Myostatin-1 Inhibits Cell Proliferation by Inhibiting the mTOR Signal Pathway and MRFs, and Activating the Ubiquitin-Proteasomal System in Skeletal Muscle Cells of Japanese Flounder Paralichthys olivaceus
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Animals
2.3. Primary Cell Cultures
2.4. mRNA-Expression of Muscle-Specific Proteins and Gene Expression
2.5. In Vitro Screening of siRNAs and mstn-1 Interfering
2.6. Overexpression of mstn-1 in Japanese Flounder Muscle Cells
2.6.1. Plasmid Construction
2.6.2. Overexpression of mstn-1
2.7. Proliferation Assay
2.8. Western Blot Analysis
2.9. Statistical Analysis
3. Results
3.1. Culture and Proliferation of Muscle Cells In Vitro
3.2. Expression of mRNA of Muscle-Specific Proteins
3.3. Knockdown of mstn-1 Expression by mstn-1-siRNA in Primary Muscle Cells
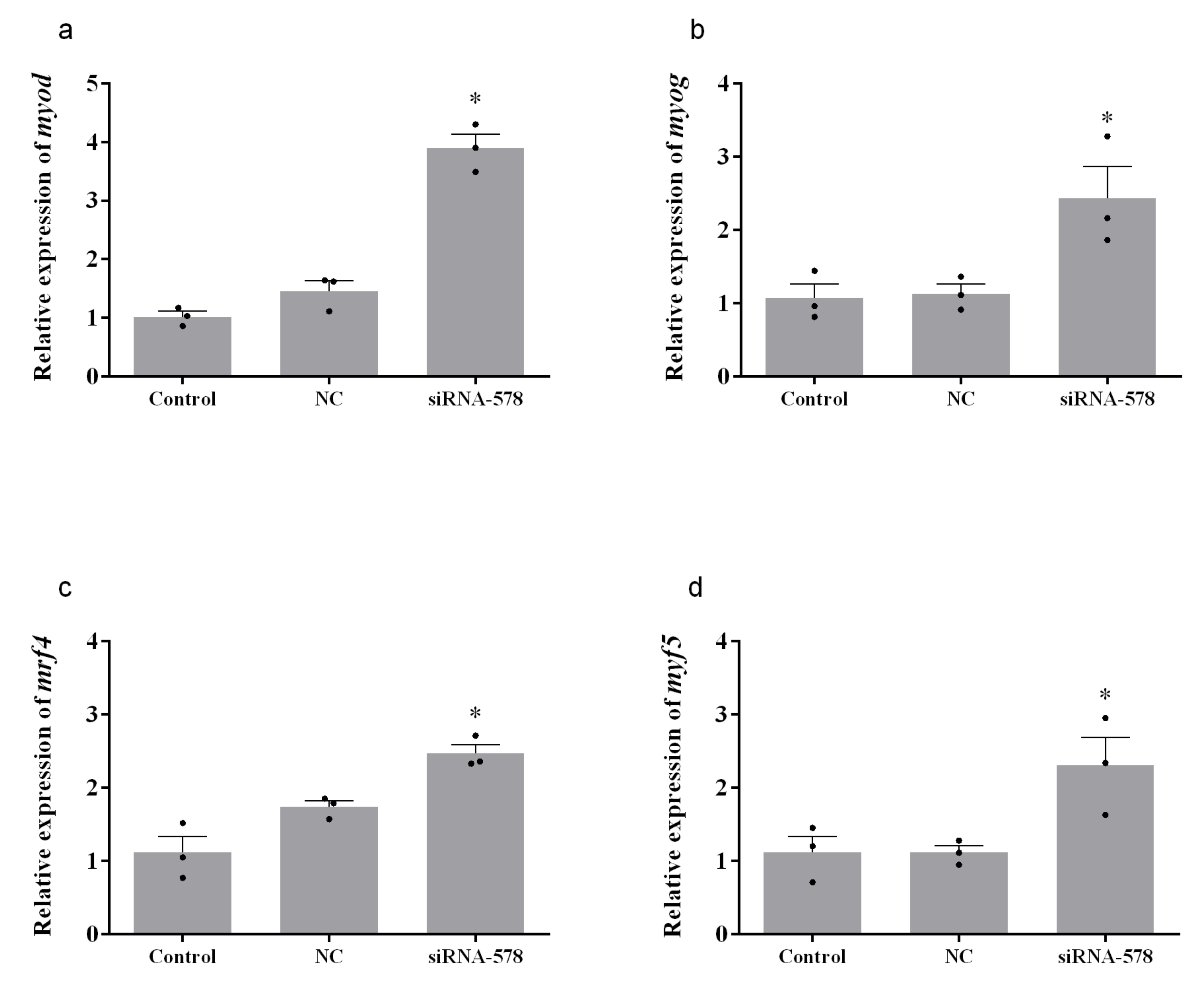
3.4. Relative Expression of Muscle Growth-Related Genes after Knockdown of mstn-1 in Muscle Cells of Japanese Flounder
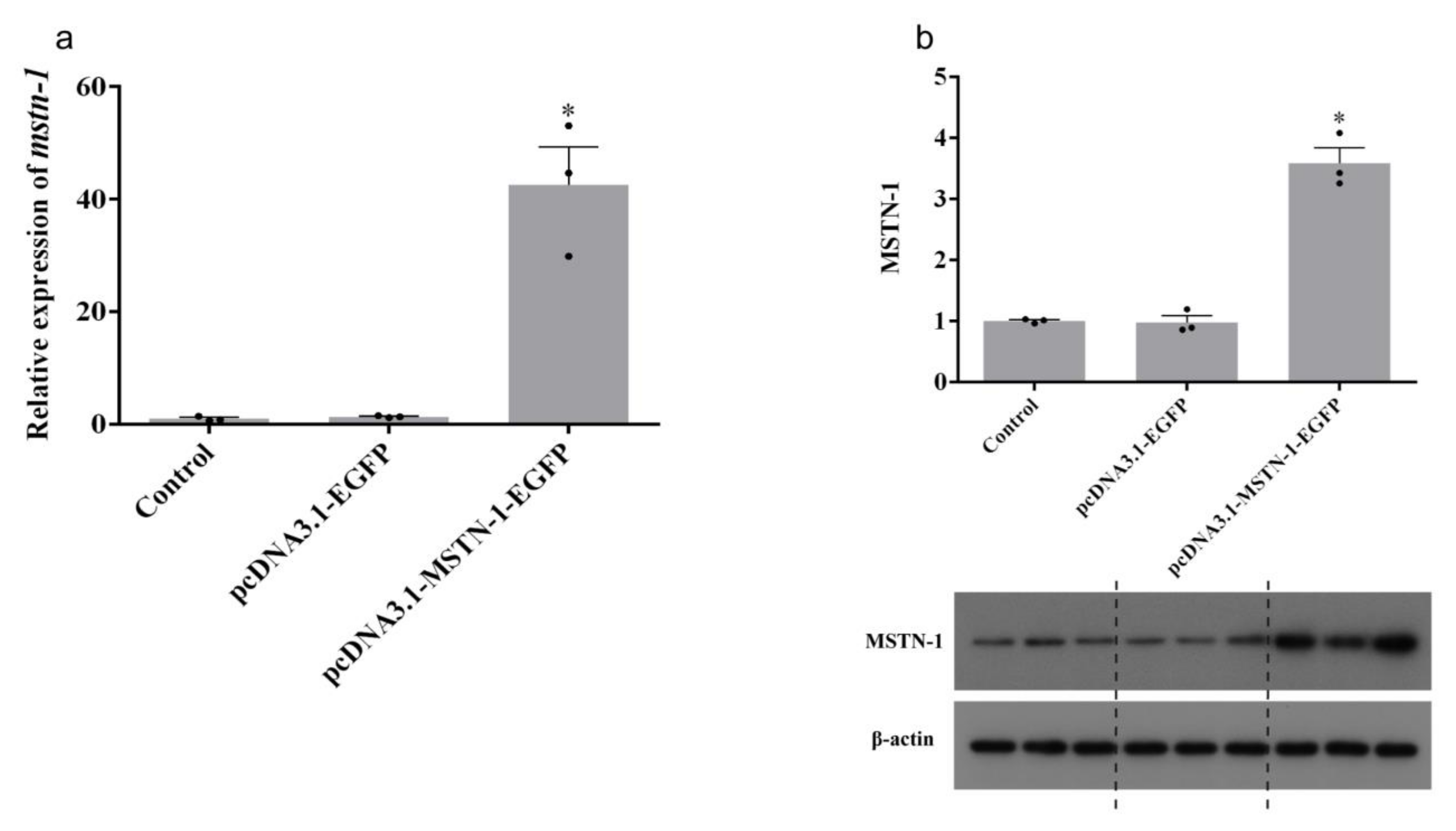
3.5. Transfection Efficiency, mstn-1 mRNA, and Protein Expression Analysis after mstn-1 Overexpression
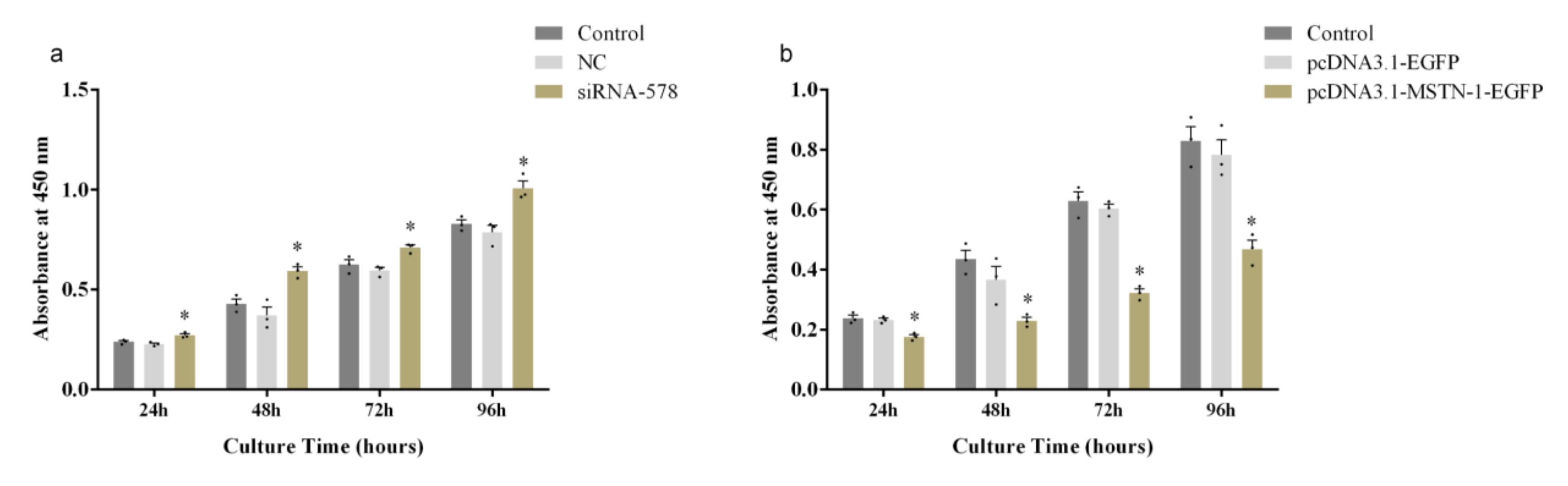
3.6. Altered Proliferation of Primary Muscle Cells by mstn-1 Knockdown and Overexpression
3.7. Relative Expression of Muscle Growth and Proteolysis-Related Genes after Overexpression of mstn-1 in Muscle Cells of Japanese Flounder
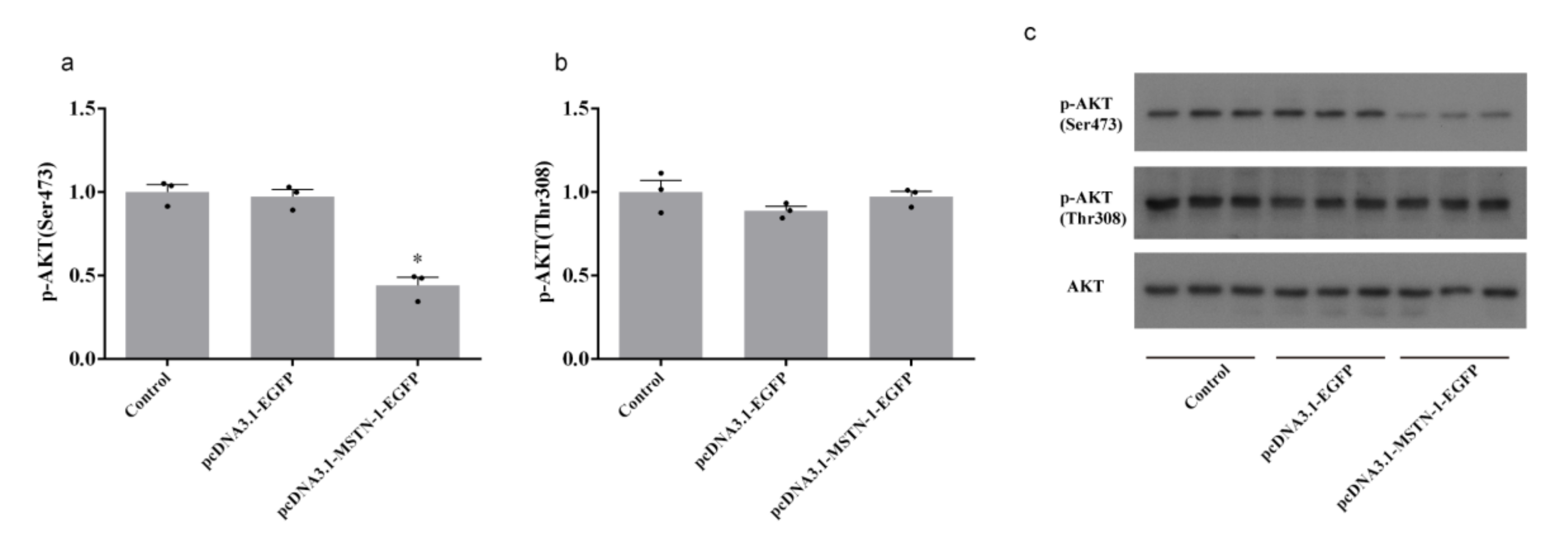
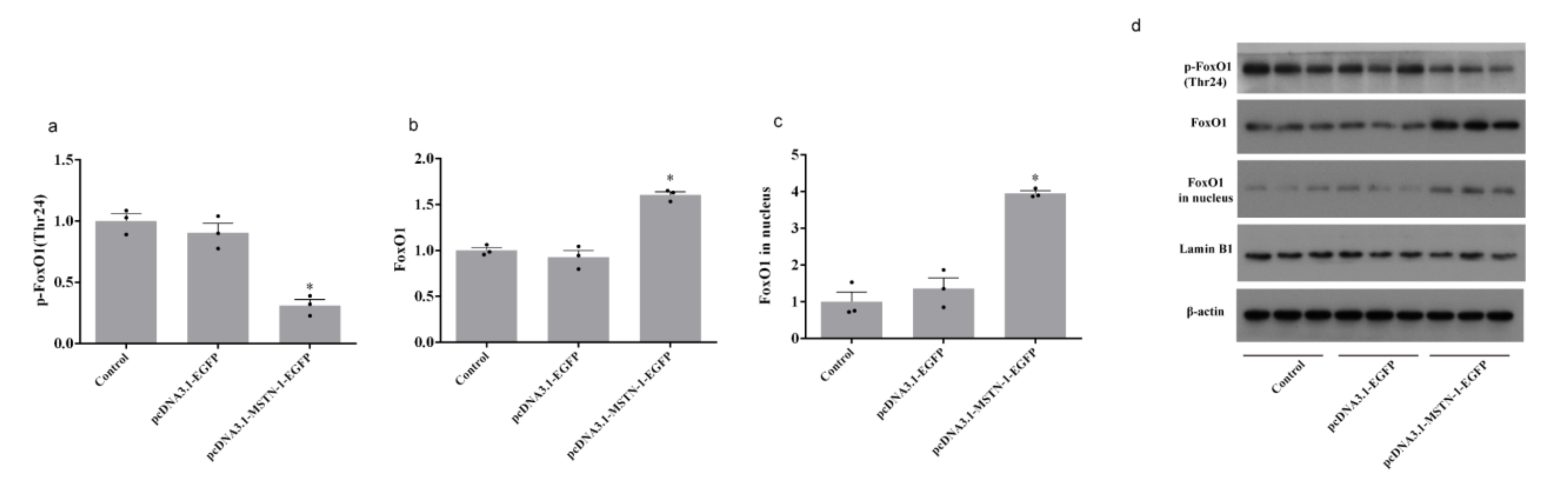
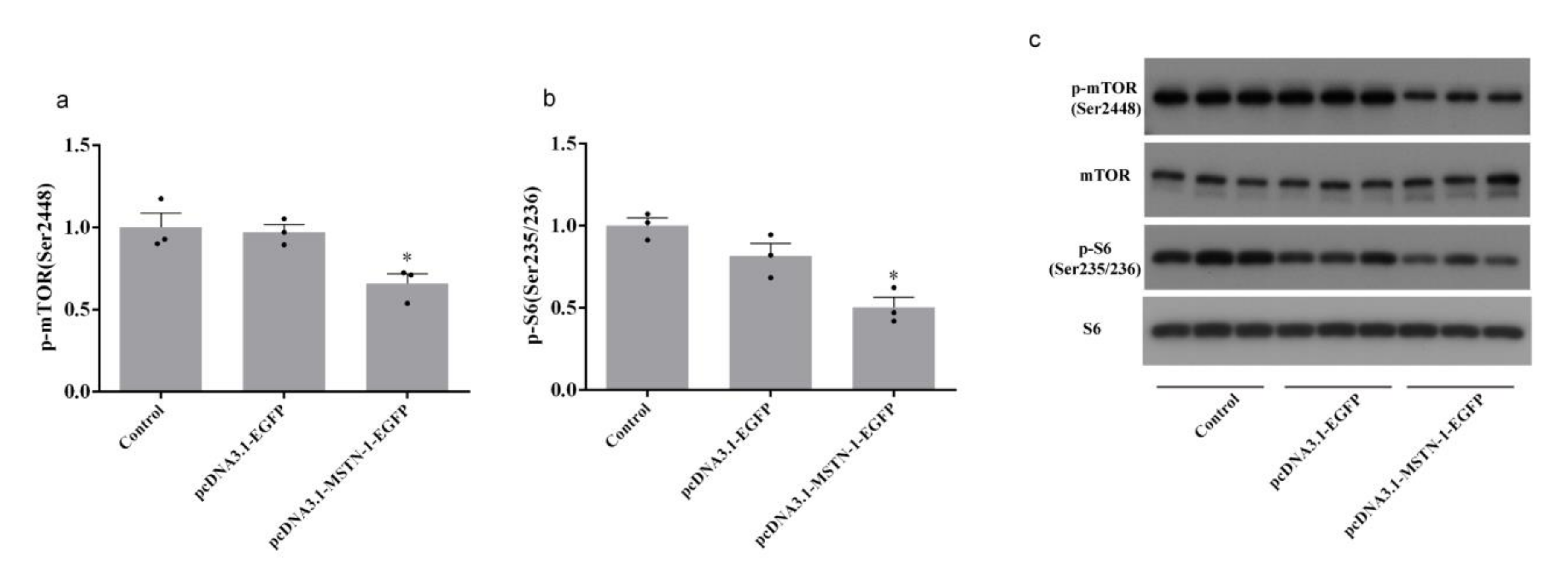
3.8. Western Blot Analysis after the Overexpression of mstn-1 in Muscle Cells of Japanese Flounder
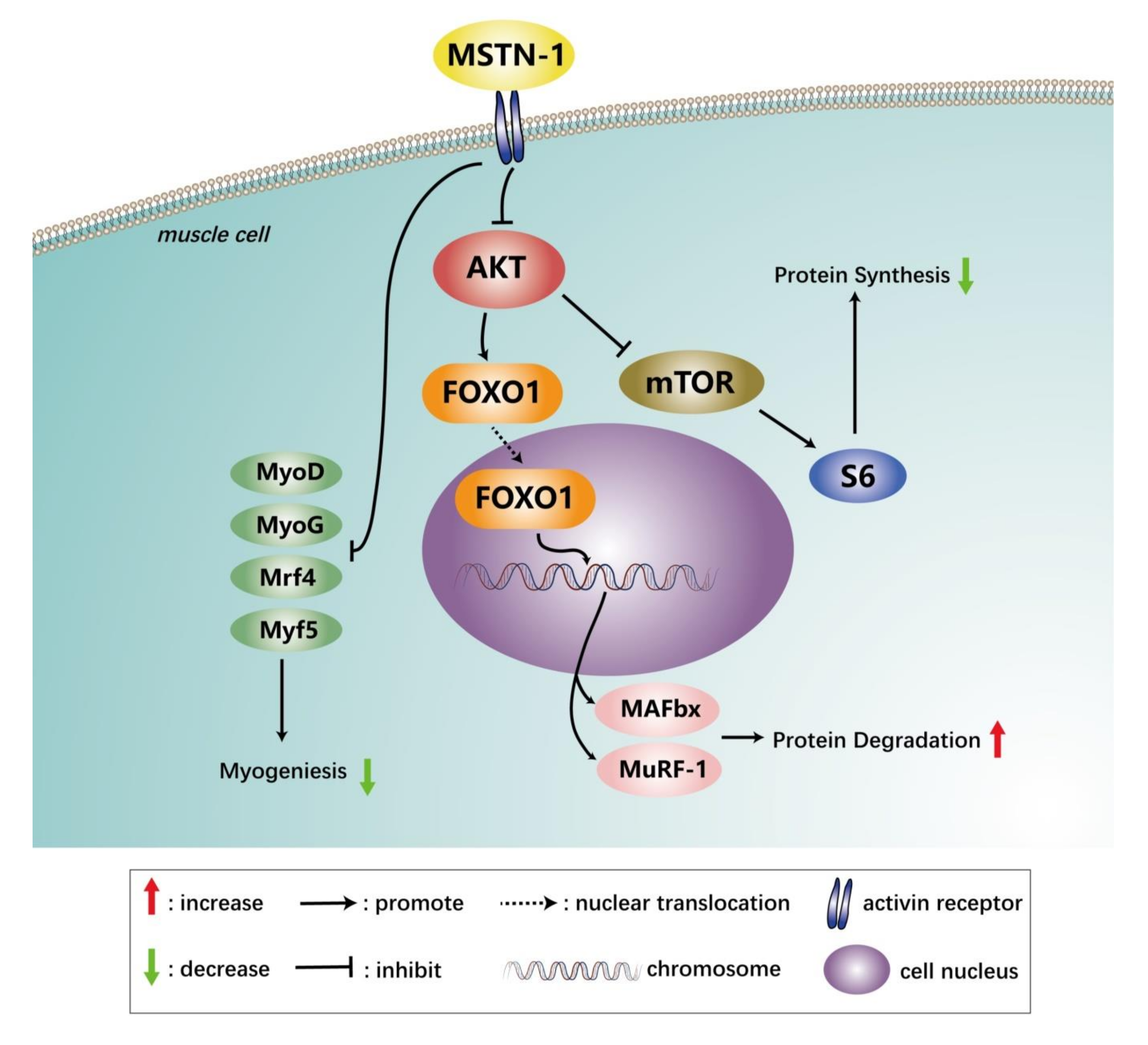
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- McPherron, A.C.; Lawler, A.M.; Lee, S.-J. Regulation of skeletal muscle mass in mice by a new TGF-p superfamily member. Nature 1997, 387, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Rios, R.; Fernandez-Nocelos, S.; Carneiro, I.; Arce, V.M.; Devesa, J. Differential response to exogenous and endogenous myostatin in myoblasts suggests that myostatin acts as an autocrine factor In Vivo. Endocrinology 2004, 145, 2795–2803. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.; Vernus, B.; Chelh, I.; Cassar-Malek, I.; Gabillard, J.