.; Collart, M.A.; Panasenko, O.O. The Not4 E3 ligase and CCR4 deadenylase play distinct roles in protein quality control. PLoS ONE 2014, 9, e86218. [Google Scholar] [CrossRef]
- Timmers, H.T.M.; Tora, L. Transcript buffering: A balancing act between mRNA synthesis and mRNA degradation. Mol. Cell 2018, 72, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Schwalb, B.; Schulz, D.; Pirkl, N.; Etzold, S.; Larivière, L.; Maier, K.C.; Seizl, M.; Tresch, A.; Cramer, P. Comparative dynamic transcriptome analysis (cDTA) reveals mutual feedback between mRNA synthesis and degradation. Genome Res. 2012, 22, 1350–1359. [Google Scholar] [CrossRef] [PubMed]
- Rambout, X.; Detiffe, C.; Bruyr, J.; Mariavelle, E.; Cherkaoui, M.; Brohée, S.; Demoitié, P.; Lebrun, M.; Soin, R.; Lesage, B.; et al. The transcription factor ERG recruits CCR4–NOT to control mRNA decay and mitotic progression. Nat. Struct. Mol. Biol. 2016, 23, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Begley, V.; Corzo, D.; Jordán-Pla, A.; Cuevas-Bermúdez, A.; Miguel-Jiménez, L.; Pérez-Aguado, D.; Mercedes, M.-O.; Francisco, N.; Chávez, M.J.; Pérez-Ortín, J.E.; et al. The mRNA degradation factor Xrn1 regulates transcription elongation in parallel to Ccr4. Nucleic Acids Res. 2019, 47, 9524–9541. [Google Scholar] [CrossRef]
- Slobodin, B.; Bahat, A.; Sehrawat, U.; Becker-Herman, S.; Zuckerman, B.; Weiss, A.N.; Han, R.; Elkon, R.; Agami, R.; Ulitsky, I.; et al. Transcription Dynamics Regulate Poly (A) Tails and Expression of the RNA Degradation Machinery to Balance mRNA Levels. Mol. Cell 2020, 78, 434–444. [Google Scholar] [CrossRef]
- Reese, J.C. The control of elongation by the yeast Ccr4–Not complex. Biochim. Biophys. Acta Gene Regul. Mech. 2013, 1829, 127–133. [Google Scholar] [CrossRef]
- Swanson, M.J.; Qiu, H.; Sumibcay, L.; Krueger, A.; Kim, S.-J.; Natarajan, K.; Yoon, S.; Hinnebusch, A.G. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol. Cell. Biol. 2003, 23, 2800–2820. [Google Scholar] [CrossRef]
- Sanders, S.L.; Jennings, J.; Canutescu, A.; Link, A.J.; Weil, P.A. Proteomics of the eukaryotic transcription machinery: Identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol. 2002, 22, 4723–4738. [Google Scholar] [CrossRef]
- Benson, J.D.; Benson, M.; Howley, P.M.; Struhl, K. Association of distinct yeast Not2 functional domains with components of Gcn5 histone acetylase and Ccr4 transcriptional regulatory complexes. EMBO J. 1998, 17, 6714–6722. [Google Scholar] [CrossRef]
- Lemaire, M.; Collart, M.A. The TATA-binding protein-associated factor yTafII19p functionally interacts with components of the global transcriptional regulator Ccr4-Not complex and physically interacts with the Not5 subunit. J. Biol. Chem. 2000, 275, 26925–26934. [Google Scholar]
- Badarinarayana, V.; Chiang, Y.-C.; Denis, C.L. Functional interaction of CCR4-NOT proteins with TATAA-binding protein (TBP) and its associated factors in yeast. Genetics 2000, 155, 1045–1054. [Google Scholar] [PubMed]
- Liu, H.-Y.; Chiang, Y.-C.; Pan, J.; Chen, J.; Salvadore, C.; Audino, D.C.; Badarinarayana, V.; Palaniswamy, V.; Anderson, B.; Denis, C.L. Characterization of CAF4 and CAF16 reveals a functional connection between the CCR4-NOT complex and a subset of SRB proteins of the RNA polymerase II holoenzyme. J. Biol. Chem. 2001, 276, 7541–7548. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.; French-Cornay, D.; Fan, H.-Y.; Klein, H.; Denis, C.L.; Jaehning, J.A. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol. Cell. Biol. 1999, 19, 1056–1067. [Google Scholar] [CrossRef]
- Oberholzer, U.; Collart, M.A. In vitro transcription of a TATA-less promoter: Negative regulation by the Not1 protein. Biol. Chem. 1999, 380, 1365–1370. [Google Scholar] [CrossRef] [PubMed]
- Wilczynska, A.; Gillen, S.L.; Schmidt, T.; Meijer, H.A.; Jukes-Jones, R.; Langlais, C.; Kopra, K.; Lu, W.; Godfrey, J.D.; Hawley, B.R.; et al. eIF4A2 drives repression of translation at initiation by Ccr4-Not through purine-rich motifs in the 5′ UTR. Genome Biol. 2019, 20, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Maillet, L.; Tu, C.; Hong, Y.K.; Shuster, E.O.; Collart, M.A. The essential function of Not1 lies within the Ccr4-Not complex. J. Mol. Biol. 2000, 303, 131–143. [Google Scholar] [CrossRef]
- Huisinga, K.L.; Pugh, B.F. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 2004, 13, 573–585. [Google Scholar] [CrossRef]
- Lenssen, E.; James, N.; Pedruzzi, I.; Dubouloz, F.; Cameroni, E.; Bisig, R.; Maillet, L.; Werner, M.; Roosen, J.; Petrovic, K.; et al. The Ccr4-Not complex independently controls both Msn2-dependent transcriptional activation—via a newly identified Glc7/Bud14 type I protein phosphatase module—and TFIID promoter distribution. Mol. Cell. Biol. 2005, 25, 488–498. [Google Scholar] [CrossRef]
- James, N.; Landrieux, E.; Collart, M.A. A SAGA-independent function of SPT3 mediates transcriptional deregulation in a mutant of the Ccr4-not complex in Saccharomyces cerevisiae. Genetics 2007, 177, 123–135. [Google Scholar] [CrossRef][Green Version]
- Baker, S.; Grant, P. The SAGA continues: Expanding the cellular role of a transcriptional co-activator complex. Oncogene 2007, 26, 5329–5340. [Google Scholar] [CrossRef]
- Rodríguez-Navarro, S. Insights into SAGA function during gene expression. EMBO Rep. 2009, 10, 843–850. [Google Scholar] [CrossRef]
- Sterner, D.E.; Grant, P.A.; Roberts, S.M.; Duggan, L.J.; Belotserkovskaya, R.; Pacella, L.A.; Winston, F.; Workman, J.L.; Berger, S.L. Functional organization of the yeast SAGA complex: Distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 1999, 19, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Kassem, S.; Villanyi, Z.; Collart, M.A. Not5-dependent co-translational assembly of Ada2 and Spt20 is essential for functional integrity of SAGA. Nucleic Acids Res. 2017, 45, 1186–1199. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Ramnarain, D.B.; Chiang, Y.-C.; Ding, L.-H.; McMahon, J.S.; Denis, C.L. Genome wide expression analysis of the CCR4-NOT complex indicates that it consists of three modules with the NOT module controlling SAGA-responsive genes. Mol. Genet. Genom. 2008, 279, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Mulder, K.W.; Brenkman, A.B.; Inagaki, A.; van den Broek, N.J.; Marc Timmers, H.T. Regulation of histone H3K4 tri-methylation and PAF complex recruitment by the Ccr4-Not complex. Nucleic Acids Res. 2007, 35, 2428–2439. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Venters, B.J.; Wachi, S.; Mavrich, T.N.; Andersen, B.E.; Jena, P.; Sinnamon, A.J.; Jain, P.; Rolleri, N.S.; Jiang, C.; Hemeryck-Walsh, C.; et al. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol. Cell 2011, 41, 480–492. [Google Scholar] [CrossRef] [PubMed]
- Struhl, K. Constitutive and inducible Saccharomyces cerevisiae promoters: Evidence for two distinct molecular mechanisms. Mol. Cell. Biol. 1986, 6, 3847–3853. [Google Scholar] [CrossRef]
- Iyer, V.; Struhl, K. Mechanism of differential utilization of the his3 TR and TC TATA elements. Mol. Cell. Biol. 1995, 15, 7059–7066. [Google Scholar] [CrossRef]
- Watanabe, K.; Kokubo, T. SAGA mediates transcription from the TATA-like element independently of Taf1p/TFIID but dependent on core promoter structures in Saccharomyces cerevisiae. PLoS ONE 2017, 12, e0188435. [Google Scholar] [CrossRef]
- Collart, M.; Struhl, K. CDC39, an essential nuclear protein that negatively regulates transcription and differentially affects the constitutive and inducible HIS3 promoters. EMBO J. 1993, 12, 177–186. [Google Scholar] [CrossRef]
- Russell, P.; Benson, J.D.; Denis, C.L. Characterization of mutations in NOT2 indicates that it plays an important role in maintaining the integrity of the CCR4–NOT complex. J. Mol. Biol. 2002, 322, 27–39. [Google Scholar] [CrossRef]
- Mohibullah, N.; Hahn, S. Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes Dev. 2008, 22, 2994–3006. [Google Scholar] [CrossRef] [PubMed]
- Collart, M.A. The NOT, SPT3, and MOT1 genes functionally interact to regulate transcription at core promoters. Mol. Cell. Biol. 1996, 16, 6668–6676. [Google Scholar] [CrossRef][Green Version]
