How Altered Ribosome Production Can Cause or Contribute to Human Disease: The Spectrum of Ribosomopathies
Abstract
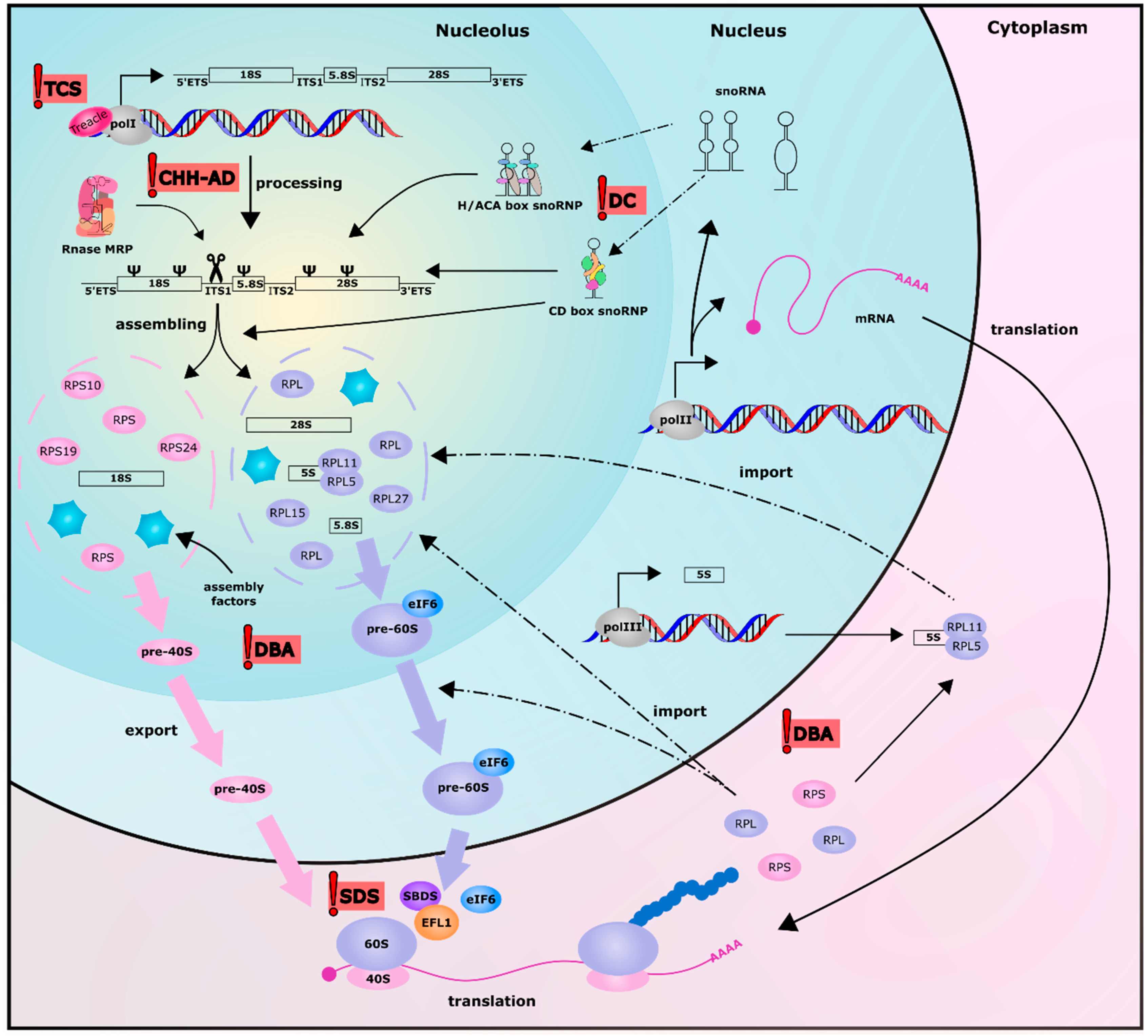
1. Introduction
2. Pure Ribosomopathies
2.1. Diamond Blackfan Anemia
2.2. Shwachman-Diamond Syndrome
2.3. Treacher Collins Syndrome
3. Mixed Ribosomopathies
3.1. Dyskeratosis Congenita
3.2. Cartilage Hair Hypoplasia-Anauxetic Dysplasia (CHH-AD) Spectrum
4. Recently Identified Ribosomopathies
5. Acquired Ribosomopathies
Funding
Acknowledgments
Conflicts of Interest
References
- Kampen, K.R.; Sulima, S.O.; Vereecke, S.; De Keersmaecker, K. Hallmarks of ribosomopathies. Nucleic Acids Res. 2019, 48, 1013–1028. [Google Scholar] [CrossRef] [PubMed]
- Farley-Barnes, K.I.; Ogawa, L.M.; Baserga, S.J. Ribosomopathies: Old Concepts, New Controversies. Trends Genet. 2019, 35, 754–767. [Google Scholar] [CrossRef] [PubMed]
- Luzzatto, L.; Karadimitris, A. Dyskeratosis and ribosomal rebellion. Nat. Genet. 1998, 19, 6–7. [Google Scholar] [CrossRef] [PubMed]
- Heiss, N.S.; Knight, S.W.; Vulliamy, T.J.; Klauck, S.M.; Wiemann, S.; Mason, P.J.; Poustka, A.; Dokal, I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat. Genet. 1998, 19, 32–38. [Google Scholar] [CrossRef]
- Ban, N.; Beckmann, R.; Cate, J.H.; Dinman, J.D.; Dragon, F.; Ellis, S.R.; Lafontaine, D.L.; Lindahl, L.; Liljas, A.; Lipton, J.M.; et al. A new system for naming ribosomal proteins. Curr. Opin. Struct. Biol. 2014, 24, 165–169. [Google Scholar] [CrossRef]
- Draptchinskaia, N.; Gustavsson, P.; Andersson, B.; Pettersson, M.; Willig, T.N.; Dianzani, I.; Ball, S.; Tchernia, G.; Klar, J.; Matsson, H.; et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anemia. Nat. Genet. 1999, 21, 169–175. [Google Scholar] [CrossRef]
- Boocock, G.R.; Morrison, J.A.; Popovic, M.; Richards, N.; Ellis, L.; Durie, P.R.; Rommens, J.M. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat. Genet. 2003, 33, 97–101. [Google Scholar] [CrossRef]
- Ridanpaa, M.; van Eenennaam, H.; Pelin, K.; Chadwick, R.; Johnson, C.; Yuan, B.; van Venrooij, W.; Pruijn, G.; Salmela, R.; Rockas, S.; et al. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell 2001, 104, 195–203. [Google Scholar] [CrossRef]
- Group, T.T.C.S.C.; Dixon, J.; Edwards, S.J.; Gladwin, A.J.; Dixon, M.J.; Loftus, S.K.; Bonner, C.A.; Koprivnikar, K.; Wasmuth, J.J. Positional cloning of a gene involved in the pathogenesis of Treacher Collins syndrome. The Treacher Collins Syndrome Collaborative Group. Nat. Genet. 1996, 12, 130–136. [Google Scholar]
- Narla, A.; Ebert, B.L. Ribosomopathies: Human disorders of ribosome dysfunction. Blood 2010, 115, 3196–3205. [Google Scholar] [CrossRef]
- Ebert, B.L.; Pretz, J.; Bosco, J.; Chang, C.Y.; Tamayo, P.; Galili, N.; Raza, A.; Root, D.E.; Attar, E.; Ellis, S.R.; et al. Identification of RPS14 as a 5q− syndrome gene by RNA interference screen. Nature 2008, 451, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Montanaro, L.; Treré, D.; Derenzini, M. Nucleolus, ribosomes, and cancer. Am. J. Pathol. 2008, 173, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Penzo, M.; Montanaro, L.; Trere, D.; Derenzini, M. The Ribosome Biogenesis-Cancer Connection. Cells 2019, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Henras, A.K.; Plisson-Chastang, C.; O’Donohue, M.F.; Chakraborty, A.; Gleizes, P.E. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA 2015, 6, 225–242. [Google Scholar] [CrossRef] [PubMed]
- Mullineux, S.T.; Lafontaine, D.L. Mapping the cleavage sites on mammalian pre-rRNAs: Where do we stand? Biochimie 2012, 94, 1521–1532. [Google Scholar] [CrossRef]
- Henras, A.K.; Soudet, J.; Gerus, M.; Lebaron, S.; Caizergues-Ferrer, M.; Mougin, A.; Henry, Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol. Life Sci. 2008, 65, 2334–2359. [Google Scholar] [CrossRef]
- McStay, B. Nucleolar organizer regions: Genomic’dark matter’ requiring illumination. Genes. Dev. 2016, 30, 1598–1610. [Google Scholar] [CrossRef]
- Paule, M.R.; White, R.J. Survey and summary: Transcription by RNA polymerases I and III. Nucleic Acids Res. 2000, 28, 1283–1298. [Google Scholar] [CrossRef]
- Bersaglieri, C.; Santoro, R. Genome Organization in and around the Nucleolus. Cells 2019, 8, 579. [Google Scholar] [CrossRef]
- Watkins, N.J.; Bohnsack, M.T. The box C/D and H/ACA snoRNPs: Key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA 2012, 3, 397–414. [Google Scholar] [CrossRef]
- Sharma, S.; Yang, J.; van Nues, R.; Watzinger, P.; Kötter, P.; Lafontaine, D.L.J.; Granneman, S.; Entian, K.D. Specialized box C/D snoRNPs act as antisense guides to target RNA base acetylation. PLoS Genet. 2017, 13, e1006804. [Google Scholar] [CrossRef]
- Massenet, S.; Bertrand, E.; Verheggen, C. Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol. 2017, 14, 680–692. [Google Scholar] [CrossRef]
- Liang, X.H.; Liu, Q.; Fournier, M.J. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA 2009, 15, 1716–1728. [Google Scholar] [CrossRef] [PubMed]