-C.; Sassi, A.H.; Seiliez, I.; Picard, B.; Bonnieu, A. Myostatin and the skeletal muscle atrophy and hypertrophy signaling pathways. Cell. Mol. Life Sci. 2014, 71, 4361–4371. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Arnold, H.B.; Della-Fera, M.A.; Azain, M.J.; Hartzell, D.L.; Baile, C.A. Myostatin knockout in mice increases myogenesis and decreases adipogenesis. Biochem. Biophys. Res. Commun. 2002, 291, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Zimmers, T.A.; Davies, M.V.; Koniaris, L.G.; Haynes, P.; Esquela, A.F.; Tomkinson, K.N.; McPherron, A.C.; Wolfman, N.M.; Lee, S.-J. Induction of cachexia in mice by systemically administered myostatin. Science 2002, 296, 1486–1488. [Google Scholar] [CrossRef]
- Trendelenburg, A.U.; Meyer, A.; Rohner, D.; Boyle, J.; Hatakeyama, S.; Glass, D.J. Myostatin reduces Akt/TORC1/p70S6K signaling, inhibiting myoblast differentiation and myotube size. Am. J. Physiol. Physiol. 2009, 296, C1258–C1270. [Google Scholar] [CrossRef] [PubMed]
- Postlethwait, J.H.; Yan, Y.-L.; Gates, M.A.; Horne, S.; Amores, A.; Brownlie, A.; Donovan, A.; Egan, E.S.; Force, A.; Gong, Z.; et al. Vertebrate genome evolution and the zebrafish gene map. Nat. Genet. 1998, 18, 345–349. [Google Scholar] [CrossRef]
- Taylor, J.S.; Braasch, I.; Frickey, T.; Meyer, A.; de Peer, Y. Genome duplication, a trait shared by 22,000 species of ray-finned fish. Genome Res. 2003, 13, 382–390. [Google Scholar] [CrossRef]
- Garikipati, D.K.; Gahr, S.A.; Rodgers, B.D. Identification, characterization, and quantitative expression analysis of rainbow trout myostatin-1a and myostatin-1b genes. Endocrinology 2006, 190, 879–888. [Google Scholar] [CrossRef] [PubMed]
- Østbye, T.-K.K.; Wetten, O.F.; Tooming-Klunderud, A.; Jakobsen, K.S.; Yafe, A.; Etzioni, S.; Moen, T.; Andersen, Ø. Myostatin (MSTN) gene duplications in Atlantic salmon (Salmo salar): Evidence for different selective pressure on teleost MSTN-1 and -2. Gene 2007, 403, 159–169. [Google Scholar] [CrossRef]
- Rescan, P.Y.; Jutel, I.; Rescan, P.Y.; Jutel, I.C.R. Two myostatin genes are differentially expressed in myotomal muscles of the trout (Oncorhynchus mykiss). J. Exp. Biol. 2001, 204, 3523–3529. [Google Scholar] [PubMed]