- Denis, C.L.; Chiang, Y.-C.; Cui, Y.; Chen, J. Genetic evidence supports a role for the yeast CCR4-NOT complex in transcriptional elongation. Genetics 2001, 158, 627–634. [Google Scholar] [PubMed]
- Hartzog, G.A.; Fu, J. The Spt4–Spt5 complex: A multi-faceted regulator of transcription elongation. Biochim. Biophys. Acta Gene Regul. Mech. 2013, 1829, 105–115. [Google Scholar] [CrossRef]
- Soutourina, J. Transcription regulation by the Mediator complex. Nat. Rev. Mol. Cell Biol. 2018, 19, 262. [Google Scholar] [CrossRef]
- Allen, B.L.; Taatjes, D.J. The Mediator complex: A central integrator of transcription. Nat. Rev. Mol. Cell Biol. 2015, 16, 155–166. [Google Scholar] [CrossRef]
- Kuchin, S.; Yeghiayan, P.; Carlson, M. Cyclin-dependent protein kinase and cyclin homologs SSN3 and SSN8 contribute to transcriptional control in yeast. Proc. Natl. Acad. Sci. USA 1995, 92, 4006–4010. [Google Scholar] [CrossRef]
- Khakhina, S.; Cooper, K.F.; Strich, R. Med13p prevents mitochondrial fission and programmed cell death in yeast through nuclear retention of cyclin C. Mol. Biol. Cell 2014, 25, 2807–2816. [Google Scholar] [CrossRef]
- Lee, T.I.; Wyrick, J.J.; Koh, S.S.; Jennings, E.G.; Gadbois, E.L.; Young, R.A. Interplay of positive and negative regulators in transcription initiation by RNA polymerase II holoenzyme. Mol. Cell. Biol. 1998, 18, 4455–4462. [Google Scholar] [CrossRef][Green Version]
- Zwartjes, C.G.; Jayne, S.; van den Berg, D.L.; Timmers, H.M. Repression of promoter activity by CNOT2, a subunit of the transcription regulatory Ccr4-not complex. J. Biol. Chem. 2004, 279, 10848–10854. [Google Scholar] [CrossRef] [PubMed]
- Winkler, G.S.; Mulder, K.W.; Bardwell, V.J.; Kalkhoven, E.; Timmers, H.T.M. Human Ccr4-Not complex is a ligand-dependent repressor of nuclear receptor-mediated transcription. EMBO J. 2006, 25, 3089–3099. [Google Scholar] [CrossRef] [PubMed]
- Jayne, S.; Zwartjes, C.G.; Van Schaik, F.M.; Timmers, H.T.M. Involvement of the SMRT/NCoR–HDAC3 complex in transcriptional repression by the CNOT2 subunit of the human Ccr4–Not complex. Biochem. J. 2006, 398, 461–467. [Google Scholar] [CrossRef]
- Peng, W.; Togawa, C.; Zhang, K.; Kurdistani, S.K. Regulators of cellular levels of histone acetylation in Saccharomyces cerevisiae. Genetics 2008, 179, 277–289. [Google Scholar] [CrossRef][Green Version]
- Neely, G.G.; Kuba, K.; Cammarato, A.; Isobe, K.; Amann, S.; Zhang, L.; Murata, M.; Elmén, L.; Gupta, V.; Arora, S.; et al. A global in vivo Drosophila RNAi screen identifies NOT3 as a conserved regulator of heart function. Cell 2010, 141, 142–153. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.; van den Hoorn, T.; Jongsma, M.L.; Bakker, M.J.; Hengeveld, R.; Janssen, L.; Murata, M.; Elmén, L.; Gupta, V.; Arora, S.; et al. A Genome-wide multidimensional RNAi screen reveals pathways controlling MHC class II antigen presentation. Cell 2011, 145, 268–283. [Google Scholar] [CrossRef]
- Bai, Y.; Salvadore, C.; Chiang, Y.-C.; Collart, M.A.; Liu, H.-Y.; Denis, C.L. The CCR4 and CAF1 proteins of the CCR4-NOT complex are physically and functionally separated from NOT2, NOT4, and NOT5. Mol. Cell. Biol. 1999, 19, 6642–6651. [Google Scholar] [CrossRef]
- Liu, H.Y.; Badarinarayana, V.; Audino, D.C.; Rappsilber, J.; Mann, M.; Denis, C.L. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 1998, 17, 1096–1106. [Google Scholar] [CrossRef]
- Neiman, A.M.; Chang, F.; Komachi, K.; Herskowitz, I. CDC36 and CDC39 are negative elements in the signal transduction pathway of yeast. Cell Regul. 1990, 1, 391–401. [Google Scholar] [CrossRef]
- de Barros Lopes, M.; Ho, J.; Reed, S.I. Mutations in cell division cycle genes CDC36 and CDC39 activate the Saccharomyces cerevisiae mating pheromone response pathway. Mol. Cell. Biol. 1990, 10, 2966–2972. [Google Scholar] [CrossRef][Green Version]
- Garapaty, S.; Mahajan, M.A.; Samuels, H.H. Components of the CCR4-NOT complex function as nuclear hormone receptor coactivators via association with the NRC-interacting Factor NIF-1. J. Biol. Chem. 2008, 283, 6806–6816. [Google Scholar] [CrossRef]
- Lin, C.Y.; Lovén, J.; Rahl, P.B.; Paranal, R.M.; Burge, C.B.; Bradner, J.E.; Lee, T.I.; Young, R.A. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 2012, 151, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Nie, Z.; Hu, G.; Wei, G.; Cui, K.; Yamane, A.; Resch, W.; Wang, R.; Green, D.R.; Tessarollo, L.; Casellas, R.; et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 2012, 151, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Venturini, G.; Rose, A.M.; Shah, A.Z.; Bhattacharya, S.S.; Rivolta, C. CNOT3 is a modifier of PRPF31 mutations in retinitis pigmentosa with incomplete penetrance. PLoS Genet. 2012, 8, e1003040. [Google Scholar] [CrossRef]
- Deery, E.C.; Vithana, E.N.; Newbold, R.J.; Gallon, V.A.; Bhattacharya, S.S.; Warren, M.J.; Hunt, D.M.; Wilkie, S.E. Disease mechanism for retinitis pigmentosa (RP11) caused by mutations in the splicing factor gene PRPF31. Hum. Mol. Genet. 2002, 11, 3209–3219. [Google Scholar] [CrossRef]
- Sohrabi-Jahromi, S. Transcriptome maps of general eukaryotic RNA degradation factors. eLife 2019, 8, e47040. [Google Scholar] [CrossRef]
- Gaillard, H.; Tous, C.; Botet, J.; González-Aguilera, C.; Quintero, M.J.; Viladevall, L.; García-Rubio, M.L.; Rodríguez-Gil, A.; Marín, A.; Ariño, J.; et al. Genome-wide analysis of factors affecting transcription elongation and DNA repair: A new role for PAF and Ccr4-not in transcription-coupled repair. PLoS Genet. 2009, 5, e1000364. [Google Scholar] [CrossRef] [PubMed]
- Bortvin, A.; Winston, F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 1996, 272, 1473–1476. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, C.D.; Laprade, L.; Winston, F. Transcription elongation factors repress transcription initiation from cryptic sites. Science 2003, 301, 1096–1099. [Google Scholar] [CrossRef]
- Verdone, L.; Cesari, F.; Denis, C.L.; Di Mauro, E.; Caserta, M. Factors Affecting Saccharomyces cerevisiae ADH2Chromatin Remodeling and Transcription. J. Biol. Chem. 1997, 272, 30828–30834. [Google Scholar] [CrossRef]
- Dronamraju, R.; Hepperla, A.J.; Shibata, Y.; Adams, A.T.; Magnuson, T.; Davis, I.J.; Strahl, B.D. Spt6 Association with RNA Polymerase II Directs mRNA Turnover During Transcription. Mol. Cell 2018, 70, 1054–1066. [Google Scholar] [CrossRef]
- Denis, C.L.; Malvar, T. The CCR4 gene from Saccharomyces cerevisiae is required for both nonfermentative and spt-mediated gene expression. Genetics 1990, 124, 283–291. [Google Scholar] [PubMed]
- Denis, C.L. Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II. Genetics 1984, 108, 833–844. [Google Scholar] [PubMed]
- Draper, M.P.; Salvadore, C.; Denis, C.L. Identification of a mouse protein whose homolog in Saccharomyces cerevisiae is a component of the CCR4 transcriptional regulatory complex. Mol. Cell. Biol. 1995, 15, 3487–3495. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.-Y.; Kim, N.; Hwang, C.-S.; Yoo, J.-Y. Protein degradation of RNA polymerase II-association factor 1 (PAF1) is controlled by CNOT4 and 26S proteasome. PLoS ONE 2015, 10, e0125599. [Google Scholar] [CrossRef]
- Kim, J.; Roeder, R.G. Direct Bre1-Paf1 complex interactions and RING finger-independent Bre1-Rad6 interactions mediate histone H2B ubiquitylation in yeast. J. Biol. Chem. 2009, 284, 20582–20592. [Google Scholar] [CrossRef]
- Mulder, K.W.; Winkler, G.S.; Timmers, H.T.M. DNA damage and replication stress induced transcription of RNR genes is dependent on the Ccr4–Not complex. Nucleic Acids Res. 2005, 33, 6384–6392. [Google Scholar] [CrossRef]
- Chen, H.; Sirupangi, T.; Wu, Z.-H.; Johnson, D.L.; Laribee, R.N. The conserved RNA recognition motif and C3H1 domain of the Not4 ubiquitin ligase regulate in vivo ligase function. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef]
- Babbarwal, V.; Fu, J.; Reese, J.C. The Rpb4/7 module of RNA polymerase II is required for carbon catabolite repressor protein 4-negative on TATA (Ccr4-not) complex to promote elongation. J. Biol. Chem. 2014, 289, 33125–33130. [Google Scholar] [CrossRef]