- Jack, K.; Bellodi, C.; Landry, D.M.; Niederer, R.O.; Meskauskas, A.; Musalgaonkar, S.; Kopmar, N.; Krasnykh, O.; Dean, A.M.; Thompson, S.R.; et al. RNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol. Cell 2011, 44, 660–666. [Google Scholar] [CrossRef]
- Ojha, S.; Malla, S.; Lyons, S.M. SnoRNPs: Functions in Ribosome Biogenesis. Biomolecules 2020, 10, 783. [Google Scholar] [CrossRef] [PubMed]
- Panse, V.G.; Johnson, A.W. Maturation of eukaryotic ribosomes: Acquisition of functionality. Trends Biochem. Sci. 2010, 35, 260–266. [Google Scholar] [CrossRef]
- Weis, F.; Giudice, E.; Churcher, M.; Jin, L.; Hilcenko, C.; Wong, C.C.; Traynor, D.; Kay, R.R.; Warren, A.J. Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nat. Struct. Mol. Biol. 2015, 22, 914–919. [Google Scholar] [CrossRef] [PubMed]
- Lebaron, S.; Schneider, C.; van Nues, R.W.; Swiatkowska, A.; Walsh, D.; Böttcher, B.; Granneman, S.; Watkins, N.J.; Tollervey, D. Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits. Nat. Struct. Mol. Biol. 2012, 19, 744–753. [Google Scholar] [CrossRef] [PubMed]
- De Keersmaecker, K.; Sulima, S.O.; Dinman, J.D. Ribosomopathies and the paradox of cellular hypo- to hyperproliferation. Blood 2015, 125, 1377–1382. [Google Scholar] [CrossRef]
- Mirabello, L.; Macari, E.R.; Jessop, L.; Ellis, S.R.; Myers, T.; Giri, N.; Taylor, A.M.; McGrath, K.E.; Humphries, J.M.; Ballew, B.J.; et al. Whole-exome sequencing and functional studies identify RPS29 as a novel gene mutated in multicase Diamond-Blackfan anemia families. Blood 2014, 124, 24–32. [Google Scholar] [CrossRef]
- Gripp, K.W.; Curry, C.; Olney, A.H.; Sandoval, C.; Fisher, J.; Chong, J.X.; Genomics UWC f, M.; Pilchman, L.; Sahraoui, R.; Stabley, D.L.; et al. Diamond-Blackfan anemia with mandibulofacial dysostosis is heterogeneous, including the novel DBA genes TSR2 and RPS28. Am. J. Med. Genet. 2014, 164A, 2240–2249. [Google Scholar] [CrossRef]
- Boria, I.; Garelli, E.; Gazda, H.T.; Aspesi, A.; Quarello, P.; Pavesi, E.; Ferrante, D.; Meerpohl, J.J.; Kartal, M.; Da Costa, L.; et al. The ribosomal basis of Diamond-Blackfan Anemia: Mutation and database update. Hum. Mutat. 2010, 31, 1269–1279. [Google Scholar] [CrossRef]
- Mirabello, L.; Khincha, P.P.; Ellis, S.R.; Giri, N.; Brodie, S.; Chandrasekharappa, S.C.; Donovan, F.X.; Zhou, W.; Hicks, B.D.; Boland, J.F.; et al. Novel and known ribosomal causes of Diamond-Blackfan anaemia identified through comprehensive genomic characterisation. J. Med. Genet. 2017, 54, 417–425. [Google Scholar] [CrossRef]
- Gazda, H.T.; Preti, M.; Sheen, M.R.; O’Donohue, M.F.; Vlachos, A.; Davies, S.M.; Kattamis, A.; Doherty, L.; Landowski, M.; Buros, C.; et al. Frameshift mutation in p53 regulator RPL26 is associated with multiple physical abnormalities and a specific pre-ribosomal RNA processing defect in diamond-blackfan anemia. Hum. Mutat. 2012, 33, 1037–1044. [Google Scholar] [CrossRef]
- Landowski, M.; O’Donohue, M.F.; Buros, C.; Ghazvinian, R.; Montel-Lehry, N.; Vlachos, A.; Sieff, C.A.; Newburger, P.E.; Niewiadomska, E.; Matysiak, M.; et al. Novel deletion of RPL15 identified by array-comparative genomic hybridization in Diamond-Blackfan anemia. Hum. Genet. 2013, 132, 1265–1274. [Google Scholar] [CrossRef]
- Wang, R.; Yoshida, K.; Toki, T.; Sawada, T.; Uechi, T.; Okuno, Y.; Sato-Otsubo, A.; Kudo, K.; Kamimaki, I.; Kanezaki, R.; et al. Loss of function mutations in RPL27 and RPS27 identified by whole-exome sequencing in Diamond-Blackfan anaemia. Br. J. Haematol. 2015, 168, 854–864. [Google Scholar] [CrossRef] [PubMed]
- Tummala, H.; Walne, A.J.; Williams, M.; Bockett, N.; Collopy, L.; Cardoso, S.; Ellison, A.; Wynn, R.; Leblanc, T.; Fitzgibbon, J.; et al. DNAJC21 Mutations Link a Cancer-Prone Bone Marrow Failure Syndrome to Corruption in 60S Ribosome Subunit Maturation. Am. J. Hum. Genet. 2016, 99, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Stepensky, P.; Chacon-Flores, M.; Kim, K.H.; Abuzaitoun, O.; Bautista-Santos, A.; Simanovsky, N.; Siliqi, D.; Altamura, D.; Mendez-Godoy, A.; Gijsbers, A.; et al. Mutations in EFL1, an SBDS partner, are associated with infantile pancytopenia, exocrine pancreatic insufficiency and skeletal anomalies in a Shwachman-Diamond like syndrome. J. Med. Genet. 2017, 54, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Carapito, R.; Konantz, M.; Paillard, C.; Miao, Z.; Pichot, A.; Leduc, M.S.; Yang, Y.; Bergstrom, K.L.; Mahoney, D.H.; Shardy, D.L.; et al. Mutations in signal recognition particle SRP54 cause syndromic neutropenia with Shwachman-Diamond-like features. J. Clin. Investig. 2017, 127, 4090–4103. [Google Scholar] [CrossRef]
- Teber, O.A.; Gillessen-Kaesbach, G.; Fischer, S.; Bohringer, S.; Albrecht, B.; Albert, A.; Arslan-Kirchner, M.; Haan, E.; Hagedorn-Greiwe, M.; Hammans, C.; et al. Genotyping in 46 patients with tentative diagnosis of Treacher Collins syndrome revealed unexpected phenotypic variation. Eur. J. Hum. Genet. 2004, 12, 879–890. [Google Scholar] [CrossRef] [PubMed]