- Rodgers, B.D.; Weber, G.M.; Sullivan, C.V.; Levine, A.M. Isolation and Characterization of Myostatin Complementary Deoxyribonucleic Acid Clones from Two Commercially Important Fish: Oreochromis mossambicusand Morone chrysops. Endocrinology 2001, 142, 1412–1418. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhong, Q.; Zhang, Q.; Chen, Y.; Sun, Y.; Qi, J.; Wang, Z.; Li, S.; Li, C.; Lan, X. The isolation and characterization of myostatin gene in Japanese flounder (Paralichthys olivaceus): Ubiquitous tissue expression and developmental specific regulation. Aquaculture 2008, 280, 247–255. [Google Scholar] [CrossRef]
- De Santis, C.; Jerry, D.R. Differential tissue-regulation of myostatin genes in the teleost fish Lates calcarifer in response to fasting. Evidence for functional differentiation. Mol. Cell. Endocrinol. 2011, 335, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Yu, X.; Tong, J. Molecular characterization of myostatin (MSTN) gene and association analysis with growth traits in the bighead carp (Aristichthys nobilis). Mol. Biol. Rep. 2012, 39, 9211–9221. [Google Scholar] [CrossRef]
- Seiliez, I.; Sabin, N.; Gabillard, J.-C. Myostatin inhibits proliferation but not differentiation of trout myoblasts. Mol. Cell. Endocrinol. 2012, 351, 220–226. [Google Scholar] [CrossRef]
- Zhu, K.C.; Yu, D.H.; Zhao, J.K.; Wang, W.M.; Wang, H.L. Morphological analysis and muscle-associated gene expression during different muscle growth phases of Megalobrama amblycephala. Genet. Mol. Res. Gmr 2015, 14, 11639–11651. [Google Scholar] [CrossRef]
- Amali, A.A.; Lin, C.J.-F.; Chen, Y.-H.; Wang, W.-L.; Gong, H.-Y.; Lee, C.-Y.; Ko, Y.-L.; Lu, J.-K.; Her, G.M.; Chen, T.T.; et al. Up-regulation of muscle-specific transcription factors during embryonic somitogenesis of zebrafish (Danio rerio) by knock-down of myostatin-1. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2004, 229, 847–856. [Google Scholar] [CrossRef]
- Acosta, J.; Carpio, Y.; Borroto, I.; González, O.; Estrada, M.P. Myostatin gene silenced by RNAi show a zebrafish giant phenotype. J. Biotechnol. 2005, 119, 324–331. [Google Scholar] [CrossRef]
- Lee, C.-Y.; Hu, S.-Y.; Gong, H.-Y.; Chen, M.H.-C.; Lu, J.-K.; Wu, J.-L. Suppression of myostatin with vector-based RNA interference causes a double-muscle effect in transgenic zebrafish. Biochem. Biophys. Res. Commun. 2009, 387, 766–771. [Google Scholar] [CrossRef]
