Cardiovascular Risk Factors and Differential Transcriptomic Profile of the Subcutaneous and Visceral Adipose Tissue and Their Resident Stem Cells
Abstract
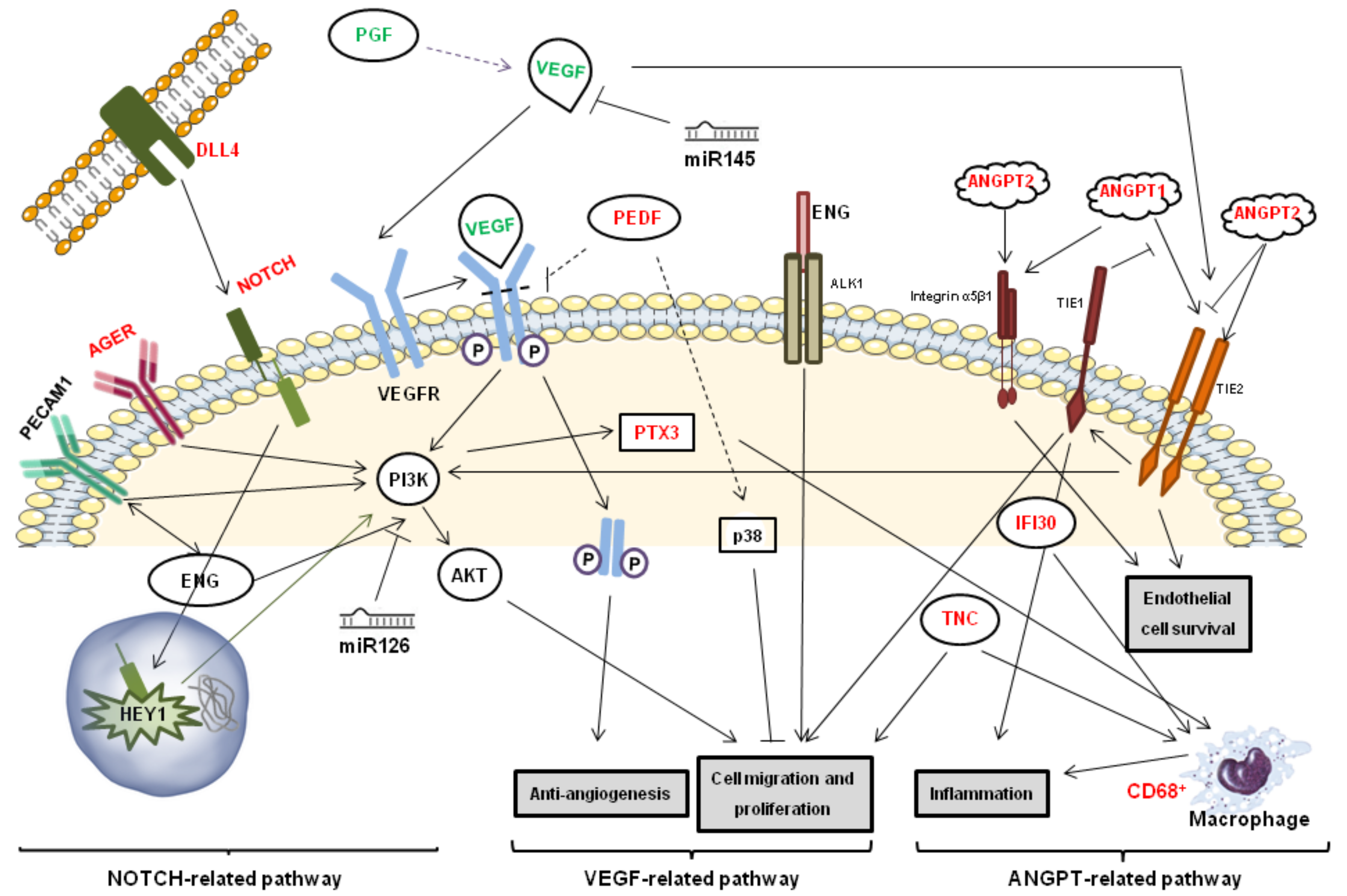
1. Introduction
2. Materials and Methods
2.1. Patient Recruitment and Adipose Tissue Sampling
2.2. RNA and cDNA Isolation from Adipose Tissue