- Dauwerse, J.G.; Dixon, J.; Seland, S.; Ruivenkamp, C.A.; van Haeringen, A.; Hoefsloot, L.H.; Peters, D.J.; Boers, A.C.; Daumer-Haas, C.; Maiwald, R.; et al. Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nat. Genet. 2011, 43, 20–22. [Google Scholar] [CrossRef]
- Sanchez, E.; Laplace-Builhe, B.; Mau-Them, F.T.; Richard, E.; Goldenberg, A.; Toler, T.L.; Guignard, T.; Gatinois, V.; Vincent, M.; Blanchet, C.; et al. POLR1B and neural crest cell anomalies in Treacher Collins syndrome type 4. Genet. Med. 2020, 22, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Elalaoui, S.C.; Laarabi, F.Z.; Mansouri, M.; Mrani, N.A.; Nishimura, G.; Sefiani, A. Further evidence of POP1 mutations as the cause of anauxetic dysplasia. Am. J. Med. Genet. A 2016, 170, 2462–2465. [Google Scholar] [CrossRef]
- Lafontaine, D.L.; Bousquet-Antonelli, C.; Henry, Y.; Caizergues-Ferrer, M.; Tollervey, D. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes. Dev. 1998, 12, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H.; Yoshikawa, H.; Izumikawa, K.; Miura, Y.; Taoka, M.; Nobe, Y.; Yamauchi, Y.; Nakayama, H.; Simpson, R.J.; Isobe, T.; et al. Poly(A)-specific ribonuclease regulates the processing of small-subunit rRNAs in human cells. Nucleic Acids Res. 2017, 45, 3437–3447. [Google Scholar] [CrossRef] [PubMed]
- Montellese, C.; Montel-Lehry, N.; Henras, A.K.; Kutay, U.; Gleizes, P.E.; O’Donohue, M.F. Poly(A)-specific ribonuclease is a nuclear ribosome biogenesis factor involved in human 18S rRNA maturation. Nucleic Acids Res. 2017, 45, 6822–6836. [Google Scholar] [CrossRef]
- Vulliamy, T.; Beswick, R.; Kirwan, M.; Marrone, A.; Digweed, M.; Walne, A.; Dokal, I. Mutations in the telomerase component NHP2 cause the premature ageing syndrome dyskeratosis congenita. Proc. Natl. Acad. Sci. USA 2008, 105, 8073–8078. [Google Scholar] [CrossRef]
- Walne, A.J.; Vulliamy, T.; Marrone, A.; Beswick, R.; Kirwan, M.; Masunari, Y.; Al-Qurashi, F.H.; Aljurf, M.; Dokal, I. Genetic heterogeneity in autosomal recessive dyskeratosis congenita with one subtype due to mutations in the telomerase-associated protein NOP10. Hum. Mol. Genet. 2007, 16, 1619–1629. [Google Scholar] [CrossRef]
- Trahan, C.; Martel, C.; Dragon, F. Effects of dyskeratosis congenita mutations in dyskerin, NHP2 and NOP10 on assembly of H/ACA pre-RNPs. Hum. Mol. Genet. 2010, 19, 825–836. [Google Scholar] [CrossRef]
- Nachmani, D.; Bothmer, A.H.; Grisendi, S.; Mele, A.; Bothmer, D.; Lee, J.D.; Monteleone, E.; Cheng, K.; Zhang, Y.; Bester, A.C.; et al. Germline NPM1 mutations lead to altered rRNA 2’-O-methylation and cause dyskeratosis congenita. Nat. Genet. 2019, 51, 1518–1529. [Google Scholar] [CrossRef]
- Vlachos, A.; Ball, S.; Dahl, N.; Alter, B.P.; Sheth, S.; Ramenghi, U.; Meerpohl, J.; Karlsson, S.; Liu, J.M.; Leblanc, T.; et al. Participants of Sixth Annual Daniella Maria Arturi International Consensus, C. Diagnosing and treating Diamond Blackfan anaemia: Results of an international clinical consensus conference. Br. J. Haematol. 2008, 142, 859–876. [Google Scholar] [CrossRef]
- Khajuria, R.K.; Munschauer, M.; Ulirsch, J.C.; Fiorini, C.; Ludwig, L.S.; McFarland, S.K.; Abdulhay, N.J.; Specht, H.; Keshishian, H.; Mani, D.R.; et al. Ribosome Levels Selectively Regulate Translation and Lineage Commitment in Human Hematopoiesis. Cell 2018, 173, 90–103. [Google Scholar] [CrossRef]
- Flygare, J.; Aspesi, A.; Bailey, J.C.; Miyake, K.; Caffrey, J.M.; Karlsson, S.; Ellis, S.R. Human RPS19, the gene mutated in Diamond-Blackfan anemia, encodes a ribosomal protein required for the maturation of 40S ribosomal subunits. Blood 2007, 109, 980–986. [Google Scholar] [CrossRef] [PubMed]
- Choesmel, V.; Bacqueville, D.; Rouquette, J.; Noaillac-Depeyre, J.; Fribourg, S.; Cretien, A.; Leblanc, T.; Tchernia, G.; Da Costa, L.; Gleizes, P.E. Impaired ribosome biogenesis in Diamond-Blackfan anemia. Blood 2007, 109, 1275–1283. [Google Scholar] [CrossRef]
- Marechal, V.; Elenbaas, B.; Piette, J.; Nicolas, J.C.; Levine, A.J. The ribosomal L5 protein is associated with mdm-2 and mdm-2-p53 complexes. Mol. Cell. Biol. 1994, 14, 7414–7420. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.S.; Lu, H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem. 2004, 279, 44475–44482. [Google Scholar] [CrossRef]
- Lohrum, M.A.; Ludwig, R.L.; Kubbutat, M.H.; Hanlon, M.; Vousden, K.H. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 2003, 3, 577–587. [Google Scholar] [CrossRef]
- Zhang, Y.; Wolf, G.W.; Bhat, K.; Jin, A.; Allio, T.; Burkhart, W.A.; Xiong, Y. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell. Biol. 2003, 23, 8902–8912. [Google Scholar] [CrossRef]