- Wang, C.; Chen, Y.L.; Bian, P.W.; Xie, S.L.; Pei, D.S. Deletion of mstna and mstnb impairs the immune system and affects growth performance in zebrafish. Fish. Shellfish Immunol. 2018, 72, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Khalil, K.; Elayat, M.; Khalifa, E.; Daghash, S.M.; Elaswad, A.; Miller, M.E.; Abdelrahman, H.; Ye, Z.; Odin, R.; Drescher, D.; et al. Generation of Myostatin Gene-Edited Channel Catfish (Ictalurus punctatus) via Zygote Injection of CRISPR/Cas9 System. Sci. Rep. 2017, 7, 7301. [Google Scholar] [CrossRef]
- Kishimoto, K.; Washio, Y.; Yoshiura, Y.; Toyoda, A.; Ueno, T.; Fukuyama, H.; Kato, K.; Kinoshita, M. Production of a breed of red sea bream Pagrus major with an increase of skeletal muscle mass and reduced body length by genome editing with CRISPR/Cas9. Aquaculture 2018, 495, 415–427. [Google Scholar] [CrossRef]
- Kim, J.; Cho, J.Y.; Kim, J.; Kim, H.C.; Noh, J.K.; Kim, Y.; Hwang, H.; Kim, W.J.; Yeo, S.; An, C.M.; et al. CRISPR/Cas9-mediated myostatin disruption enhances muscle mass in the olive flounder Paralichthys olivaceus. Aquaculture 2019, 512, 734336. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, F.; Dong, Z.; Dong, X.; Chi, J.; Chen, H.; Zhao, Q.; Li, K. A new strain of yellow catfish carrying genome edited myostatin alleles exhibits double muscling phenotype with hyperplasia. Aquaculture 2020, 735187. [Google Scholar] [CrossRef]
- Xu, C.; Wu, G.; Zohar, Y.; Du, S.J. Analysis of myostatin gene structure, expression and function in zebrafish. J. Exp. Biol. 2003, 206, 4067–4079. [Google Scholar] [CrossRef]
- Carpio, Y.; Acosta, J.; Morales, R.; Santisteban, Y.; Sanchéz, A.; Estrada, M.P. Regulation of body mass growth through activin type IIB receptor in teleost fish. Endocrinology 2009, 160, 158–167. [Google Scholar] [CrossRef]
- Medeiros, E.F.; Phelps, M.P.; Fuentes, F.D.; Bradley, T.M. Overexpression of follistatin in trout stimulates increased muscling. Am. J. Physiol. Integr. Comp. Physiol. 2009, 297, R235–R242. [Google Scholar] [CrossRef]