- Reines, D.; Chamberlin, M.; Kane, C. Transcription elongation factor SII (TFIIS) enables RNA polymerase II to elongate through a block to transcription in a human gene in vitro. J. Biol. Chem. 1989, 264, 10799–10809. [Google Scholar]
- Hausner, W.; Lange, U.; Musfeldt, M. Transcription factor S, a cleavage induction factor of the archaeal RNA polymerase. J. Biol. Chem. 2000, 275, 12393–12399. [Google Scholar] [CrossRef] [PubMed]
- Nudler, E. RNA polymerase backtracking in gene regulation and genome instability. Cell 2012, 149, 1438–1445. [Google Scholar] [CrossRef] [PubMed]
- Dutta, A.; Babbarwal, V.; Fu, J.; Brunke-Reese, D.; Libert, D.M.; Willis, J.; Reese, J.C. Ccr4-Not and TFIIS function cooperatively to rescue arrested RNA polymerase II. Mol. Cell. Biol. 2015, 35, 1915–1925. [Google Scholar] [CrossRef] [PubMed]
- Wind, M.; Reines, D. Transcription elongation factor SII. Bioessays 2000, 22, 327–336. [Google Scholar] [CrossRef]
- Choder, M. Rpb4 and Rpb7: Subunits of RNA polymerase II and beyond. Trends Biochem. Sci. 2004, 29, 674–681. [Google Scholar] [CrossRef]
- Verma-Gaur, J.; Rao, S.N.; Taya, T.; Sadhale, P. Genomewide recruitment analysis of Rpb4, a subunit of polymerase II in Saccharomyces cerevisiae, reveals its involvement in transcription elongation. Eukaryot. Cell 2008, 7, 1009–1018. [Google Scholar] [CrossRef]
- Kordyukova, M.; Sokolova, O.; Morgunova, V.; Ryazansky, S.; Akulenko, N.; Glukhov, S.; Kalmykova, A. Nuclear Ccr4-Not mediates the degradation of telomeric and transposon transcripts at chromatin in the Drosophila germline. Nucleic Acids Res. 2020, 48, 141–156. [Google Scholar] [CrossRef]
- Shalem, O.; Dahan, O.; Levo, M.; Martinez, M.R.; Furman, I.; Segal, E.; Pilpel, Y. Transient transcriptional responses to stress are generated by opposing effects of mRNA production and degradation. Mol. Syst. Biol. 2008, 4, 223. [Google Scholar] [CrossRef]
- Dori-Bachash, M.; Shema, E.; Tirosh, I. Coupled evolution of transcription and mRNA degradation. PLoS Biol. 2011, 9, e1001106. [Google Scholar] [CrossRef]
- Shalem, O.; Groisman, B.; Choder, M.; Dahan, O.; Pilpel, Y. Transcriptome kinetics is governed by a genome-wide coupling of mRNA production and degradation: A role for RNA Pol II. PLoS Genet. 2011, 7, e1002273. [Google Scholar] [CrossRef]
- Aguilera, A. The connection between transcription and genomic instability. EMBO J. 2002, 21, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Gottipati, P.; Helleday, T. Transcription-associated recombination in eukaryotes: Link between transcription, replication and recombination. Mutagenesis 2009, 24, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Reaban, M.E.; Griffin, J.A. Induction of RNA-stabilized DMA conformers by transcription of an immunoglobulin switch region. Nature 1990, 348, 342–344. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, J.; Alt, F.W. Class-switch recombination: Interplay of transcription, DNA deamination and DNA repair. Nat. Rev. Immunol. 2004, 4, 541–552. [Google Scholar] [CrossRef]
- Mizuta, R.; Iwai, K.; Shigeno, M.; Mizuta, M.; Uemura, T.; Ushiki, T.; Kitamura, D. Molecular visualization of immunoglobulin switch region RNA/DNA complex by atomic force microscope. J. Biol. Chem. 2003, 278, 4431–4434. [Google Scholar] [CrossRef]
- Yu, K.; Chedin, F.; Hsieh, C.-L.; Wilson, T.E.; Lieber, M.R. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 2003, 4, 442–451. [Google Scholar] [CrossRef]
- Biddick, R.; Young, E.T. The disorderly study of ordered recruitment. Yeast 2009, 26, 205–220. [Google Scholar] [CrossRef]
- Saponaro, M.; Kantidakis, T.; Mitter, R.; Kelly, G.P.; Heron, M.; Williams, H.; Johannes, S.; Stewart, A.; Svejstrup, J.Q. RECQL5 controls transcript elongation and suppresses genome instability associated with transcription stress. Cell 2014, 157, 1037–1049. [Google Scholar] [CrossRef]
- Kotsantis, P.; Silva, L.M.; Irmscher, S.; Jones, R.M.; Folkes, L.; Gromak, N.; Petermann, E. Increased global transcription activity as a mechanism of replication stress in cancer. Nat. Commun. 2016, 7, 1–13. [Google Scholar] [CrossRef]
- El Hage, A.; French, S.L.; Beyer, A.L.; Tollervey, D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010, 24, 1546–1558. [Google Scholar] [CrossRef]
- Manzo, S.G.; Hartono, S.R.; Sanz, L.A.; Marinello, J.; De Biasi, S.; Cossarizza, A.; Capranico, G.; Chedin, F. DNA Topoisomerase I differentially modulates R-loops across the human genome. Genome Biol. 2018, 19, 100. [Google Scholar] [CrossRef] [PubMed]
- Zatreanu, D.; Han, Z.; Mitter, R.; Tumini, E.; Williams, H.; Gregersen, L.; Dirac-Svejstrup, A.B.; Roma, S.; Stewart, A.; Aguilera, A.; et al. Elongation factor TFIIS prevents transcription stress and r-loop accumulation to maintain genome stability. Mol. Cell 2019, 76, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Skourti-Stathaki, K.; Proudfoot, N.J. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014, 28, 1384–1396. [Google Scholar] [CrossRef]
- Mischo, H.E.; Gómez-González, B.; Grzechnik, P.; Rondón, A.G.; Wei, W.; Steinmetz, L.; Aguilera, A.; Proudfoot, N.J. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol. Cell 2011, 41, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Wahba, L.; Amon, J.D.; Koshland, D.; Vuica-Ross, M. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA: DNA hybrids from generating genome instability. Mol. Cell 2011, 44, 978–988. [Google Scholar] [CrossRef]
- Cipres-Palacin, G.; Kane, C.M. Cleavage of the nascent transcript induced by TFIIS is insufficient to promote read-through of intrinsic blocks to elongation by RNA polymerase II. Proc. Natl. Acad. Sci. USA 1994, 91, 8087–8091. [Google Scholar] [CrossRef]
- Davie, J.K.; Kane, C.M. Genetic interactions between TFIIS and the Swi-Snf chromatin-remodeling complex. Mol. Cell. Biol. 2000, 20, 5960–5973. [Google Scholar] [CrossRef]
- Sharma, V.M.; Li, B.; Reese, J.C. SWI/SNF-dependent chromatin remodeling of RNR3 requires TAFIIs and the general transcription machinery. Genes Dev. 2003, 17, 502–515. [Google Scholar] [CrossRef][Green Version]
- Gómez-González, B.; García-Rubio, M.; Bermejo, R.; Gaillard, H.; Shirahige, K.; Marín, A.; Foiani, M.; Aguilera, A. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J. 2011, 30, 3106–3119. [Google Scholar] [CrossRef]
- Domínguez-Sánchez, M.S.; Barroso, S.; Gómez-González, B.; Luna, R.; Aguilera, A. Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLoS Genet. 2011, 7, e1002386. [Google Scholar] [CrossRef]
- Santos-Pereira, J.M.; Herrero, A.B.; García-Rubio, M.L.; Marín, A.; Moreno, S.; Aguilera, A. The Npl3 hnRNP prevents R-loop-mediated transcription–replication conflicts and genome instability. Genes Dev. 2013, 27, 2445–2458. [Google Scholar] [CrossRef]
- Chávez, S.; Aguilera, A. The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev. 1997, 11, 3459–3470. [Google Scholar] [CrossRef]
- Piruat, J.I.; Aguilera, A. A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J. 1998, 17, 4859–4872. [Google Scholar] [CrossRef]
- Chávez, S.; Beilharz, T.; Rondón, A.G.; Erdjument-Bromage, H.; Tempst, P.; Svejstrup, J.Q.; Lithgow, T.; Aguilera, A. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J. 2000, 19, 5824–5834. [Google Scholar] [CrossRef]
- Freedman, J.A.; Jinks-Robertson, S. Effects of mismatch repair and Hpr1 on transcription-stimulated mitotic recombination in the yeast Saccharomyces cerevisiae. DNA Repair 2004, 3, 1437–1446. [Google Scholar] [CrossRef] [PubMed]
- Huertas, P.; Aguilera, A. Cotranscriptionally formed DNA: RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 2003, 12, 711–721. [Google Scholar] [CrossRef]
- Kerr, S.C.; Azzouz, N.; Fuchs, S.M.; Collart, M.A.; Strahl, B.D.; Corbett, A.H.; Laribee, R.N. The Ccr4-Not complex interacts with the mRNA export machinery. PLoS ONE 2011, 6, e18302. [Google Scholar] [CrossRef]
- Sollier, J.; Stork, C.T.; García-Rubio, M.L.; Paulsen, R.D.; Aguilera, A.; Cimprich, K.A. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol. Cell 2014, 56, 777–785. [Google Scholar] [CrossRef]