2.3. Transcriptomic Analysis and RT-PCR
2.4. Bioinformatic Analysis
2.5. ASCs Isolation and Characterization
2.6. MTS Viability/Proliferation Analysis
2.7. Microvesicles Isolation
2.8. Cell Migration
2.9. Gene Expression Analysis from ASCs
2.10. Western Blot Analysis
2.11. Statistical Analysis
3. Results
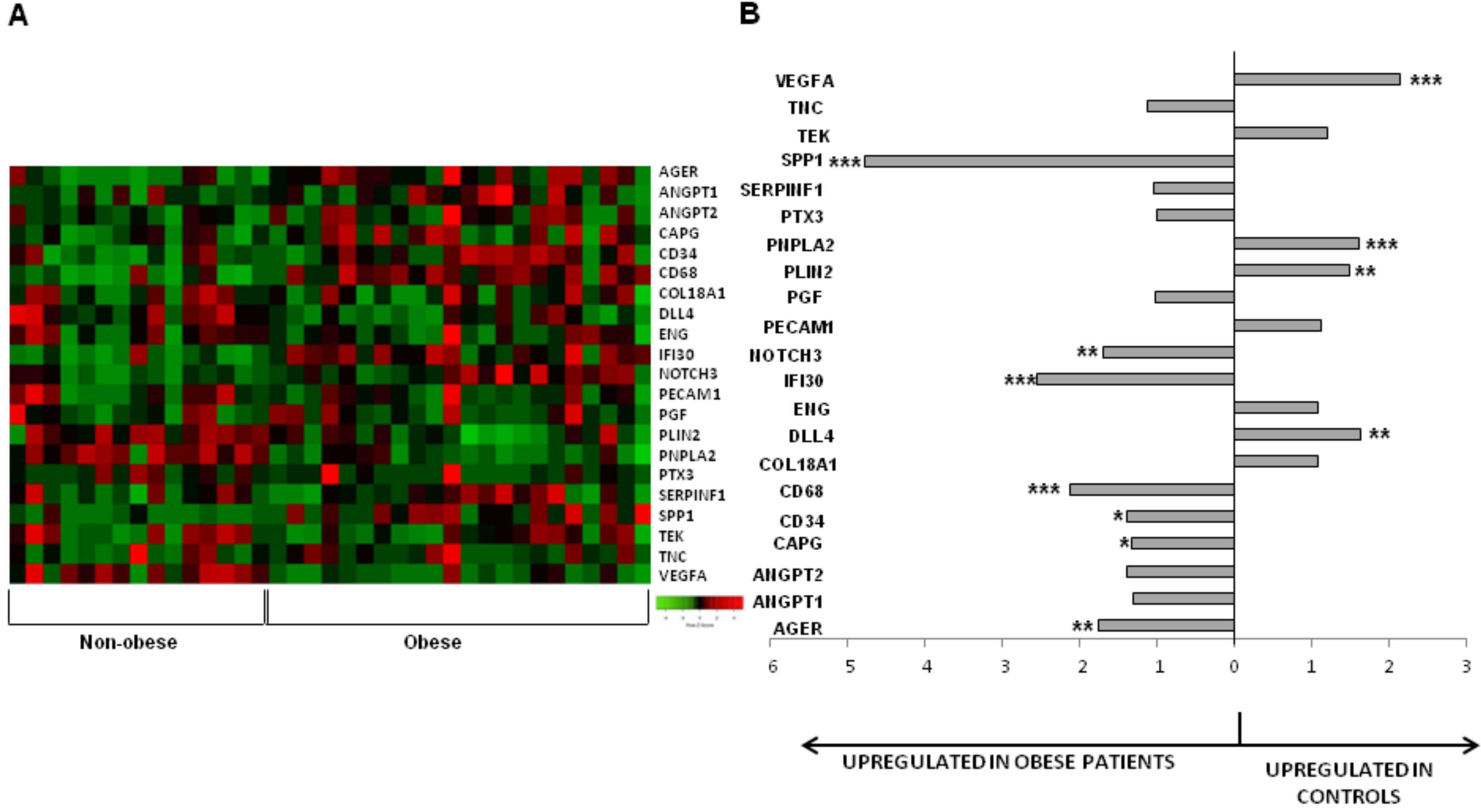
3.1. Influence of Obesity on the Subcutaneous Adipose Tissue
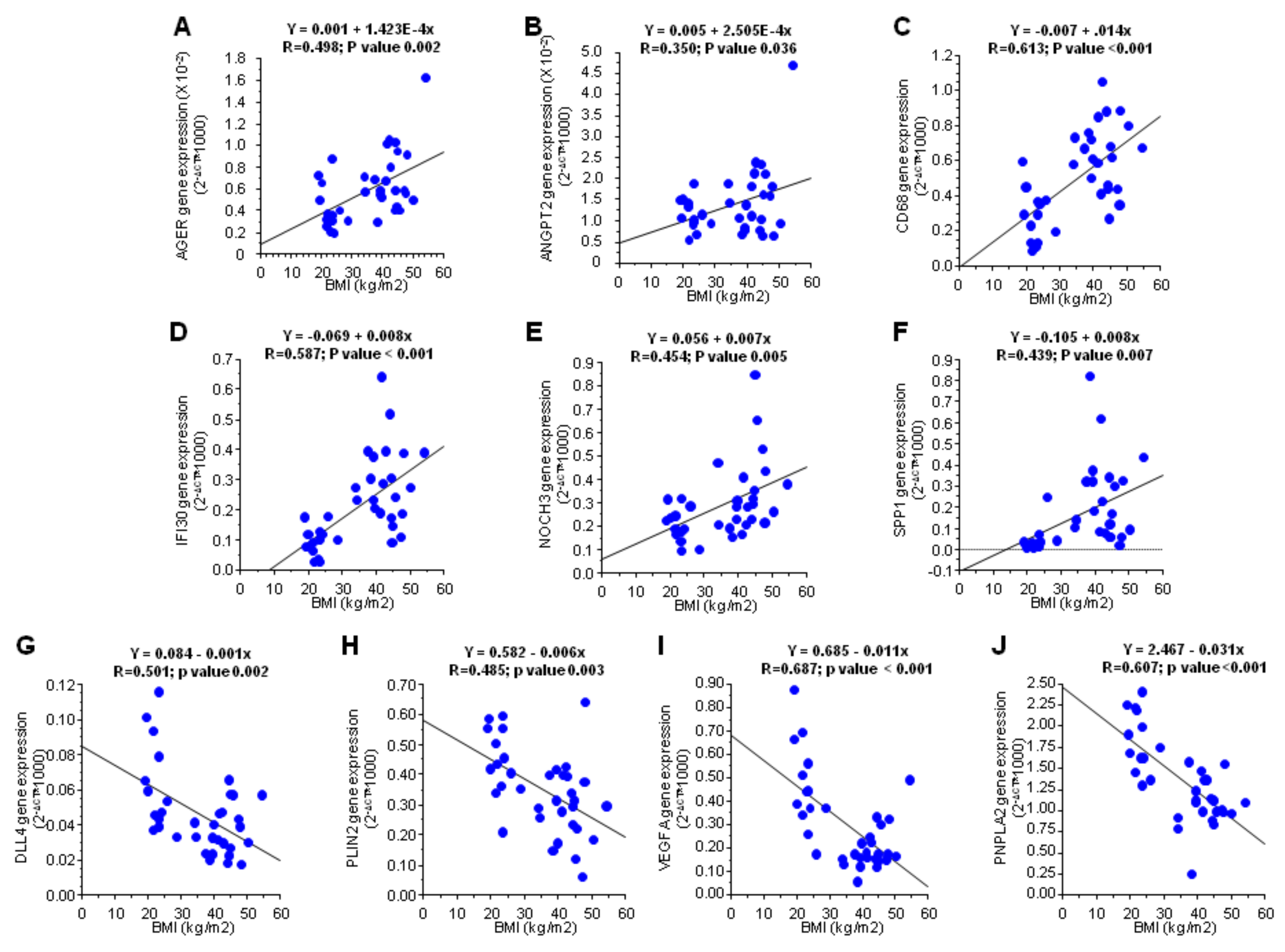