- Seiliez, I.; Taty, G.C.; Bugeon, J.; Dias, K.; Sabin, N.; Gabillard, J.-C. Myostatin induces atrophy of trout myotubes through inhibiting the TORC1 signaling and promoting Ubiquitin-Proteasome and Autophagy-Lysosome degradative pathways. Gen. Comp. Endocrinol. 2013, 186, 9–15. [Google Scholar] [CrossRef]
- Lee, S.B.; Kim, Y.S.; Oh, M.-Y.; Jeong, I.; Seong, K.-B.; Jin, H.-J. Improving rainbow trout (Oncorhynchus mykiss) growth by treatment with a fish (Paralichthys olivaceus) myostatin prodomain expressed in soluble forms in E. coli. Aquaculture 2010, 302, 270–278. [Google Scholar] [CrossRef]
- Vegusdal, A.; Østbye, T.K.; Tran, T.-N.; Gjøen, T.; Ruyter, B. β-oxidation, esterification, and secretion of radiolabeled fatty acids in cultivated Atlantic salmon skeletal muscle cells. Lipids 2004, 39, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Amilburu, V.; Salmerón, C.; Codina, M.; Navarro, I.; Capilla, E.; Gutiérrez, J. Insulin-like growth factors effects on the expression of myogenic regulatory factors in gilthead sea bream muscle cells. Gen. Comp. Endocrinol. 2013, 188, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Joulia, D.; Bernardi, H.; Garandel, V.; Rabenoelina, F.; Vernus, B.; Cabello, G. Mechanisms involved in the inhibition of myoblast proliferation and differentiation by myostatin. Exp. Cell Res. 2003, 286, 263–275. [Google Scholar] [CrossRef]
- Lokireddy, S.; Mouly, V.; Butlerbrowne, G.; Gluckman, P.D.; Sharma, M.; Kambadur, R.; Mcfarlane, C. Myostatin promotes the wasting of human myoblast cultures through promoting ubiquitin-proteasome pathway-mediated loss of sarcomeric proteins. Am. J. Physiol. Physiol. 2011, 301, C1316–C1324. [Google Scholar] [CrossRef] [PubMed]
- McFarlane, C.; Plummer, E.; Thomas, M.; Hennebry, A.; Ashby, M.; Ling, N.; Smith, H.; Sharma, M.; Kambadur, R. Myostatin induces cachexia by activating the ubiquitin proteolytic system through an NF-κ-independent, FoxO1-dependent mechanism. J. Cell. Physiol. 2006, 209, 501–514. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.; Vernus, B.; Toubiana, M.; Jublanc, E.; Tintignac, L.; Leibovitch, S.; Bonnieu, A. Myostatin inactivation increases myotube size through regulation of translational initiation machinery. J. Cell. Biochem. 2011, 112, 3531–3542. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ma, S.; Ding, Q.; Wang, X.; Chen, Y. CRISPR/Cas9-mediated MSTN gene editing induced mitochondrial alterations in C2C12 myoblast cells. Electron. J. Biotechnol. 2019, 40, 30–39. [Google Scholar] [CrossRef]