- Ulirsch, J.C.; Verboon, J.M.; Kazerounian, S.; Guo, M.H.; Yuan, D.; Ludwig, L.S.; Handsaker, R.E.; Abdulhay, N.J.; Fiorini, C.; Genovese, G.; et al. The Genetic Landscape of Diamond-Blackfan Anemia. Am. J. Hum. Genet. 2018, 103, 930–947. [Google Scholar] [CrossRef]
- Bezzerri, V.; Cipolli, M. Shwachman-Diamond Syndrome: Molecular Mechanisms and Current Perspectives. Mol. Diagn. 2019, 23, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Warren, A.J. Molecular basis of the human ribosomopathy Shwachman-Diamond syndrome. Adv. Biol. Regul. 2018, 67, 109–127. [Google Scholar] [CrossRef] [PubMed]
- Shwachman, H.; Diamond, L.K.; Oski, F.A.; Khaw, K.T. The Syndrome of Pancreatic Insufficiency and Bone Marrow Dysfunction. J. Pediatr. 1964, 65, 645–663. [Google Scholar] [CrossRef]
- Finch, A.J.; Hilcenko, C.; Basse, N.; Drynan, L.F.; Goyenechea, B.; Menne, T.F.; Gonzalez Fernandez, A.; Simpson, P.; D’Santos, C.S.; Arends, M.J.; et al. Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman-Diamond syndrome. Genes. Dev. 2011, 25, 917–929. [Google Scholar] [CrossRef] [PubMed]
- Tourlakis, M.E.; Zhang, S.; Ball, H.L.; Gandhi, R.; Liu, H.; Zhong, J.; Yuan, J.S.; Guidos, C.J.; Durie, P.R.; Rommens, J.M. In Vivo Senescence in the Sbds-Deficient Murine Pancreas: Cell-Type Specific Consequences of Translation Insufficiency. PLoS Genet. 2015, 11, e1005288. [Google Scholar] [CrossRef] [PubMed]
- Valdez, B.C.; Henning, D.; So, R.B.; Dixon, J.; Dixon, M.J. The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc. Natl. Acad. Sci. USA 2004, 101, 10709–10714. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.; Dixon, M.J. Genetic background has a major effect on the penetrance and severity of craniofacial defects in mice heterozygous for the gene encoding the nucleolar protein Treacle. Dev. DYN 2004, 229, 907–914. [Google Scholar] [CrossRef] [PubMed]
- Noack Watt, K.E.; Achilleos, A.; Neben, C.L.; Merrill, A.E.; Trainor, P.A. The Roles of RNA Polymerase I and III Subunits Polr1c and Polr1d in Craniofacial Development and in Zebrafish Models of Treacher Collins Syndrome. PLoS Genet. 2016, 12, e1006187. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.; Jones, N.C.; Sandell, L.L.; Jayasinghe, S.M.; Crane, J.; Rey, J.P.; Dixon, M.J.; Trainor, P.A. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc. Natl. Acad. Sci. USA 2006, 103, 13403–13408. [Google Scholar] [CrossRef]
- Jones, N.C.; Lynn, M.L.; Gaudenz, K.; Sakai, D.; Aoto, K.; Rey, J.P.; Glynn, E.F.; Ellington, L.; Du, C.; Dixon, J.; et al. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat. Med. 2008, 14, 125–133. [Google Scholar] [CrossRef]
- Ciccia, A.; Huang, J.W.; Izhar, L.; Sowa, M.E.; Harper, J.W.; Elledge, S.J. Treacher Collins syndrome TCOF1 protein cooperates with NBS1 in the DNA damage response. Proc. Natl. Acad. Sci. USA 2014, 111, 18631–18636. [Google Scholar] [CrossRef]
- Larsen, D.H.; Hari, F.; Clapperton, J.A.; Gwerder, M.; Gutsche, K.; Altmeyer, M.; Jungmichel, S.; Toledo, L.I.; Fink, D.; Rask, M.B.; et al. The NBS1-Treacle complex controls ribosomal RNA transcription in response to DNA damage. Nat. Cell. Biol. 2014, 16, 792–803. [Google Scholar] [CrossRef] [PubMed]
- Sakai, D.; Dixon, J.; Achilleos, A.; Dixon, M.; Trainor, P.A. Prevention of Treacher Collins syndrome craniofacial anomalies in mouse models via maternal antioxidant supplementation. Nat. Commun. 2016, 7, 10328. [Google Scholar] [CrossRef] [PubMed]
- Zinsser, F. Atropha cutis reticularis pigmentation, dystrophia ungiumet leukoplakia oris. Ikonogr. Derm. 1906, 5, 219–223. [Google Scholar]
- Engman, M.F. A unique case of reticular pigmentation of the skin with atrophy. Arch. Bleg. Derm. Syphil. 1926, 13, 685–687. [Google Scholar]
- Cole, H.N.; Rauschkolb, J.E.; Toomey, J. Dyskeratosis congenita with pigmentation, dystrophia unguis and leukokeratosis oris. Arch. Dermatol. Syphilol. 1930, 21, 71–95. [Google Scholar] [CrossRef]
- Dokal, I. Dyskeratosis congenita. Hematol. Am. Soc. Hematol. Educ. Program 2011, 2011, 480–486. [Google Scholar] [CrossRef]
- Alter, B.P.; Giri, N.; Savage, S.A.; Rosenberg, P.S. Cancer in dyskeratosis congenita. Blood 2009, 113, 6549–6557. [Google Scholar] [CrossRef]
- Ganot, P.; Bortolin, M.L.; Kiss, T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 1997, 89, 799–809. [Google Scholar] [CrossRef]
- Wang, C.; Meier, U.T. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 2004, 23, 1857–1867. [Google Scholar] [CrossRef]
- Yu, Y.T.; Meier, U.T. RNA-guided isomerization of uridine to pseudouridine—Pseudouridylation. RNA Biol. 2014, 11, 1483–1494. [Google Scholar] [CrossRef]
- Mitchell, J.R.; Wood, E.; Collins, K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 1999, 402, 551–555. [Google Scholar] [CrossRef] [PubMed]
- Vulliamy, T.; Marrone, A.; Goldman, F.; Dearlove, A.; Bessler, M.; Mason, P.J.; Dokal, I. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature 2001, 413, 432–435. [Google Scholar] [CrossRef]