- Stork, C.T.; Bocek, M.; Crossley, M.P.; Sollier, J.; Sanz, L.A.; Chedin, F.; Swigut, T.; Cimprich, K.A. Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. eLife 2016, 5, e17548. [Google Scholar] [CrossRef] [PubMed]
- Morillo-Huesca, M.; Vanti, M.; Chávez, S. A simple in vivo assay for measuring the efficiency of gene length-dependent processes in yeast mRNA biogenesis. FEBS J. 2006, 273, 756–769. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Morita, M.; Hijikata, A.; Fukuda-Yuzawa, Y.; Adachi, S.; Isono, K.; Ikawa, T.; Kawamoto, H.; Koseki, H.; Natsume, T.; et al. CNOT3 contributes to early B cell development by controlling Igh rearrangement and p53 mRNA stability. J. Exp. Med. 2015, 212, 1465–1479. [Google Scholar] [CrossRef] [PubMed]
- Chapat, C.; Chettab, K.; Simonet, P.; Wang, P.; De La Grange, P.; Le Romancer, M.; Corbo, L. Alternative splicing of CNOT7 diversifies CCR4–NOT functions. Nucleic Acids Res. 2017, 45, 8508–8523. [Google Scholar] [CrossRef] [PubMed]
- Robin-Lespinasse, Y.; Sentis, S.; Kolytcheff, C.; Rostan, M.-C.; Corbo, L.; Le Romancer, M. hCAF1, a new regulator of PRMT1-dependent arginine methylation. J. Cell Sci. 2007, 120, 638–647. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.-J.; Gary, J.D.; Yang, M.C.; Clarke, S.; Herschman, H.R. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J. Biol. Chem. 1996, 271, 15034–15044. [Google Scholar] [CrossRef]
- Choi, K.-S.; Kim, J.Y.; Lim, S.-K.; Choi, Y.W.; Kim, Y.H.; Kang, S.Y.; Park, T.J.; Lim, I.K. TIS21/BTG2/PC3 accelerates the repair of DNA double strand breaks by enhancing Mre11 methylation and blocking damage signal transfer to the Chk2T68–p53S20 pathway. DNA Repair 2012, 11, 965–975. [Google Scholar] [CrossRef]
- Rouault, J.-P.; Prévôt, D.; Berthet, C.; Birot, A.-M.; Billaud, M.; Magaud, J.-P.; Corbo, L. Interaction of BTG1 and p53-regulatedBTG2 Gene Products with mCaf1, the Murine Homolog of a Component of the Yeast CCR4 Transcriptional Regulatory Complex. J. Biol. Chem. 1998, 273, 22563–22569. [Google Scholar] [CrossRef]
- Liu, Q.; Dreyfuss, G. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol. Cell. Biol. 1995, 15, 2800–2808. [Google Scholar] [CrossRef]
- Coˆté, J.; Boisvert, F.o.-M.; Boulanger, M.-C.; Bedford, M.T.; Richard, S. Sam68 RNA binding protein is an in vivo substrate for protein arginine N-methyltransferase 1. Mol. Biol. Cell 2003, 14, 274–287. [Google Scholar] [CrossRef]
- Shen, E.C.; Henry, M.F.; Weiss, V.H.; Valentini, S.R.; Silver, P.A.; Lee, M.S. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 1998, 12, 679–691. [Google Scholar] [CrossRef]
- Michael, C.Y.; Bachand, F.; McBride, A.E.; Komili, S.; Casolari, J.M.; Silver, P.A. Arginine methyltransferase affects interactions and recruitment of mRNA processing and export factors. Genes Dev. 2004, 18, 2024–2035. [Google Scholar]
- Azzouz, N.; Panasenko, O.O.; Colau, G.; Collart, M.A. The CCR4-NOT complex physically and functionally interacts with TRAMP and the nuclear exosome. PLoS ONE 2009, 4. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.T.; Wang, X. Nuclear RNA surveillance: No sign of substrates tailing off. Crit. Rev. Biochem. Mol. Biol. 2009, 44, 16–24. [Google Scholar] [CrossRef]
- Houseley, J.; Tollervey, D. The nuclear RNA surveillance machinery: The link between ncRNAs and genome structure in budding yeast? Biochim. Biophys. Acta Gene Regul. Mech. 2008, 1779, 239–246. [Google Scholar] [CrossRef] [PubMed]
- LaCava, J.; Houseley, J.; Saveanu, C.; Petfalski, E.; Thompson, E.; Jacquier, A.; Tollervey, D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 2005, 121, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.; Petfalski, E.; Shevchenko, A.; Mann, M.; Tollervey, D. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′ → 5′ exoribonucleases. Cell 1997, 91, 457–466. [Google Scholar] [CrossRef]
- Fasken, M.B.; Corbett, A.H. Mechanisms of nuclear mRNA quality control. RNA Biol. 2009, 6, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Jensen, T.H. The exosome: A multipurpose RNA-decay machine. Trends Biochem. Sci. 2008, 33, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Assenholt, J.; Mouaikel, J.; Saguez, C.; Rougemaille, M.; Libri, D.; Jensen, T.H. Implication of Ccr4-Not complex function in mRNA quality control in Saccharomyces cerevisiae. RNA 2011, 17, 1788–1794. [Google Scholar] [CrossRef]
- Cotobal, C.; Rodríguez-López, M.; Duncan, C.; Hasan, A.; Yamashita, A.; Yamamoto, M.; Jürg, B.; Mata, J. Role of Ccr4-Not complex in heterochromatin formation at meiotic genes and subtelomeres in fission yeast. Epigenet. Chromatin 2015, 8, 28. [Google Scholar] [CrossRef]
- Simonetti, F.; Candelli, T.; Leon, S.; Libri, D.; Rougemaille, M. Ubiquitination-dependent control of sexual differentiation in fission yeast. eLife 2017, 6, e28046. [Google Scholar] [CrossRef]
- Cheng, D.D.; Li, J.; Li, S.J.; Yang, Q.C.; Fan, C.Y. CNOT 1 cooperates with LMNA to aggravate osteosarcoma tumorigenesis through the Hedgehog signaling pathway. Mol. Oncol. 2017, 11, 388–404. [Google Scholar] [CrossRef]
- Mittal, S.; Aslam, A.; Doidge, R.; Medica, R.; Winkler, G.S. The Ccr4a (CNOT6) and Ccr4b (CNOT6L) deadenylase subunits of the human Ccr4–Not complex contribute to the prevention of cell death and senescence. Mol. Biol. Cell 2011, 22, 748–758. [Google Scholar] [CrossRef] [PubMed]
- Kadyrova, L.Y.; Habara, Y.; Lee, T.H.; Wharton, R.P. Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development 2007, 134, 1519–1527. [Google Scholar] [CrossRef]
- Kim, J.H.; Richter, J.D. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell 2006, 24, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Voeltz, G.K.; Ongkasuwan, J.; Standart, N.; Steitz, J.A. A novel embryonic poly (A) binding protein, ePAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes Dev. 2001, 15, 774–788. [Google Scholar] [CrossRef] [PubMed]
- Martins, J.P.S.; Liu, X.; Oke, A.; Arora, R.; Franciosi, F.; Viville, S.; Laird, D.J.; Fung, J.C.; Conti, M. DAZL and CPEB1 regulate mRNA translation synergistically during oocyte maturation. J. Cell Sci. 2016, 129, 1271–1282. [Google Scholar] [CrossRef]
- Sha, Q.-Q.; Dai, X.-X.; Dang, Y.; Tang, F.; Liu, J.; Zhang, Y.-L.; Fan, H.-Y. A MAPK cascade couples maternal mRNA translation and degradation to meiotic cell cycle progression in mouse oocytes. Development 2017, 144, 452–463. [Google Scholar] [CrossRef]
- Ma, J.; Fukuda, Y.; Schultz, R.M. Mobilization of dormant Cnot7 mRNA promotes deadenylation of maternal transcripts during mouse oocyte maturation. Biol. Reprod. 2015, 93, 1–12. [Google Scholar] [CrossRef]
- Yu, C.; Ji, S.-Y.; Sha, Q.-Q.; Dang, Y.; Zhou, J.-J.; Zhang, Y.-L.; Liu, Y.; Wang, Z.; Hu, B.; Sun, Q.; et al. BTG4 is a meiotic cell cycle–coupled maternal-zygotic-transition licensing factor in oocytes. Nat. Struct. Mol. Biol. 2016, 23, 387–394. [Google Scholar] [CrossRef]
- Piqué, M.; López, J.M.; Foissac, S.; Guigó, R.; Méndez, R. A combinatorial code for CPE-mediated translational control. Cell 2008, 132, 434–448. [Google Scholar] [CrossRef]
- Sha, Q.Q.; Yu, J.L.; Guo, J.X.; Dai, X.X.; Jiang, J.C.; Zhang, Y.L.; Yu, C.; Ji, S.; Jiang, Y.; Zhang, S.; et al. CNOT 6L couples the selective degradation of maternal transcripts to meiotic cell cycle progression in mouse oocyte. EMBO J. 2018, 37, e99333. [Google Scholar] [CrossRef] [PubMed]
- Tennyson, C.N.; Lee, J.; Andrews, B.J. A role for the Pcl9-Pho85 cyclin–cdk complex at the M/G1 boundary in Saccharomyces cerevisiae. Mol. Microbiol 1998, 28, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Friesen, H.; Andrews, B. Pho85, a multifunctional cyclin-dependent protein kinase in budding yeast. Mol. Microbiol 2007, 66, 303–314. [Google Scholar] [CrossRef]