- Watabe, S. Myogenic regulatory factors. Fish. Physiol. 2001, 18, 19–41. [Google Scholar]
- De Almeida, F.L.A.; Pessotti, N.S.; Pinhal, D.; Padovani, C.R.; de Jesus Leitão, N.; Carvalho, R.F.; Martins, C.; Portella, M.C.; Dal Pai-Silva, M. Quantitative expression of myogenic regulatory factors MyoD and myogenin in pacu (Piaractus mesopotamicus) skeletal muscle during growth. Micron 2010, 41, 997–1004. [Google Scholar] [CrossRef]
- Sabourin, L.A.; Rudnicki, M.A. The molecular regulation of myogenesis. Clin. Genet. 2001, 57, 16–25. [Google Scholar] [CrossRef]
- Zhu, X.; Topouzis, S.; Liang, L.; Stotish, R.L. Myostatin signaling through Smad2, Smad3 and Smad4 is regulated by the inhibitory Smad7 by a negative feedback mechanism. Cytokine 2004, 26, 262–272. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.A.; Kinoshita, M.; Maekawa, S.; Kulkarni, A.; Aoki, T. TALENs-mediated gene disruption of myostatin produces a larger phenotype of medaka with an apparently compromised immune system. Fish. Shellfish Immunol. 2015, 48, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Ohanna, M.; Sobering, A.K.; Lapointe, T.; Lorenzo, L.; Praud, C.; Petroulakis, E.; Sonenberg, N.; Kelly, P.A.; Sotiropoulos, A.; Pende, M. Atrophy of S6K1-/- skeletal muscle cells reveals distinct mTOR effectors for cell cycle and size control. Nat. Cell Biol. 2005, 7, 286. [Google Scholar] [CrossRef] [PubMed]
- Ruvinsky, I.; Sharon, N.; Lerer, T.; Cohen, H.; Stolovich-Rain, M.; Nir, T.; Dor, Y.; Zisman, P.; Meyuhas, O. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 2005, 19, 2199–2211. [Google Scholar] [CrossRef] [PubMed]
- Ruvinsky, I.; Meyuhas, O. Ribosomal protein S6 phosphorylation: From protein synthesis to cell size. Trends Biochem. Sci. 2006, 31, 342–348. [Google Scholar] [CrossRef]
- Amirouche, A.; Durieux, A.-C.; Banzet, S.; Koulmann, N.; Bonnefoy, R.; Mouret, C.; Bigard, X.; Peinnequin, A.; Freyssenet, D. Down-regulation of Akt/mammalian target of rapamycin signaling pathway in response to myostatin overexpression in skeletal muscle. Endocrinology 2009, 150, 286–294. [Google Scholar] [CrossRef]