- Marrone, A.; Walne, A.; Tamary, H.; Masunari, Y.; Kirwan, M.; Beswick, R.; Vulliamy, T.; Dokal, I. Telomerase reverse-transcriptase homozygous mutations in autosomal recessive dyskeratosis congenita and Hoyeraal-Hreidarsson syndrome. Blood 2007, 110, 4198–4205. [Google Scholar] [CrossRef]
- Savage, S.A.; Giri, N.; Baerlocher, G.M.; Orr, N.; Lansdorp, P.M.; Alter, B.P. TINF2, a component of the shelterin telomere protection complex, is mutated in dyskeratosis congenita. Am. J. Hum. Genet. 2008, 82, 501–509. [Google Scholar] [CrossRef]
- Savage, S.A. Gene Reviews; Adam, M.P., Ed.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Giordano, E.; Peluso, I.; Senger, S.; Furia, M. minifly, a Drosophila gene required for ribosome biogenesis. J. Cell Biol. 1999, 144, 1123–1133. [Google Scholar] [CrossRef]
- Ruggero, D.; Grisendi, S.; Piazza, F.; Rego, E.; Mari, F.; Rao, P.H.; Cordon-Cardo, C.; Pandolfi, P.P. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science 2003, 299, 259–262. [Google Scholar] [CrossRef]
- Blasco, M.A.; Lee, H.W.; Hande, M.P.; Samper, E.; Lansdorp, P.M.; DePinho, R.A.; Greider, C.W. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 1997, 91, 25–34. [Google Scholar] [CrossRef]
- McKusick, V.A.; Eldridge, R.; Hostetler, J.A.; Ruangwit, U.; Egeland, J.A. Dwarfism in the Amish. Ii. Cartilage-Hair Hypoplasia. Bull. Johns Hopkins Hosp. 1965, 116, 285–326. [Google Scholar]
- Makitie, O.; Kaitila, I. Cartilage-hair hypoplasia—clinical manifestations in 108 Finnish patients. Eur. J. Pediatr. 1993, 152, 211–217. [Google Scholar] [CrossRef]
- Taskinen, M.; Ranki, A.; Pukkala, E.; Jeskanen, L.; Kaitila, I.; Makitie, O. Extended follow-up of the Finnish cartilage-hair hypoplasia cohort confirms high incidence of non-Hodgkin lymphoma and basal cell carcinoma. Am. J. Med. Genet. A 2008, 146, 2370–2375. [Google Scholar] [CrossRef] [PubMed]
- Makitie, O.; Heikkinen, M.; Kaitila, I.; Rintala, R. Hirschsprung’s disease in cartilage-hair hypoplasia has poor prognosis. J. Pediatr. Surg. 2002, 37, 1585–1588. [Google Scholar] [CrossRef] [PubMed]
- Kostjukovits, S.; Klemetti, P.; Fohr, A.; Kajosaari, M.; Valta, H.; Taskinen, M.; Toiviainen-Salo, S.; Makitie, O. High prevalence of bronchiectasis in patients with cartilage-hair hypoplasia. J. Allergy Clin. Immunol. 2017, 139, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Makitie, O.M.; Tapanainen, P.J.; Dunkel, L.; Siimes, M.A. Impaired spermatogenesis: An unrecognized feature of cartilage-hair hypoplasia. Ann. Med. 2001, 33, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Vakkilainen, S.; Makitie, R.; Klemetti, P.; Valta, H.; Taskinen, M.; Husebye, E.S.; Makitie, O. A Wide Spectrum of Autoimmune Manifestations and Other Symptoms Suggesting Immune Dysregulation in Patients with Cartilage-Hair Hypoplasia. Front. Immunol. 2018, 9, 2468. [Google Scholar] [CrossRef]
- Vakkilainen, S.; Taskinen, M.; Klemetti, P.; Pukkala, E.; Makitie, O. A 30-Year Prospective Follow-Up Study Reveals Risk Factors for Early Death in Cartilage-Hair Hypoplasia. Front. Immunol. 2019, 10, 1581. [Google Scholar] [CrossRef]
- Welting, T.J.; van Venrooij, W.J.; Pruijn, G.J. Mutual interactions between subunits of the human RNase MRP ribonucleoprotein complex. Nucleic Acids Res. 2004, 32, 2138–2146. [Google Scholar] [CrossRef]
- Chang, D.D.; Clayton, D.A. A mammalian mitochondrial RNA processing activity contains nucleus-encoded RNA. Science 1987, 235, 1178–1184. [Google Scholar] [CrossRef]
- Thiel, C.T.; Horn, D.; Zabel, B.; Ekici, A.B.; Salinas, K.; Gebhart, E.; Ruschendorf, F.; Sticht, H.; Spranger, J.; Muller, D.; et al. Severely incapacitating mutations in patients with extreme short stature identify RNA-processing endoribonuclease RMRP as an essential cell growth regulator. Am. J. Hum. Genet. 2005, 77, 795–806. [Google Scholar] [CrossRef]
- Mattijssen, S.; Hinson, E.R.; Onnekink, C.; Hermanns, P.; Zabel, B.; Cresswell, P.; Pruijn, G.J. Viperin mRNA is a novel target for the human RNase MRP/RNase P endoribonuclease. Cell Mol. Life Sci. 2011, 68, 2469–2480. [Google Scholar] [CrossRef]
- Maida, Y.; Yasukawa, M.; Furuuchi, M.; Lassmann, T.; Possemato, R.; Okamoto, N.; Kasim, V.; Hayashizaki, Y.; Hahn, W.C.; Masutomi, K. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature 2009, 461, 230–235. [Google Scholar] [CrossRef]
- Thiel, C.T.; Mortier, G.; Kaitila, I.; Reis, A.; Rauch, A. Type and level of RMRP functional impairment predicts phenotype in the cartilage hair hypoplasia-anauxetic dysplasia spectrum. Am. J. Hum. Genet. 2007, 81, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Goldfarb, K.C.; Cech, T.R. Targeted CRISPR disruption reveals a role for RNase MRP RNA in human preribosomal RNA processing. Genes Dev. 2017, 31, 59–71. [Google Scholar] [CrossRef]