- Bömeke, K.; Pries, R.; Korte, V.; Scholz, E.; Herzog, B.; Schulze, F.; Braus, G.H. Yeast Gcn4p stabilization is initiated by the dissociation of the nuclear Pho85p/Pcl5p complex. Mol. Biol. Cell 2006, 17, 2952–2962. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shemer, R.; Meimoun, A.; Holtzman, T.; Kornitzer, D. Regulation of the transcription factor Gcn4 by Pho85 cyclin PCL5. Mol. Cell. Biol. 2002, 22, 5395–5404. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, M.; Kawasumi, M.; Fujino, M.; Toh-e, A. Phosphorylation of sic1, a cyclin-dependent kinase (Cdk) inhibitor, by Cdk including Pho85 kinase is required for its prompt degradation. Mol. Biol. Cell 1998, 9, 2393–2405. [Google Scholar] [CrossRef] [PubMed]
- Toyn, J.H.; Johnson, A.L.; Donovan, J.D.; Toone, W.M.; Johnston, L.H. The Swi5 transcription factor of Saccharomyces cerevisiae has a role in exit from mitosis through induction of the cdk-inhibitor Sic1 in telophase. Genetics 1997, 145, 85–96. [Google Scholar] [PubMed]
- Schwob, E.; Böhm, T.; Mendenhall, M.D.; Nasmyth, K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 1994, 79, 233–244. [Google Scholar] [CrossRef]
- Epstein, C.B.; Cross, F.R. CLB5: A novel B cyclin from budding yeast with a role in S phase. Genes Dev. 1992, 6, 1695–1706. [Google Scholar] [CrossRef]
- Schwob, E.; Nasmyth, K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 1993, 7, 1160–1175. [Google Scholar] [CrossRef]
- Liu, H.Y.; Toyn, J.H.; Chiang, Y.C.; Draper, M.P.; Johnston, L.H.; Denis, C.L. DBF2, a cell cycle-regulated protein kinase, is physically and functionally associated with the CCR4 transcriptional regulatory complex. EMBO J. 1997, 16, 5289–5298. [Google Scholar] [CrossRef] [PubMed]
- Trcek, T.; Larson, D.R.; Moldón, A.; Query, C.C.; Singer, R.H. Single-molecule mRNA decay measurements reveal promoter-regulated mRNA stability in yeast. Cell 2011, 147, 1484–1497. [Google Scholar] [CrossRef] [PubMed]
- Donovan, J.D.; Toyn, J.H.; Johnson, A.L.; Johnston, L.H. P40SDB25, a putative CDK inhibitor, has a role in the M/G1 transition in Saccharomyces cerevisiae. Genes Dev. 1994, 8, 1640–1653. [Google Scholar] [CrossRef] [PubMed]
- Barberis, M.; De Gioia, L.; Ruzzene, M.; Sarno, S.; Coccetti, P.; Fantucci, P.; Vanoni, M.; Alberghina, L. The yeast cyclin-dependent kinase inhibitor Sic1 and mammalian p27Kip1 are functional homologues with a structurally conserved inhibitory domain. Biochem. J. 2005, 387, 639–647. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kedde, M.; Van Kouwenhove, M.; Zwart, W.; Vrielink, J.A.O.; Elkon, R.; Agami, R. A Pumilio-induced RNA structure switch in p27–3′ UTR controls miR-221 and miR-222 accessibility. Nat. Cell Biol. 2010, 12, 1014–1020. [Google Scholar] [CrossRef]
- Van Etten, J.; Schagat, T.L.; Hrit, J.; Weidmann, C.A.; Brumbaugh, J.; Coon, J.J.; Goldstrohm, A.C. Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. J. Biol. Chem. 2012, 287, 36370–36383. [Google Scholar] [CrossRef]
- Bennin, D.A.; Don, A.S.A.; Brake, T.; McKenzie, J.L.; Rosenbaum, H.; Ortiz, L.; DePaoli-Roach, A.A.; Horne, M.C. Cyclin G2 associates with protein phosphatase 2A catalytic and regulatory B′ subunits in active complexes and induces nuclear aberrations and a G1/S phase cell cycle arrest. J. Biol. Chem. 2002, 277, 27449–27467. [Google Scholar] [CrossRef]
- Manukyan, A.; Zhang, J.; Thippeswamy, U.; Yang, J.; Zavala, N.; Mudannayake, M.P.; Asmussen, M.; Schneider, C.; Schneider, B.L. Ccr4 alters cell size in yeast by modulating the timing of CLN1 and CLN2 expression. Genetics 2008, 179, 345–357. [Google Scholar] [CrossRef]
- Levin, D.E.; Fields, F.O.; Kunisawa, R.; Bishop, J.M.; Thorner, J.A. candidate protein kinase C gene, PKC1, is required for the S. cerevisiae cell cycle. Cell 1990, 62, 213–224. [Google Scholar] [CrossRef]
- Gray, J.V.; Ogas, J.P.; Kamada, Y.; Stone, M.; Levin, D.E.; Herskowitz, I. A role for the Pkc1 MAP kinase pathway of Saccharomyces cerevisiae in bud emergence and identification of a putative upstream regulator. EMBO J. 1997, 16, 4924–4937. [Google Scholar] [CrossRef]
- Marini, N.; Meldrum, E.; Buehrer, B.; Hubberstey, A.V.; Stone, D.; Traynor-Kaplan, A.; Reed, S.I. A pathway in the yeast cell division cycle linking protein kinase C (Pkc1) to activation of Cdc28 at START. EMBO J. 1996, 15, 3040–3052. [Google Scholar] [CrossRef] [PubMed]
- Oehlen, L.; Cross, F.R. G1 cyclins CLN1 and CLN2 repress the mating factor response pathway at Start in the yeast cell cycle. Genes Dev. 1994, 8, 1058–1070. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, W.; Nie, Q.; Yi, T.-M.; Chou, C.-S. Modelling of yeast mating reveals robustness strategies for cell-cell interactions. PLoS Comput. Biol. 2016, 12, e1004988. [Google Scholar] [CrossRef]
- Trueheart, J.; Fink, G.R. The yeast cell fusion protein FUS1 is O-glycosylated and spans the plasma membrane. Proc. Natl. Acad. Sci. USA 1989, 86, 9916–9920. [Google Scholar] [CrossRef]
- Pope, P.A.; Pryciak, P.M. Functional overlap among distinct G1/S inhibitory pathways allows robust G1 arrest by yeast mating pheromones. Mol. Biol. Cell 2013, 24, 3675–3688. [Google Scholar] [CrossRef]
- Mei, Q.; Huang, J.; Chen, W.; Tang, J.; Xu, C.; Yu, Q.; Cheng, Y.; Ma, L.; Yu, X.; Li, S. Regulation of DNA replication-coupled histone gene expression. Oncotarget 2017, 8, 95005–95022. [Google Scholar] [CrossRef]
- Alabert, C.; Bukowski-Wills, J.-C.; Lee, S.-B.; Kustatscher, G.; Nakamura, K.; de Lima Alves, F.; Menard, P.; Mejlvang, J.; Rappsilber, J.; Groth, A. Chromatin dynamics during DNA replication and uncharacterized replication factors determined by nascent chromatin capture (NCC) proteomics. Nat. Cell Biol. 2014, 16, 281–293. [Google Scholar] [CrossRef]
- Raynaud, C.; Mallory, A.C.; Latrasse, D.; Jégu, T.; Bruggeman, Q.; Delarue, M.; Bergounioux, C.; Benhamed, M. Chromatin meets the cell cycle. J. Exp. Bot. 2014, 65, 2677–2689. [Google Scholar] [CrossRef] [PubMed]
- Burgess, R.J.; Zhou, H.; Han, J.; Zhang, Z. A role for Gcn5 in replication-coupled nucleosome assembly. Mol. Cell 2010, 37, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Howe, L.; Auston, D.; Grant, P.; John, S.; Cook, R.G.; Workman, J.L.; Pillus, L. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev. 2001, 15, 3144–3154. [Google Scholar] [CrossRef]
- Zhang, W.; Bone, J.R.; Edmondson, D.G.; Turner, B.M.; Roth, S.Y. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 1998, 17, 3155–3167. [Google Scholar] [CrossRef] [PubMed]
- Biswas, D.; Yu, Y.; Mitra, D.; Stillman, D.J. Genetic interactions between Nhp6 and Gcn5 with Mot1 and the Ccr4–Not complex that regulate binding of TATA-binding protein in Saccharomyces cerevisiae. Genetics 2006, 172, 837–849. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Goldstrohm, A.C.; Seay, D.J.; Hook, B.A.; Wickens, M. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J. Biol. Chem. 2007, 282, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Goldstrohm, A.C.; Hook, B.A.; Seay, D.J.; Wickens, M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 2006, 13, 533–539. [Google Scholar] [CrossRef]
- Gerber, A.P.; Herschlag, D.; Brown, P.O. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004, 2. [Google Scholar] [CrossRef]
- Huang, S.; Litt, M.; Felsenfeld, G. Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes Dev. 2005, 19, 1885–1893. [Google Scholar] [CrossRef]
- Vicencio, J.M.; Galluzzi, L.; Tajeddine, N.; Ortiz, C.; Criollo, A.; Tasdemir, E.; Morselli, E.; Younes, A.B.; Maiuri, M.C.; Lavandero, S.; et al. Senescence, apoptosis or autophagy? Gerontology 2008, 54, 92–99. [Google Scholar] [CrossRef]
- Pawlowska, E.; Szczepanska, J.; Szatkowska, M.; Blasiak, J. An interplay between senescence, apoptosis and autophagy in glioblastoma multiforme—role in pathogenesis and therapeutic perspective. Int. J. Mol. Sci. 2018, 19, 889. [Google Scholar] [CrossRef]
- Hayflick, L.; Moorhead, P.S. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961, 25, 585–621. [Google Scholar] [CrossRef]
- Hayflick, L. The limited in vitro lifetime of human diploid cell strains. Exp. Cell Res. 1965, 37, 614–636. [Google Scholar] [CrossRef]
- Grimes, A.; Chandra, S.B. Significance of cellular senescence in aging and cancer. Cancer Res. Treat. Off. J. Korean Cancer Assoc. 2009, 41, 187. [Google Scholar] [CrossRef]