- Li, X.; Liu, H.; Wang, H.; Sun, L.; Ding, F.; Sun, W.; Han, C.; Wang, J. Follistatin could promote the proliferation of duck primary myoblasts by activating PI3K/Akt/mTOR signalling. Biosci. Rep. 2014, 34. [Google Scholar] [CrossRef]
- Belghit, I.; Skiba-Cassy, S.; Geurden, I.; Dias, K.; Surget, A.; Kaushik, S.; Panserat, S.; Seiliez, I. Dietary methionine availability affects the main factors involved in muscle protein turnover in rainbow trout (Oncorhynchus mykiss). Br. J. Nutr. 2014, 112, 493–503. [Google Scholar] [CrossRef]
- Bodine, S.C.; Baehr, L.M. Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1. Am. J. Physiol. Metab. 2014, 307, E469–E484. [Google Scholar] [CrossRef]
- Sandri, M.; Sandri, C.; Gilbert, A.; Skurk, C.; Calabria, E.; Picard, A.; Walsh, K.; Schiaffino, S.; Lecker, S.H.; Goldberg, A.L. Foxo Transcription Factors Induce the Atrophy-Related Ubiquitin Ligase Atrogin-1 and Cause Skeletal Muscle Atrophy. Cell 2004, 117, 399–412. [Google Scholar] [CrossRef]
- Stitt, T.; Drujan, D.; Clarke, B.A.; Panaro, F.; Timofeyva, Y.; Kline, W.O.; Gonzalez, M.; Yancopoulos, G.D.; Glass, D.J. The IGF-1/PI3K/Akt Pathway Prevents Expression of Muscle Atrophy-Induced Ubiquitin Ligases by Inhibiting FOXO Transcription Factors. Mol. Cell 2004, 14, 395–403. [Google Scholar] [CrossRef]
- Ramaswamy, S.; Nakamura, N.; Sansal, I.; Bergeron, L.; Sellers, W.R. A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell 2002, 2, 81–91. [Google Scholar] [CrossRef]
- Gilson, H.; Schakman, O.; Combaret, L.; Lause, P.; Grobet, L.; Attaix, D.; Ketelslegers, J.; Thissen, J. Myostatin gene deletion prevents glucocorticoid-induced muscle atrophy. Endocrinology 2007, 148, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Taylor, W.E.; Bhasin, S.; Artaza, J.N.; Byhower, F.; Azam, M.; Willard, D.H.; Kull, F.C.; Gonzalezcadavid, N.F. Myostatin inhibits cell proliferation and protein synthesis in C2C12 muscle cells. Am. J. Physiol. Metab. 2001, 280, E221–E228. [Google Scholar] [CrossRef]





| Genes | Forward (5’-3’) | Reverse (5’-3’) | Accession No. |
|---|---|---|---|
| myf5 | GCAACGCCATCCACTACATCG | TGCATTCAACTGGTGCCACACT | DQ872515 |
| myod | GCAACGCCATCAGCTACATCG | CGTTTGGAGTCTGGGAGAAATAAG | DQ184914 |