- Thiel, C.T.; Rauch, A. The molecular basis of the cartilage-hair hypoplasia-anauxetic dysplasia spectrum. Best Pr. Res. Clin. Endocrinol. Metab. 2011, 25, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Mattijssen, S.; Welting, T.J.; Pruijn, G.J. RNase MRP and disease. Wiley Interdiscip. Rev. RNA 2010, 1, 102–116. [Google Scholar] [CrossRef]
- Kavadas, F.D.; Giliani, S.; Gu, Y.; Mazzolari, E.; Bates, A.; Pegoiani, E.; Roifman, C.M.; Notarangelo, L.D. Variability of clinical and laboratory features among patients with ribonuclease mitochondrial RNA processing endoribonuclease gene mutations. J. Allergy Clin. Immunol. 2008, 122, 1178–1184. [Google Scholar] [CrossRef]
- Welting, T.J.; Mattijssen, S.; Peters, F.M.; van Doorn, N.L.; Dekkers, L.; van Venrooij, W.J.; Heus, H.A.; Bonafé, L.; Pruijn, G.J. Cartilage-hair hypoplasia-associated mutations in the RNase MRP P3 domain affect RNA folding and ribonucleoprotein assembly. Biochim. Biophys. Acta 2008, 1783, 455–466. [Google Scholar] [CrossRef][Green Version]
- Nakashima, E.; Tran, J.R.; Welting, T.J.; Pruijn, G.J.; Hirose, Y.; Nishimura, G.; Ohashi, H.; Schurman, S.H.; Cheng, J.; Candotti, F.; et al. Cartilage hair hypoplasia mutations that lead to RMRP promoter inefficiency or RNA transcript instability. Am. J. Med. Genet. A 2007, 143, 2675–2681. [Google Scholar] [CrossRef] [PubMed]
- Hermanns, P.; Bertuch, A.A.; Bertin, T.K.; Dawson, B.; Schmitt, M.E.; Shaw, C.; Zabel, B.; Lee, B. Consequences of mutations in the non-coding RMRP RNA in cartilage-hair hypoplasia. Hum. Mol. Genet. 2005, 14, 3723–3740. [Google Scholar] [CrossRef]
- Steinbusch, M.M.F.; Caron, M.M.J.; Surtel, D.A.M.; Friedrich, F.; Lausch, E.; Pruijn, G.J.M.; Verhesen, W.; Schroen, B.L.M.; van Rhijn, L.W.; Zabel, B.; et al. Expression of RMRP RNA is regulated in chondrocyte hypertrophy and determines chondrogenic differentiation. Sci. Rep. 2017, 7, 6440. [Google Scholar] [CrossRef]
- Steinbusch, M.M.F.; Caron, M.M.J.; Surtel, D.A.M.; van den Akker, G.G.H.; van Dijk, P.J.; Friedrich, F.; Zabel, B.; van Rhijn, L.W.; Peffers, M.J.; Welting, T.J.M. The antiviral protein viperin regulates chondrogenic differentiation via CXCL10 protein secretion. J. Biol. Chem. 2019, 294, 5121–5136. [Google Scholar] [CrossRef]
- Vakkilainen, S.; Skoog, T.; Einarsdottir, E.; Middleton, A.; Pekkinen, M.; Ohman, T.; Katayama, S.; Krjutskov, K.; Kovanen, P.E.; Varjosalo, M.; et al. The human long non-coding RNA gene RMRP has pleiotropic effects and regulates cell-cycle progression at G2. Sci. Rep. 2019, 9, 13758. [Google Scholar] [CrossRef]
- Rogler, L.E.; Kosmyna, B.; Moskowitz, D.; Bebawee, R.; Rahimzadeh, J.; Kutchko, K.; Laederach, A.; Notarangelo, L.D.; Giliani, S.; Bouhassira, E.; et al. Small RNAs derived from lncRNA RNase MRP have gene-silencing activity relevant to human cartilage-hair hypoplasia. Hum. Mol. Genet. 2014, 23, 368–382. [Google Scholar] [CrossRef]
- Bowen, P.; Conradi, G.J. Syndrome of skeletal and genitourinary anomalies with unusual facies and failure to thrive in Hutterite sibs. Birth Defects Orig. Artic. Ser. 1976, 12, 101–108. [Google Scholar]
- Lowry, R.B.; Innes, A.M.; Bernier, F.P.; McLeod, D.R.; Greenberg, C.R.; Chudley, A.E.; Chodirker, B.; Marles, S.L.; Crumley, M.J.; Loredo-Osti, J.C.; et al. Bowen-Conradi syndrome: A clinical and genetic study. Am. J. Med. Genet. A 2003, 120a, 423–428. [Google Scholar] [CrossRef]
- Armistead, J.; Khatkar, S.; Meyer, B.; Mark, B.L.; Patel, N.; Coghlan, G.; Lamont, R.E.; Liu, S.; Wiechert, J.; Cattini, P.A.; et al. Mutation of a gene essential for ribosome biogenesis, EMG1, causes Bowen-Conradi syndrome. Am. J. Hum. Genet. 2009, 84, 728–739. [Google Scholar] [CrossRef]
- Armistead, J.; Hemming, R.; Patel, N.; Triggs-Raine, B. Mutation of EMG1 causing Bowen-Conradi syndrome results in reduced cell proliferation rates concomitant with G2/M arrest and 18S rRNA processing delay. BBA Clin. 2014, 1, 33–43. [Google Scholar] [CrossRef]
- Nieminen, T.T.; O’Donohue, M.F.; Wu, Y.; Lohi, H.; Scherer, S.W.; Paterson, A.D.; Ellonen, P.; Abdel-Rahman, W.M.; Valo, S.; Mecklin, J.P.; et al. Germline mutation of RPS20, encoding a ribosomal protein, causes predisposition to hereditary nonpolyposis colorectal carcinoma without DNA mismatch repair deficiency. Gastroenterology 2014, 147, 595–598. [Google Scholar] [CrossRef]
- Broderick, P.; Dobbins, S.E.; Chubb, D.; Kinnersley, B.; Dunlop, M.G.; Tomlinson, I.; Houlston, R.S. Validation of Recently Proposed Colorectal Cancer Susceptibility Gene Variants in an Analysis of Families and Patients—A Systematic Review. Gastroenterology 2017, 152, 75–77. [Google Scholar] [CrossRef]
- Klauck, S.M.; Felder, B.; Kolb-Kokocinski, A.; Schuster, C.; Chiocchetti, A.; Schupp, I.; Wellenreuther, R.; Schmotzer, G.; Poustka, F.; Breitenbach-Koller, L.; et al. Mutations in the ribosomal protein gene RPL10 suggest a novel modulating disease mechanism for autism. Mol. Psychiatry 2006, 11, 1073–1084. [Google Scholar] [CrossRef] [PubMed]