- O’sullivan, R.J.; Kubicek, S.; Schreiber, S.L.; Karlseder, J. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat. Struct. Mol. Biol. 2010, 17, 1218. [Google Scholar] [CrossRef] [PubMed]
- Funayama, R.; Saito, M.; Tanobe, H.; Ishikawa, F. Loss of linker histone H1 in cellular senescence. J. Cell Biol. 2006, 175, 869–880. [Google Scholar] [CrossRef] [PubMed]
- Criscione, S.W.; Teo, Y.V.; Neretti, N. The chromatin landscape of cellular senescence. Trends Genet. 2016, 32, 751–761. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Sen, P. The senescent cell epigenome. Aging 2018, 10, 3590. [Google Scholar] [CrossRef]
- Shah, P.P.; Donahue, G.; Otte, G.L.; Capell, B.C.; Nelson, D.M.; Cao, K.; Aggarwala, V.; Cruickshanks, H.A.; Rai, T.S.; McBryan, T.; et al. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev. 2013, 27, 1787–1799. [Google Scholar] [CrossRef]
- Ivanov, A.; Pawlikowski, J.; Manoharan, I.; van Tuyn, J.; Nelson, D.M.; Rai, T.S.; Shah, P.P.; Hewitt, G.; Korolchuk, V.I.; Passos, J.F.; et al. Lysosome-mediated processing of chromatin in senescence. J. Cell Biol. 2013, 202, 129–143. [Google Scholar] [CrossRef]
- Sun, L.; Yu, R.; Dang, W. Chromatin architectural changes during cellular senescence and aging. Genes 2018, 9, 211. [Google Scholar] [CrossRef]
- Saito, N.; Araya, J.; Ito, S.; Tsubouchi, K.; Minagawa, S.; Hara, H.; Ito, A.; Nakano, T.; Hosaka, Y.; Ichikawa, A.; et al. Involvement of Lamin B1 Reduction in Accelerated Cellular Senescence during Chronic Obstructive Pulmonary Disease Pathogenesis. J. Immunol. 2019, 202, 1428–1440. [Google Scholar] [CrossRef]
- Scaffidi, P.; Misteli, T. Lamin A-dependent nuclear defects in human aging. Science 2006, 312, 1059–1063. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef]
- Yuan, Y.-Y.; Xie, K.-X.; Wang, S.-L.; Yuan, L.-W. Inflammatory caspase-related pyroptosis: Mechanism, regulation and therapeutic potential for inflammatory bowel disease. Gastroenterol. Rep. 2018, 6, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Raju, K.K.; Natarajan, S.; Kumar, N.S.; Kumar, D.A.; Raghavendra, N.M. Role of cytoplasmic deadenylation and mRNA decay factors in yeast apoptosis. FEMS Yeast Res. 2015, 15. [Google Scholar] [CrossRef] [PubMed]
- Ericsson, J. Studies on induced cellular autophagy: I. Electron microscopy of cells with in vivo labelled lysosomes. Exp. Cell Res. 1969, 55, 95–106. [Google Scholar] [CrossRef]
- De Duve, C.; Wattiaux, R. Functions of lysosomes. Annu. Rev. Physiol. 1966, 28, 435–492. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Ohsumi, Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999, 15, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Nakatogawa, H.; Suzuki, K.; Kamada, Y.; Ohsumi, Y. Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat. Rev. Mol. Cell Biol. 2009, 10, 458–467. [Google Scholar] [CrossRef]
- Hata, H.; Mitsui, H.; Liu, H.; Bai, Y.; Denis, C.L.; Shimizu, Y.; Sakai, A. Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics 1998, 148, 571–579. [Google Scholar]
- Yamaguchi, T.; Suzuki, T.; Sato, T.; Takahashi, A.; Watanabe, H.; Kadowaki, A.; Natsui, M.; Inagaki, H.; Arakawa, S.; Nakaoka, S.; et al. The CCR4-NOT deadenylase complex controls Atg7-dependent cell death and heart function. Sci. Signal. 2018, 11. [Google Scholar] [CrossRef]
- Lenssen, E.; Oberholzer, U.; Labarre, J.; De Virgilio, C.; Collart, M. Saccharomyces cerevisiae Ccr4–Not complex contributes to the control of Msn2p-dependent transcription by the Ras/cAMP pathway. Mol. Microbiol 2002, 43, 1023–1037. [Google Scholar] [CrossRef][Green Version]
- Vlahakis, A.; Lopez Muniozguren, N.; Powers, T. Stress-response transcription factors Msn2 and Msn4 couple TORC2-Ypk1 signaling and mitochondrial respiration to ATG8 gene expression and autophagy. Autophagy 2017, 13, 1804–1812. [Google Scholar] [CrossRef] [PubMed]
- Jeong, K.; Kwon, H.Y.; Jeong, M.S.; Sohn, E.J.; Kim, S.-H. CNOT2 promotes degradation of p62/SQSTM1 as a negative regulator in ATG5 dependent autophagy. Oncotarget 2017, 8, 46034. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xu, X.; Zhi, T.; Chao, H.; Jiang, K.; Liu, Y.; Bao, Z.; Fan, L.; Wang, D.; Li, Z.; Liu, N.; et al. ERK1/2/mTOR/Stat3 pathway-mediated autophagy alleviates traumatic brain injury-induced acute lung injury. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 1663–1674. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Zhang, W.; Liu, S.; Wu, W.; Qin, M.; Huang, J. Activation of the SphK1/ERK/p-ERK pathway promotes autophagy in colon cancer cells. Oncol. Lett. 2018, 15, 9719–9724. [Google Scholar] [CrossRef]
- Martinez-Lopez, N.; Athonvarangkul, D.; Mishall, P.; Sahu, S.; Singh, R. Autophagy proteins regulate ERK phosphorylation. Nat. Commun. 2013, 4, 1–14. [Google Scholar] [CrossRef]
- Will, M.; Qin, A.C.R.; Toy, W.; Yao, Z.; Rodrik-Outmezguine, V.; Schneider, C.; Huang, X.; Monian, P.; Jiang, X.; de Stanchina, E.; et al. Rapid induction of apoptosis by PI3K inhibitors is dependent upon their transient inhibition of RAS–ERK signaling. Cancer Discov. 2014, 4, 334–347. [Google Scholar] [CrossRef]
- Guo, Y.J.; Pan, W.W.; Liu, S.B.; Shen, Z.F.; Xu, Y.; Hu, L.L. ERK/MAPK signalling pathway and tumorigenesis. Exp. Therapeut. Med. 2020, 19, 1997–2007. [Google Scholar] [CrossRef]
- Bermudez, O.; Jouandin, P.; Rottier, J.; Bourcier, C.; Pagès, G.; Gimond, C. Post-transcriptional regulation of the DUSP6/MKP-3 phosphatase by MEK/ERK signaling and hypoxia. J. Cell. Physiol. 2011, 226, 276–284. [Google Scholar] [CrossRef]
- Galgano, A.; Forrer, M.; Jaskiewicz, L.; Kanitz, A.; Zavolan, M.; Gerber, A.P. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS ONE 2008, 3, e3164. [Google Scholar] [CrossRef]
- Jafarnejad, S.M.; Chapat, C.; Matta-Camacho, E.; Gelbart, I.A.; Hesketh, G.G.; Arguello, M.; Garzia, A.; Kim, S.; Attig, J.; Shapiro, M. Translational control of ERK signaling through miRNA/4EHP-directed silencing. eLife 2018, 7, e35034. [Google Scholar] [CrossRef]
- Cook, K.L.; Shajahan, A.N.; Clarke, R. Autophagy and endocrine resistance in breast cancer. Expert Rev. Anticancer Ther. 2011, 11, 1283–1294. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.; Liu, S.Y.; Van Hasselt, C.A.; Vlantis, A.C.; Ng, E.K.; Zhang, H.; Dong, Y.; Ng, S.K.; Chu, R.; Chan, A.B.W.; et al. Estrogen receptor α induces prosurvival autophagy in papillary thyroid cancer via stimulating reactive oxygen species and extracellular signal regulated kinases. J. Clin. Endocrinol. Metab. 2015, 100, E561–E571. [Google Scholar] [CrossRef] [PubMed]
- Felzen, V.; Hiebel, C.; Koziollek-Drechsler, I.; Reißig, S.; Wolfrum, U.; Kögel, D.; Brandts, C.; Behl, C.; Morawe, T. Estrogen receptor α regulates non-canonical autophagy that provides stress resistance to neuroblastoma and breast cancer cells and involves BAG3 function. Cell Death Dis. 2015, 6, e1812. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhang, L.; Zhang, K.; Zhou, B.; Kuo, M.-L.; Hu, S.; Chen, L.; Tang, M.; Chen, Y.; Yang, L.; et al. Reciprocal regulation of autophagy and dNTP pools in human cancer cells. Autophagy 2014, 10, 1272–1284. [Google Scholar] [CrossRef]
- Bellot, G.; Garcia-Medina, R.; Gounon, P.; Chiche, J.; Roux, D.; Pouysségur, J.; Mazure, N.M. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol. Cell. Biol. 2009, 29, 2570–2581. [Google Scholar] [CrossRef]
- Tan, Q.; Wang, M.; Yu, M.; Zhang, J.; Bristow, R.G.; Hill, R.P.; Tannock, I.F. Role of autophagy as a survival mechanism for hypoxic cells in tumors. Neoplasia 2016, 18, 347–355. [Google Scholar] [CrossRef]
- Ciccia, A.; Elledge, S.J. The DNA damage response: Making it safe to play with knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef]
- Deshpande, G.P.; Hayles, J.; Hoe, K.-L.; Kim, D.-U.; Park, H.-O.; Hartsuiker, E. Screening a genome-wide S. pombe deletion library identifies novel genes and pathways involved in genome stability maintenance. DNA Repair 2009, 8, 672–679. [Google Scholar] [CrossRef]
- Westmoreland, T.J.; Marks, J.R.; Olson, J.A.; Thompson, E.M.; Resnick, M.A.; Bennett, C.B. Cell cycle progression in G1 and S phases is CCR4 dependent following ionizing radiation or replication stress in Saccharomyces cerevisiae. Eukaryot. Cell 2004, 3, 430–446. [Google Scholar] [CrossRef]