| myog | GTCTGGGGGTGTTGGAGTTGG | GACGCCTCTTCTCCCTCATCG | EF144128 |
| mrf4 | AGAGCAGCGGGGAGGAACAC | GACCTTGCAGGCCCACATGA | MK453386 |
| mstn-1 | TTTGAGGACTTTGGCTGGGACT | GCGACATCTTGGTGGGGGTA | DQ412048 |
| murf-1 | TTGTGCCGTAGTTGTGCTAGTGAC | CATGGCGATCAAGCACGACCTC | MK292717 |
| mafbx | GCTGGGTGAAAACCGAGGAG | CTTCTTGGCAGCCATGTCGT | MK453387 |
| β-actin | GGAAATCGTGCGTGACATTAAG | CCTCTGGACAACGGAACCTCT | HQ386788 |
| Genes | Forward (5’-3’) | Reverse (5’-3’) | Accession No. |
|---|---|---|---|
| Pax7b | AGCTAGCGGCATTCAACCAT | GTGTTGTGGCTGTGAGGAGA | KP323416 |
| myod | GCAACGCCATCAGCTACATCG | CGTTTGGAGTCTGGGAGAAATAAG | DQ184914 |
| myog | GAGTCTGTCTGGGGGTGTTG | ACTGCAGAGATGCTGTCCAC | EF144128 |
| mstn-1 | TTTGAGGACTTTGGCTGGGACT | GCGACATCTTGGTGGGGGTA | MK453386 |
| β-actin | GAGCGTGGCTACTCCTTCAC | TACGCTCAGGTGGGGCAAT | HQ386788 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Pan, M.; Huang, D.; Guo, Y.; Yang, M.; Zhang, W.; Mai, K. Myostatin-1 Inhibits Cell Proliferation by Inhibiting the mTOR Signal Pathway and MRFs, and Activating the Ubiquitin-Proteasomal System in Skeletal Muscle Cells of Japanese Flounder Paralichthys olivaceus. Cells 2020, 9, 2376. https://doi.org/10.3390/cells9112376
Liu J, Pan M, Huang D, Guo Y, Yang M, Zhang W, Mai K. Myostatin-1 Inhibits Cell Proliferation by Inhibiting the mTOR Signal Pathway and MRFs, and Activating the Ubiquitin-Proteasomal System in Skeletal Muscle Cells of Japanese Flounder Paralichthys olivaceus. Cells. 2020; 9(11):2376. https://doi.org/10.3390/cells9112376
Chicago/Turabian StyleLiu, Jiahuan, Mingzhu Pan, Dong Huang, Yanlin Guo, Mengxi Yang, Wenbing Zhang, and Kangsen Mai. 2020. "Myostatin-1 Inhibits Cell Proliferation by Inhibiting the mTOR Signal Pathway and MRFs, and Activating the Ubiquitin-Proteasomal System in Skeletal Muscle Cells of Japanese Flounder Paralichthys olivaceus" Cells 9, no. 11: 2376. https://doi.org/10.3390/cells9112376
APA StyleLiu, J., Pan, M., Huang, D., Guo, Y., Yang, M., Zhang, W., & Mai, K. (2020). Myostatin-1 Inhibits Cell Proliferation by Inhibiting the mTOR Signal Pathway and MRFs, and Activating the Ubiquitin-Proteasomal System in Skeletal Muscle Cells of Japanese Flounder Paralichthys olivaceus. Cells, 9(11), 2376. https://doi.org/10.3390/cells9112376