- Brooks, S.S.; Wall, A.L.; Golzio, C.; Reid, D.W.; Kondyles, A.; Willer, J.R.; Botti, C.; Nicchitta, C.V.; Katsanis, N.; Davis, E.E. A novel ribosomopathy caused by dysfunction of RPL10 disrupts neurodevelopment and causes X-linked microcephaly in humans. Genetics 2014, 198, 723–733. [Google Scholar] [CrossRef]
- Bourque, D.K.; Hartley, T.; Nikkel, S.M.; Pohl, D.; Tetreault, M.; Kernohan, K.D.; Care4Rare Canada, C.; Dyment, D.A. A de novo mutation in RPL10 causes a rare X-linked ribosomopathy characterized by syndromic intellectual disability and epilepsy: A new case and review of the literature. Eur. J. Med. Genet. 2018, 61, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Chen, Z.; Liu, N.; Chen, Y. Ribosomal protein L10 in mitochondria serves as a regulator for ROS level in pancreatic cancer cells. Redox. Biol. 2018, 19, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Monteclaro, F.S.; Vogt, P.K. A Jun-binding protein related to a putative tumor suppressor. Proc. Natl. Acad. Sci. USA 1993, 90, 6726–6730. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.L.; Diaz, J.J.; Denoroy, L.; Madjar, J.J.; Wool, I.G. The primary structure of rat ribosomal protein L10: Relationship to a Jun-binding protein and to a putative Wilms’ tumor suppressor. Biochem. Biophys. Res. Commun. 1996, 225, 952–956. [Google Scholar] [CrossRef]
- Damianov, A.; Kann, M.; Lane, W.S.; Bindereif, A. Human RBM28 protein is a specific nucleolar component of the spliceosomal snRNPs. Biol. Chem. 2006, 387, 1455–1460. [Google Scholar] [CrossRef]
- Chagnon, P.; Michaud, J.; Mitchell, G.; Mercier, J.; Marion, J.F.; Drouin, E.; Rasquin-Weber, A.; Hudson, T.J.; Richter, A. A missense mutation (R565W) in cirhin (FLJ14728) in North American Indian childhood cirrhosis. Am. J. Hum. Genet. 2002, 71, 1443–1449. [Google Scholar] [CrossRef]
- Freed, E.F.; Prieto, J.L.; McCann, K.L.; McStay, B.; Baserga, S.J. NOL11, implicated in the pathogenesis of North American Indian childhood cirrhosis, is required for pre-rRNA transcription and processing. PLoS Genet. 2012, 8, e1002892. [Google Scholar] [CrossRef]
- Bolze, A.; Mahlaoui, N.; Byun, M.; Turner, B.; Trede, N.; Ellis, S.R.; Abhyankar, A.; Itan, Y.; Patin, E.; Brebner, S.; et al. Ribosomal protein SA haploinsufficiency in humans with isolated congenital asplenia. Science 2013, 340, 976–978. [Google Scholar] [CrossRef]
- Marneros, A.G. BMS1 is mutated in aplasia cutis congenita. PLoS Genet. 2013, 9, e1003573. [Google Scholar] [CrossRef]
- Paolini, N.A.; Attwood, M.; Sondalle, S.B.; Vieira, C.; van Adrichem, A.M.; di Summa, F.M.; O’Donohue, M.F.; Gleizes, P.E.; Rachuri, S.; Briggs, J.W.; et al. A Ribosomopathy Reveals Decoding Defective Ribosomes Driving Human Dysmorphism. Am. J. Hum. Genet. 2017, 100, 506–522. [Google Scholar] [CrossRef]
- Jenkinson, E.M.; Rodero, M.P.; Kasher, P.R.; Uggenti, C.; Oojageer, A.; Goosey, L.C.; Rose, Y.; Kershaw, C.J.; Urquhart, J.E.; Williams, S.G.; et al. Mutations in SNORD118 cause the cerebral microangiopathy leukoencephalopathy with calcifications and cysts. Nat. Genet. 2016, 48, 1185–1192. [Google Scholar] [CrossRef] [PubMed]
- Arancio, W.; Genovese, S.I.; Bongiovanni, L.; Tripodo, C. A ceRNA approach may unveil unexpected contributors to deletion syndromes, the model of 5q− syndrome. Oncoscience 2015, 2, 872–879. [Google Scholar] [CrossRef] [PubMed]
- Falini, B.; Nicoletti, I.; Martelli, M.F.; Mecucci, C. Acute myeloid leukemia carrying cytoplasmic/mutated nucleophosmin (NPMc + AML): Biologic and clinical features. Blood 2007, 109, 874–885. [Google Scholar] [CrossRef] [PubMed]
- De Keersmaecker, K.; Atak, Z.K.; Li, N.; Vicente, C.; Patchett, S.; Girardi, T.; Gianfelici, V.; Geerdens, E.; Clappier, E.; Porcu, M.; et al. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat. Genet. 2013, 45, 186–190. [Google Scholar] [CrossRef]
- Ajore, R.; Raiser, D.; McConkey, M.; Jöud, M.; Boidol, B.; Mar, B.; Saksena, G.; Weinstock, D.M.; Armstrong, S.; Ellis, S.R.; et al. Deletion of ribosomal protein genes is a common vulnerability in human cancer, especially in concert with TP53 mutations. EMBO Mol. Med. 2017, 9, 498–507. [Google Scholar] [CrossRef]
- Gong, J.; Li, Y.; Liu, C.J.; Xiang, Y.; Li, C.; Ye, Y.; Zhang, Z.; Hawke, D.H.; Park, P.K.; Diao, L.; et al. A Pan-cancer Analysis of the Expression and Clinical Relevance of Small Nucleolar RNAs in Human Cancer. Cell Rep. 2017, 21, 1968–1981. [Google Scholar] [CrossRef]
- Penzo, M.; Clima, R.; Trerè, D.; Montanaro, L. Separated Siamese Twins: Intronic Small Nucleolar RNAs and Matched Host Genes May be Altered in Conjunction or Separately in Multiple Cancer Types. Cells 2020, 9, 387. [Google Scholar] [CrossRef]
- Falini, B.; Mecucci, C.; Tiacci, E.; Alcalay, M.; Rosati, R.; Pasqualucci, L.; La Starza, R.; Diverio, D.; Colombo, E.; Santucci, A.; et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N. Engl. J. Med. 2005, 352, 254–266. [Google Scholar] [CrossRef]