- Abreu, C.M.; Kumar, R.; Hamilton, D.; Dawdy, A.W.; Creavin, K.; Eivers, S.; Finn, K.; Balsbaugh, J.L.; O’Connor, R.; Kiely, P.A.; et al. Site-specific phosphorylation of the DNA damage response mediator rad9 by cyclin-dependent kinases regulates activation of checkpoint kinase 1. PLoS Genet. 2013, 9, e1003310. [Google Scholar] [CrossRef]
- Sanchez, Y.; Bachant, J.; Wang, H.; Hu, F.; Liu, D.; Tetzlaff, M. Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science 1999, 286, 1166–1171. [Google Scholar] [CrossRef] [PubMed]
- Woolstencroft, R.N.; Beilharz, T.H.; Cook, M.A.; Preiss, T.; Durocher, D.; Tyers, M. Ccr4 contributes to tolerance of replication stress through control of CRT1 mRNA poly (A) tail length. J. Cell Sci. 2006, 119, 5178–5192. [Google Scholar] [CrossRef]
- Sanchez-Perez, I.; Manguan-Garcia, C.; Menacho-Marquez, M.; Murguía, J.R.; Perona, R. hCCR4/cNOT6 targets DNA-damage response proteins. Cancer Lett. 2009, 273, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Zhou, Z.; Elledge, S.J. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 1998, 94, 595–605. [Google Scholar] [CrossRef]
- Basrai, M.A.; Velculescu, V.E.; Kinzler, K.W.; Hieter, P. NORF5/HUG1 is a component of theMEC1-mediated checkpoint response to DNA damage and replication arrest in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999, 19, 7041–7049. [Google Scholar] [CrossRef]
- Takahashi, S.; Kontani, K.; Araki, Y.; Katada, T. Caf1 regulates translocation of ribonucleotide reductase by releasing nucleoplasmic Spd1–Suc22 assembly. Nucleic Acids Res. 2007, 35, 1187–1197. [Google Scholar] [CrossRef]
- Niida, H.; Shimada, M.; Murakami, H.; Nakanishi, M. Mechanisms of dNTP supply that play an essential role in maintaining genome integrity in eukaryotic cells. Cancer Sci. 2010, 101, 2505–2509. [Google Scholar] [CrossRef]
- Xie, M.; Yen, Y.; Owonikoko, T.K.; Ramalingam, S.S.; Khuri, F.R.; Curran, W.J.; Doetsch, P.W.; Deng, X. Bcl2 induces DNA replication stress by inhibiting ribonucleotide reductase. Cancer Res. 2014, 74, 212–223. [Google Scholar] [CrossRef]
- Lau, N.-C.; Mulder, K.W.; Brenkman, A.B.; Mohammed, S.; van den Broek, N.J.; Heck, A.J.; Timmers, H.M. Phosphorylation of Not4p functions parallel to BUR2 to regulate resistance to cellular stresses in Saccharomyces cerevisiae. PLoS ONE 2010, 5, e9864. [Google Scholar] [CrossRef]
- Zou, Z.; Tao, T.; Li, H.; Zhu, X. mTOR signaling pathway and mTOR inhibitors in cancer: Progress and challenges. Cell Biosci. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Weiler, M.; Blaes, J.; Pusch, S.; Sahm, F.; Czabanka, M.; Luger, S.; Bunse, L.; Solecki, G.; Eichwald, V.; Jugold, M.; et al. mTOR target NDRG1 confers MGMT-dependent resistance to alkylating chemotherapy. Proc. Natl. Acad. Sci. USA 2014, 111, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Dominick, G.; Bowman, J.; Li, X.; Miller, R.A.; Garcia, G.G. mTOR regulates the expression of DNA damage response enzymes in long-lived Snell dwarf, GHRKO, and PAPPA-KO mice. Aging Cell 2017, 16, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Boylston, W.; DeFord, J.H.; Papaconstantinou, J. Identification of longevity-associated genes in long-lived Snell and Ames dwarf mice. Age 2006, 28, 125. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Velimezi, G.; Robinson-Garcia, L.; Muñoz-Martínez, F.; Wiegant, W.W.; da Silva, J.F.; Owusu, M.; Moder, M.; Wiedner, M.; Rosenthal, S.B.; Fisch, K.M.; et al. Map of synthetic rescue interactions for the Fanconi anemia DNA repair pathway identifies USP48. Nat. Commun. 2018, 9, 2280. [Google Scholar] [CrossRef]
- Wu, H.I.; Brown, J.A.; Dorie, M.J.; Lazzeroni, L.; Brown, J.M. Genome-wide identification of genes conferring resistance to the anticancer agents cisplatin, oxaliplatin, and mitomycin C. Cancer Res. 2004, 64, 3940–3948. [Google Scholar] [CrossRef] [PubMed]
- Wook Choi, D.; Yong Choi, C. HIPK2 modification code for cell death and survival. Mol. Cell. Oncol. 2014, 1, e955999. [Google Scholar] [CrossRef]
- Moriya, H.; Shimizu-Yoshida, Y.; Omori, A.; Iwashita, S.; Katoh, M.; Sakai, A. Yak1p, a DYRK family kinase, translocates to the nucleus and phosphorylates yeast Pop2p in response to a glucose signal. Genes Dev. 2001, 15, 1217–1228. [Google Scholar] [CrossRef]
- Sellam, A.; van het Hoog, M.; Tebbji, F.; Beaurepaire, C.; Whiteway, M.; Nantel, A. Modeling the transcriptional regulatory network that controls the early hypoxic response in Candida albicans. Eukaryot. Cell 2014, 13, 675–690. [Google Scholar] [CrossRef]
- Dagley, M.J.; Gentle, I.E.; Beilharz, T.H.; Pettolino, F.A.; Djordjevic, J.T.; Lo, T.L.; Uwamahoro, N.; Rupasinghe, T.; Tull, D.L.; McConville, M.; et al. Cell wall integrity is linked to mitochondria and phospholipid homeostasis in Candida albicans through the activity of the post-transcriptional regulator Ccr4-Pop2. Mol. Microbiol 2011, 79, 968–989. [Google Scholar] [CrossRef]
- Vergara, R.; Parada, F.; Rubio, S.; Pérez, F.J. Hypoxia induces H2O2 production and activates antioxidant defence system in grapevine buds through mediation of H2O2 and ethylene. J. Exp. Bot. 2012, 63, 4123–4131. [Google Scholar] [CrossRef]
- Lenssen, E.; Azzouz, N.; Michel, A.; Landrieux, E.; Collart, M.A. The Ccr4-not complex regulates Skn7 through Srb10 kinase. Eukaryot. Cell 2007, 6, 2251–2259. [Google Scholar] [CrossRef]
- Zhou, X.; Sun, H.; Chen, H.; Zavadil, J.; Kluz, T.; Arita, A.; Max, C. Hypoxia induces trimethylated H3 lysine 4 by inhibition of JARID1A demethylase. Cancer Res. 2010, 70, 4214–4221. [Google Scholar] [CrossRef] [PubMed]
- Gasch, A.P.; Spellman, P.T.; Kao, C.M.; Carmel-Harel, O.; Eisen, M.B.; Storz, G.; David, B.; Patrick, O.B.; Pamela, A.S. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 2000, 11, 4241–4257. [Google Scholar] [CrossRef] [PubMed]
- Carmel-Harel, O.; Stearman, R.; Gasch, A.P.; Botstein, D.; Brown, P.O.; Storz, G. Role of thioredoxin reductase in the Yap1p-dependent response to oxidative stress in Saccharomyces cerevisiae. Mol. Microbiol 2001, 39, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Gulshan, K.; Thommandru, B.; Moye-Rowley, W.S. Proteolytic degradation of the Yap1 transcription factor is regulated by subcellular localization and the E3 ubiquitin ligase Not4. J. Biol. Chem. 2012, 287, 26796–26805. [Google Scholar] [CrossRef]
- Amer, Y.O.; Hebert-Chatelain, E. Mitochondrial cAMP-PKA signaling: What do we really know? Biochim. Biophys. Acta Bioenerg. 2018, 1859, 868–877. [Google Scholar] [CrossRef]
- Simko, V.; Iuliano, F.; Sevcikova, A.; Labudova, M.; Barathova, M.; Radvak, P.; Pastorekova, S.; Pastorek, J.; Csaderova, L. Hypoxia induces cancer-associated cAMP/PKA signalling through HIF-mediated transcriptional control of adenylyl cyclases VI and VII. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Varela, J.; Praekelt, U.M.; Meacock, P.A.; Planta, R.J.; Mager, W.H. The Saccharomyces cerevisiae HSP12 gene is activated by the high-osmolarity glycerol pathway and negatively regulated by protein kinase A. Mol. Cell. Biol. 1995, 15, 6232–6245. [Google Scholar] [CrossRef]
- Chen, J.; Chiang, Y.C.; Denis, C.L. CCR4, a 3′–5′ poly (A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 2002, 21, 1414–1426. [Google Scholar] [CrossRef]
- Li, X.; Jiang, Y.; Meisenhelder, J.; Yang, W.; Hawke, D.H.; Zheng, Y.; Xia, Y.; Aldape, K.; He, J.; Hunter, T.; et al. Mitochondria-translocated PGK1 functions as a protein kinase to coordinate glycolysis and the TCA cycle in tumorigenesis. Mol. Cell 2016, 61, 705–719. [Google Scholar] [CrossRef]
- Yang, W.; Lu, Z. Regulation and function of pyruvate kinase M2 in cancer. Cancer Lett. 2013, 339, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Sakai, A.; Chibazakura, T.; Shimizu, Y.; Hishinuma, F. Molecular analysis of POP2 gene, a gene required for glucose-derepression of gene expression in Saccharomyces cerevisiae. Nucleic Acids Res. 1992, 20, 6227–6233. [Google Scholar] [CrossRef][Green Version]
- Zou, L.-H.; Shang, Z.-F.; Tan, W.; Liu, X.-D.; Xu, Q.-Z.; Song, M.; Wang, Y.; Guan, H.; Zhang, S.; Yu, L.; et al. TNKS1BP1 functions in DNA double-strand break repair though facilitating DNA-PKcs autophosphorylation dependent on PARP-1. Oncotarget 2015, 6, 7011. [Google Scholar] [CrossRef]
- Matsuoka, S.; Ballif, B.A.; Smogorzewska, A.; McDonald, E.R.; Hurov, K.E.; Luo, J.; Bakalarski, C.E.; Zhao, Z.; Solimini, N.; Lerenthal, Y.; et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007, 316, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Camino, A.; Lopez-Lopez, E.; Martin-Guerrero, I.; Piñan, M.A.; Garcia-Miguel, P.; Sanchez-Toledo, J.; Ana, C.B.; Javier, U.; Aurora, N.; Africa, G.-O.; et al. Noncoding RNA–related polymorphisms in pediatric acute lymphoblastic leukemia susceptibility. Pediatr. Res. 2014, 75, 767–773. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ito, K.; Takahashi, A.; Wang, G.; Suzuki, T.; Nakazawa, T.; Yamamoto, T.; Yokoyama, K. Distinct expression patterns of the subunits of the CCR4–NOT deadenylase complex during neural development. Biochem. Biophys. Res. Commun. 2011, 411, 360–364. [Google Scholar] [CrossRef]
- Kruszka, P.; Berger, S.I.; Weiss, K.; Everson, J.L.; Martinez, A.F.; Hong, S.; Anyane-Yeboa, K.; Lipinski, R.J.; Muenke, M. A CCR4-NOT transcription complex, subunit 1, CNOT1, variant associated with Holoprosencephaly. Am. J. Hum. Genet. 2019, 104, 990–993. [Google Scholar] [CrossRef]
- De Franco, E.; Watson, R.A.; Weninger, W.J.; Wong, C.C.; Flanagan, S.E.; Caswell, R.; Green, A.; Tudor, C.; Lelliott, C.J.; Geyer, S.H.; et al. A specific CNOT1 mutation results in a novel syndrome of pancreatic agenesis and holoprosencephaly through impaired pancreatic and neurological development. Am. J. Hum. Genet. 2019, 104, 985–989. [Google Scholar] [CrossRef]
- Vissers, L.E.; Kalvakuri, S.; de Boer, E.; Geuer, S.; Oud, M.; van Outersterp, I.; Kwint, M.; Witmond, M.; Kersten, S.; Polla, D.L.; et al. De Novo Variants in CNOT1, a Central Component of the CCR4-NOT Complex Involved in Gene Expression and RNA and Protein Stability, Cause Neurodevelopmental Delay. Am. J. Hum. Genet. 2020, 107, 164–172. [Google Scholar] [CrossRef]
- Arking, D.E.; Pulit, S.L.; Crotti, L.; Van Der Harst, P.; Munroe, P.B.; Koopmann, T.T.; Sotoodehnia, N.; Rossin, E.J.; Morley, M.; Wang, X.; et al. Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat. Genet. 2014, 46, 826–836. [Google Scholar] [CrossRef]
- Elmén, L.; Volpato, C.B.; Kervadec, A.; Pineda, S.; Kalvakuri, S.; Alayari, N.N.; Foco, L.; Pramstaller, P.P.; Ocorr, K.; Rossini, A.; et al. Silencing of CCR4-NOT complex subunits affect heart structure and function. Dis. Models Mech. 2020, 13. [Google Scholar] [CrossRef]
- Díaz-Peña, R.; Aransay, A.M.; Suárez-Álvarez, B.; Bruges-Armas, J.; Rodríguez-Ezpeleta, N.; Regueiro, M.; Pimentel-Santos, F.M.; Mulero, J.; Sánchez, A.; Collantes, E.; et al. A high density SNP genotyping approach within the 19q13 chromosome region identifies an association of a CNOT3 polymorphism with ankylosing spondylitis. Ann. Rheum. Dis. 2012, 71, 714–717. [Google Scholar] [CrossRef] [PubMed]
- De Keersmaecker, K.; Atak, Z.K.; Li, N.; Vicente, C.; Patchett, S.; Girardi, T.; Gianfelici, V.; Geerdens, E.; Clappier, E.; Porcu, M.; et al. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat. Genet. 2013, 45, 186–190. [Google Scholar] [CrossRef] [PubMed]
- La Starza, R.; Barba, G.; Demeyer, S.; Pierini, V.; Di Giacomo, D.; Gianfelici, V.; Schwab, C.; Matteucci, C.; Vicente, C.; Cools, J.; et al. Deletions of the long arm of chromosome 5 define subgroups of T-cell acute lymphoblastic leukemia. Haematologica 2016, 101, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Tay, Y.; Kats, L.; Salmena, L.; Weiss, D.; Tan, S.M.; Ala, U.; Karreth, F.; Poliseno, L.; Provero, P.; di Cunto, F.; et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 2011, 147, 344–357. [Google Scholar] [CrossRef] [PubMed]
- Maragozidis, P.; Karangeli, M.; Labrou, M.; Dimoulou, G.; Papaspyrou, K.; Salataj, E.; Pournaras, S.; Matsouka, P.; Gourgoulianis, K.I.; Balatsos, N.A.A. Alterations of deadenylase expression in acute leukemias: Evidence for poly (a)-specific ribonuclease as a potential biomarker. Acta Haematol. 2012, 128, 39–46. [Google Scholar] [CrossRef]
- Zhou, F.; Wang, Y.; Liu, H.; Ready, N.; Han, Y.; Hung, R.J.; Brhane, Y.; McLaughlin, J.; Brennan, P.; Bickeböller, H.; et al. Susceptibility loci of CNOT6 in the general mRNA degradation pathway and lung cancer risk—A re-analysis of eight GWASs. Mol. Carcinog. 2017, 56, 1227–1238. [Google Scholar] [CrossRef]
- Seiden-Long, I.; Brown, K.; Shih, W.; Wigle, D.A.; Radulovich, N.; Jurisica, I.; Tsao, M. Transcriptional targets of hepatocyte growth factor signaling and Ki-ras oncogene activation in colorectal cancer. Oncogene 2006, 25, 91–102. [Google Scholar] [CrossRef]
- Li, X.; Morita, M.; Kikuguchi, C.; Takahashi, A.; Suzuki, T.; Yamamoto, T. Adipocyte-specific disruption of mouse Cnot3 causes lipodystrophy. FEBS Lett. 2017, 591, 358–368. [Google Scholar] [CrossRef]
- Mostafa, D.; Yanagiya, A.; Georgiadou, E.; Wu, Y.; Stylianides, T.; Rutter, G.A.; Suzuki, T.; Yamamoto, T. Loss of β-cell identity and diabetic phenotype in mice caused by disruption of CNOT3-dependent mRNA deadenylation. Commun. Biol. 2020, 3, 1–16. [Google Scholar] [CrossRef]
- Morita, M.; Oike, Y.; Nagashima, T.; Kadomatsu, T.; Tabata, M.; Suzuki, T.; Nakamura, T.; Yoshida, N.; Okada, M.; Yamamoto, T. Obesity resistance and increased hepatic expression of catabolism-related mRNAs in Cnot3+/− mice. EMBO J. 2011, 30, 4678–4691. [Google Scholar] [CrossRef] [PubMed]
- Morita, M.; Siddiqui, N.; Katsumura, S.; Rouya, C.; Larsson, O.; Nagashima, T.; Hekmatnejad, B.; Takahashi, A.; Kiyonari, H.; Zang, M.; et al. Hepatic posttranscriptional network comprised of CCR4–NOT deadenylase and FGF21 maintains systemic metabolic homeostasis. Proc. Natl. Acad. Sci. USA 2019, 116, 7973–7981. [Google Scholar] [CrossRef]
- Takahashi, A.; Suzuki, T.; Soeda, S.; Takaoka, S.; Kobori, S.; Yamaguchi, T.; Mohamed, H.M.A.; Yanagiya, A.; Abe, T.; Shigeta, M.; et al. The CCR4–NOT complex maintains liver homeostasis through mRNA deadenylation. Life Sci. Alliance 2020, 3. [Google Scholar] [CrossRef]
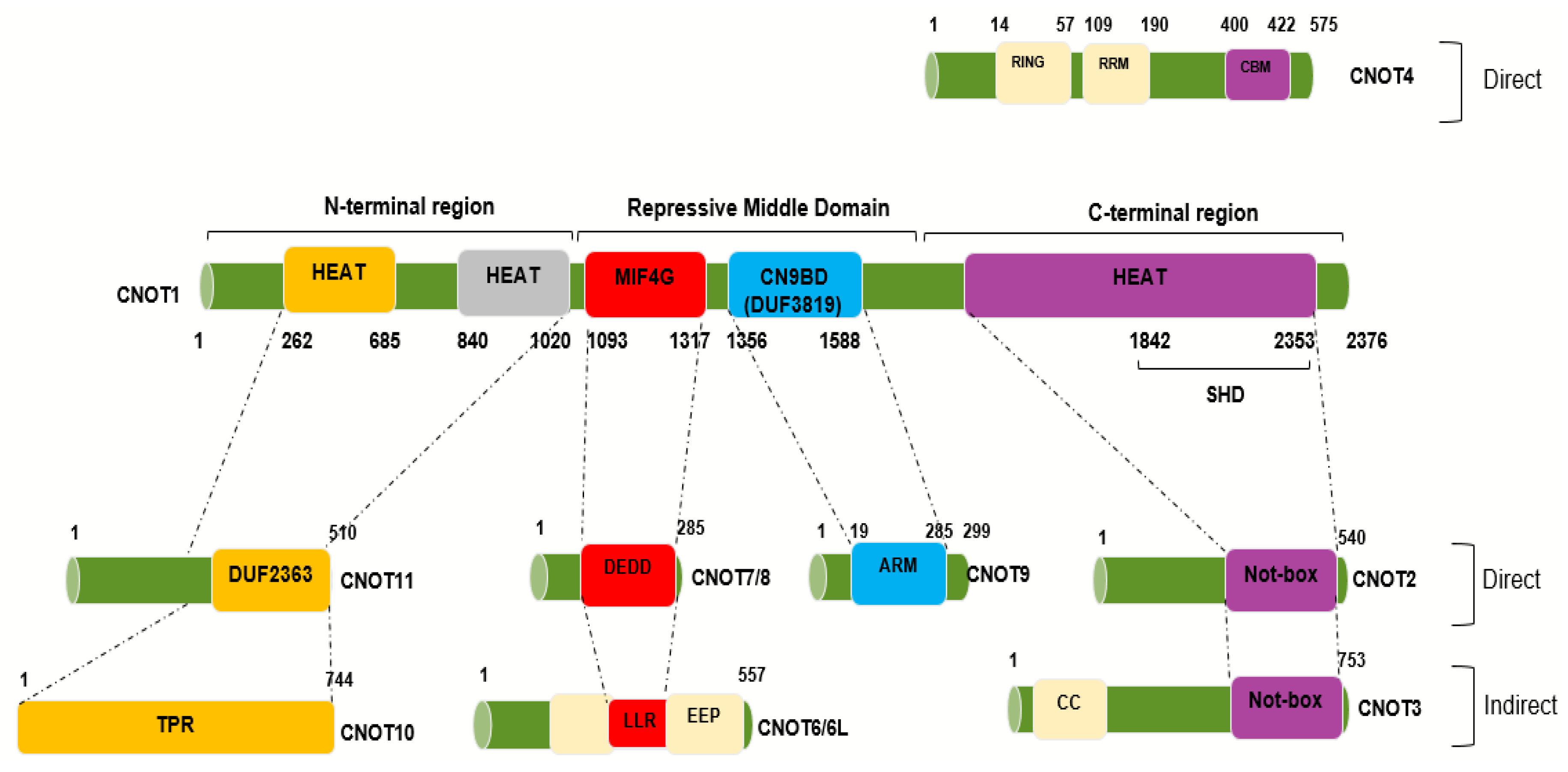
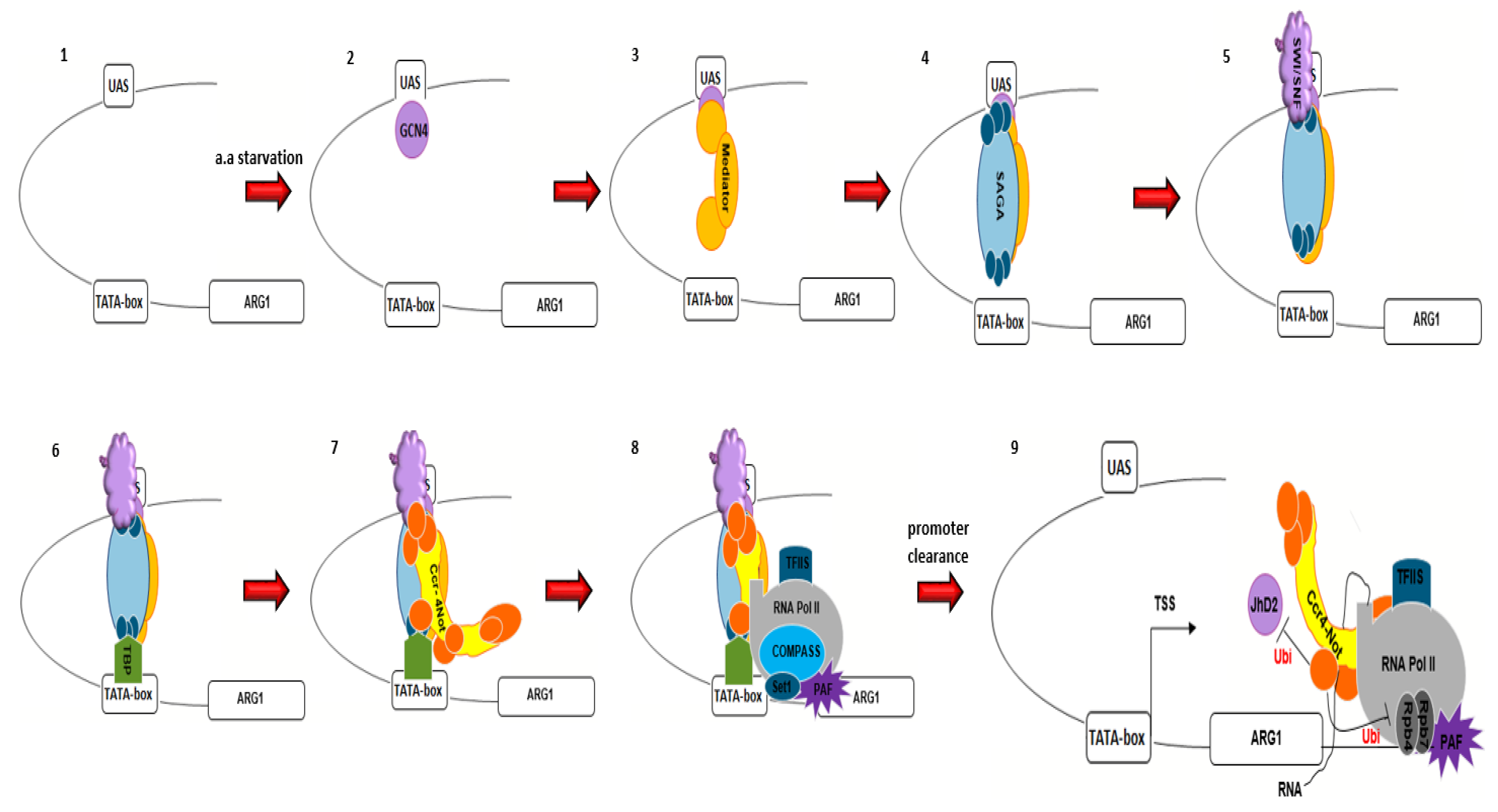
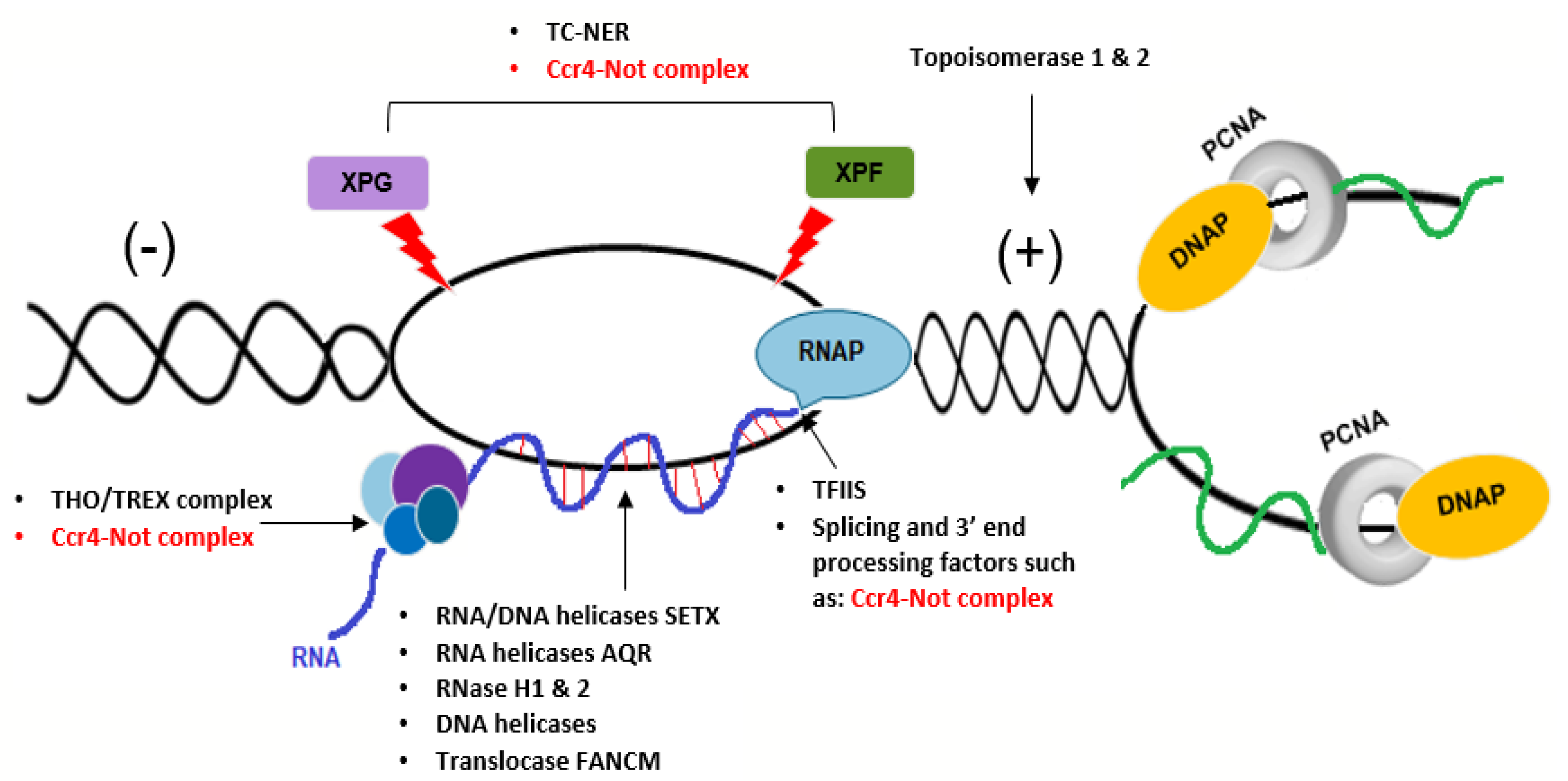
| Saccharomyces Cerevisiae | Homo Sapiens | Drosophila Melanogaster | Function |
|---|---|---|---|
| NOT1/CDC39 | CNOT1 | NOT1 | Scaffold |
| NOT2/CDC36 | CNOT2 | ReginaNOT2 | Unknown but contributes to stabilization of the complex and RNA substrate recruitment |
| NOT3 | Not present | Not present | Unknown but contributes to stabilization of the complex and RNA substrate recruitment |
| NOT5 | CNOT3 | NOT3 | Interaction with ribosomes |
| NOT4/MOT2/SIG1 | CNOT4 | NOT4 | Ubiquitin E3-ligase activity |
| CCR4 | CNOT6, CNOT6L | Twin, CCR4 | Deadenylase |
| CAF1 | CNOT7, CNOT8 | P0P2, CAF1 | Deadenylase |
| CAF40 | CNOT9 (RQCD1) | RCD1 | Transcriptional cofactor |
| Not present | CNOT10 | NOT10 | Unknown but contributes to stabilization of the complex and RNA substrate recruitment |
| Not present | CNOT11 (C2orf29) | NOT11 | Unknown but contributes to stabilization of the complex and RNA substrate recruitment |
| CAF130 | Not present | Not present | Unknown |
| Not present | TNKS1BP1 (Tab182) | Not present | Multifunctional |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chalabi Hagkarim, N.; Grand, R.J. The Regulatory Properties of the Ccr4–Not Complex. Cells 2020, 9, 2379. https://doi.org/10.3390/cells9112379
Chalabi Hagkarim N, Grand RJ. The Regulatory Properties of the Ccr4–Not Complex. Cells. 2020; 9(11):2379. https://doi.org/10.3390/cells9112379
Chicago/Turabian StyleChalabi Hagkarim, Nafiseh, and Roger J. Grand. 2020. "The Regulatory Properties of the Ccr4–Not Complex" Cells 9, no. 11: 2379. https://doi.org/10.3390/cells9112379
APA StyleChalabi Hagkarim, N., & Grand, R. J. (2020). The Regulatory Properties of the Ccr4–Not Complex. Cells, 9(11), 2379. https://doi.org/10.3390/cells9112379