- Ljungström, V.; Cortese, D.; Young, E.; Pandzic, T.; Mansouri, L.; Plevova, K.; Ntoufa, S.; Baliakas, P.; Clifford, R.; Sutton, L.A.; et al. Whole-exome sequencing in relapsing chronic lymphocytic leukemia: Clinical impact of recurrent RPS15 mutations. Blood 2016, 127, 1007–1016. [Google Scholar] [CrossRef]
| Congenital/Inherited Ribosomopathies | ||||
|---|---|---|---|---|
| Disease | Gene Mutated | Role in Ribosome Biogenesis | Clinical Manifestations | Type of Ribosomopathy |
| Diamond Blackfan anemia | RPS19, RPS26, RPS17, RPS29 [30] RPS28 [31] RPS24, RPL5, RPL11 [32] RPL35, RPL18 [33] RPL26 [34] RPL15 [35] RPS27, RPL27 [36] | 40S and 60S subunits protein | Macrocytic anemia, skeletal abnormalities, short stature, cardiac and genitourinary malformations, cancer predisposition | Pure |
| Shwachman-Diamond syndrome | SBDS [7] DNAJC21 [37] EFL1 [38] SRP54 [39] | Assembly of 60S and 40S subunits in active 80S ribosomes | Bone marrow failure, skeletal dysplasia, cognitive impairment, and risk of developing myelodysplastic syndrome | Pure |
| Treacher Collins syndrome | TCOF1 [40] POLR1C, POLR1D [41] POLR1B [42] | Ribosomal RNA transcription | Severe craniofacial defects and mental retardation | Pure |
| Cartilage Hair Hypoplasia-Anauxetic dysplasia spectrum | RMRP [8] POP1 [43] | Ribosomal RNA processing | Short-limbed dwarfism, sparse hypoplastic hair, immunodeficiency, hypoplastic anemia, and predisposition to cancer | Mixed |
| Dyskeratosis Congenita | DKC1 [4,44] PARN [45,46] NHP2, NOP10 [47,48,49] NPM1 [50] | Ribosomal RNA pseudouridylation and processing | Abnormal skin pigmentation, dystrophy of the nails, oral leukoplakia, bone marrow failure, and cancer predisposition | Mixed |
| Recently Identified Ribosomopathies | |||
|---|---|---|---|
| Disease | Gene Mutated | Role in Ribosome Biogenesis | Clinical Manifestations |
| Alopecia, neurologic defects, and endocrinopathy syndrome | RBM28 [126] | Ribosomal RNA processing | Alopecia, mental retardation, progressive motor deterioration, central hypogonadotropic hypogonadism and short stature, microcephaly, gynecomastia, and hypodontia |
| North American Indian Childhood Cirrhosis | CIRHIN [127] NOL11 [128] | 18S rRNA processing | Transient neonatal jaundice that evolves into biliary cirrhosis requiring hepatic transplantation |
| Bowen-Conradi syndrome | EMG1 [116] | Ribosome assembly | Mental retardation, microcephaly, micrognathia, rocker bottom feet, and flexion contractures of the joints; causes early death |
| Familial colorectal cancer type X | RPS20 [118] | 40S subunit protein | Hereditary colorectal cancer without mutations in mismatch repair genes |
| Congenital asplenia | RPSA [129] | 40S subunit protein | Absence of spleen |
| Aplasia cutis congenita | BMS1 [130] | Ribosomal GTPase, 18S rRNA processing | Skin defect and alopecia of the scalp |
| RPS23-related ribosomopathy | RPS23 [131] | 40S subunit protein | Microcephaly, hearing loss, dysmorphic features, intellectual disability, and autism spectrum disorder |
| Leukoencephalopathy, intracranial calcifications, and cysts (LCC) | SNORD118 [132] | C/D box snoRNA U8 involved in ribosome biogenesis | Neurological disorder with leukoencephalopathy, intracranial calcifications, and cysts |
| Autism | RPL10 [120] | 60S subunit protein | Autism spectrum disorder |
| Microcephaly | RPL10 [121,122] | 60S subunit protein | Microcephaly, intellectual disability, epilepsy, and growth retardation |
| Acquired Ribosomopathies | |||
|---|---|---|---|
| Disease | Gene Mutated | Role in Ribosome Biogenesis | Clinical Manifestations |
| 5q− syndrome | RPS14 [11] | 40S subunit protein | Macrocytic anemia and erythroid hypoplasia; may progress to AML |
| Acute myeloid leukemia (AML) | NPM1 [139] | Ribosome processing | AML with normal karyotype |
| Pediatric acute lymphoblastic leukemia (T-ALL) | RPL5, RPL10, RPL22 [135] | 60S subunit proteins | T-ALL |
| Relapsed CLL | RPS15, RPSA, RPS20 [140] | 40S subunit proteins | Relapse after first-line treatment |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Venturi, G.; Montanaro, L. How Altered Ribosome Production Can Cause or Contribute to Human Disease: The Spectrum of Ribosomopathies. Cells 2020, 9, 2300. https://doi.org/10.3390/cells9102300
Venturi G, Montanaro L. How Altered Ribosome Production Can Cause or Contribute to Human Disease: The Spectrum of Ribosomopathies. Cells. 2020; 9(10):2300. https://doi.org/10.3390/cells9102300
Chicago/Turabian StyleVenturi, Giulia, and Lorenzo Montanaro. 2020. "How Altered Ribosome Production Can Cause or Contribute to Human Disease: The Spectrum of Ribosomopathies" Cells 9, no. 10: 2300. https://doi.org/10.3390/cells9102300
APA StyleVenturi, G., & Montanaro, L. (2020). How Altered Ribosome Production Can Cause or Contribute to Human Disease: The Spectrum of Ribosomopathies. Cells, 9(10), 2300. https://doi.org/10.3390/cells9102300