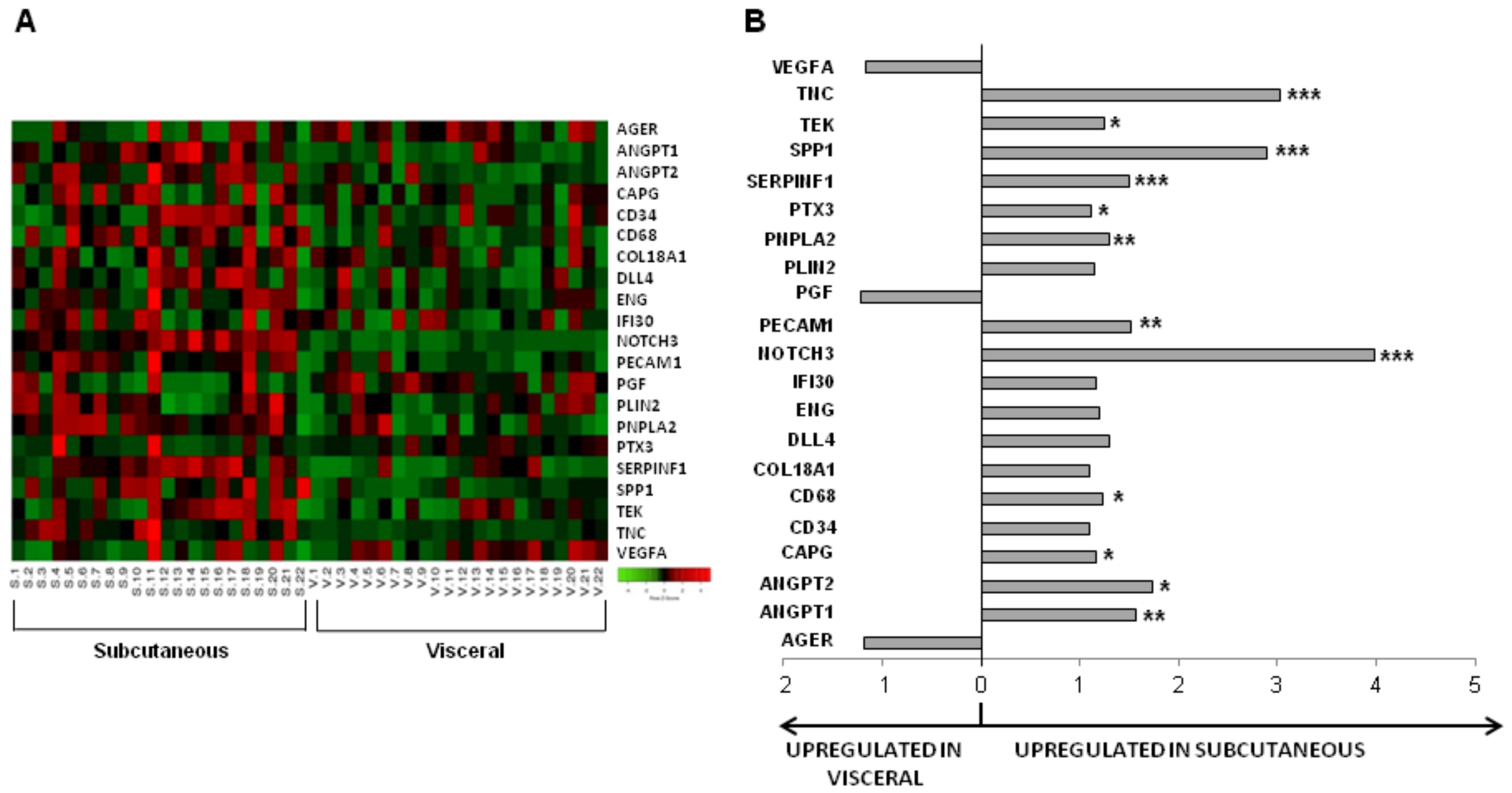
3.2. Influence of Obesity in Fat Depot Genomic Profile
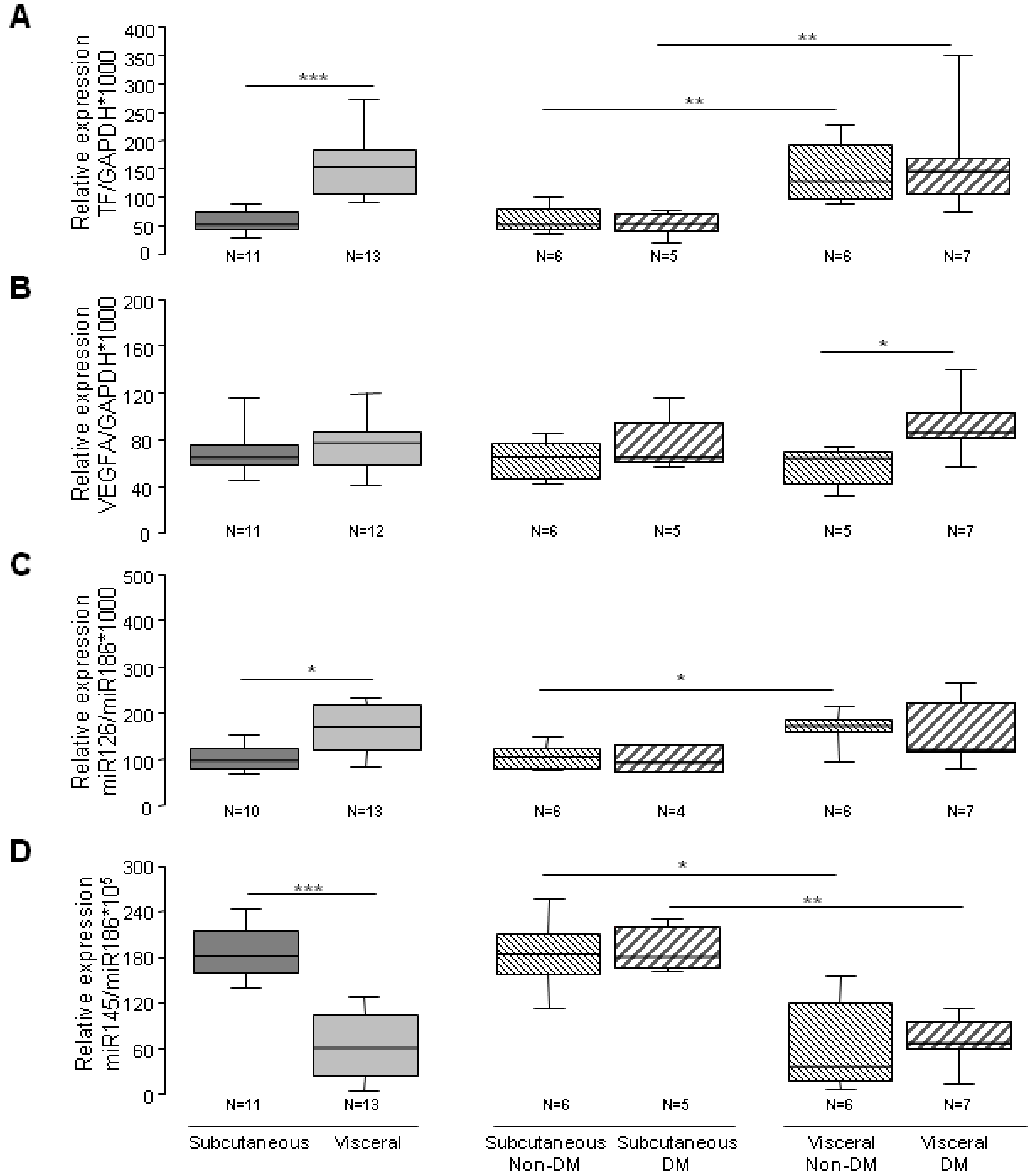
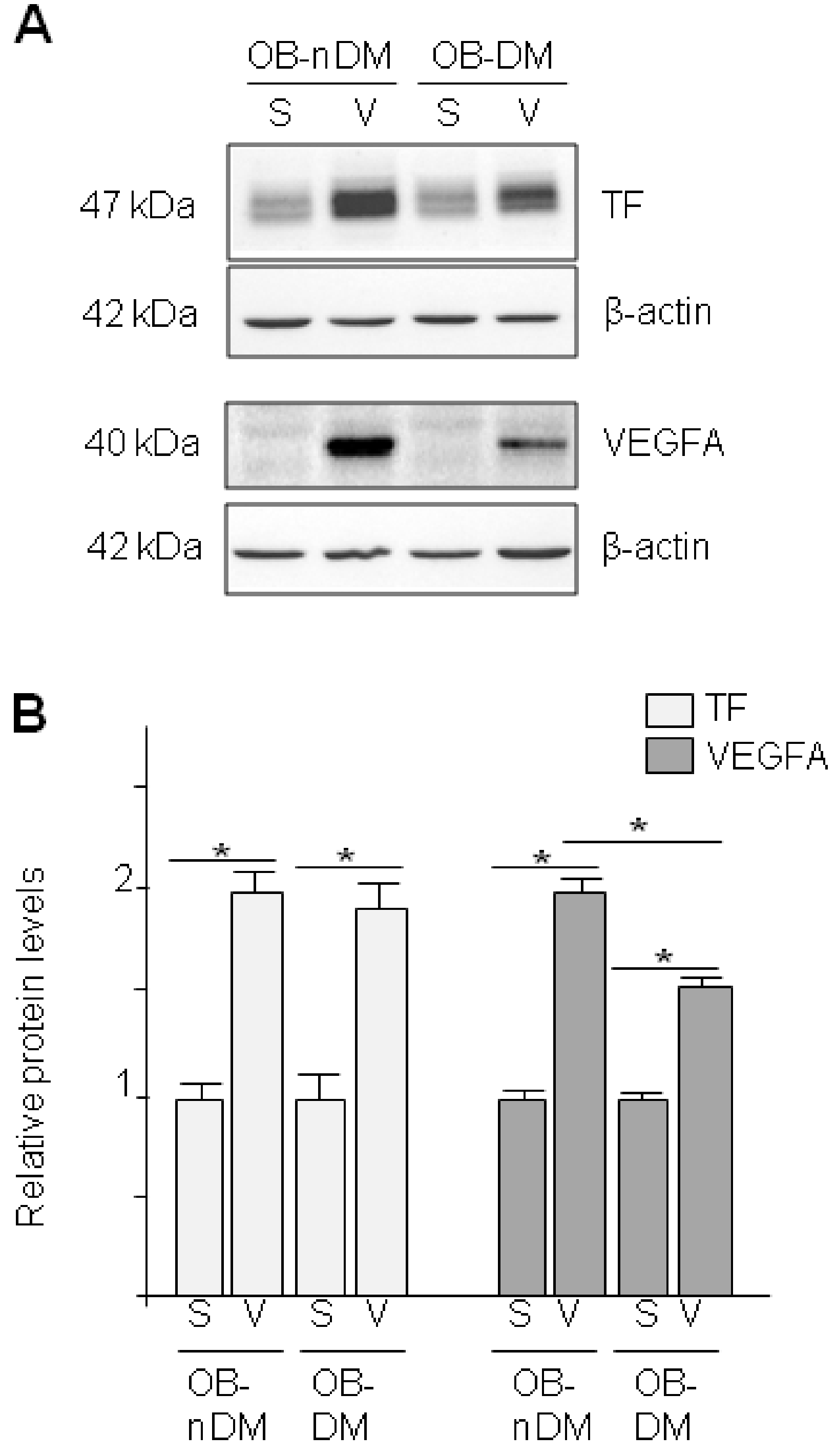
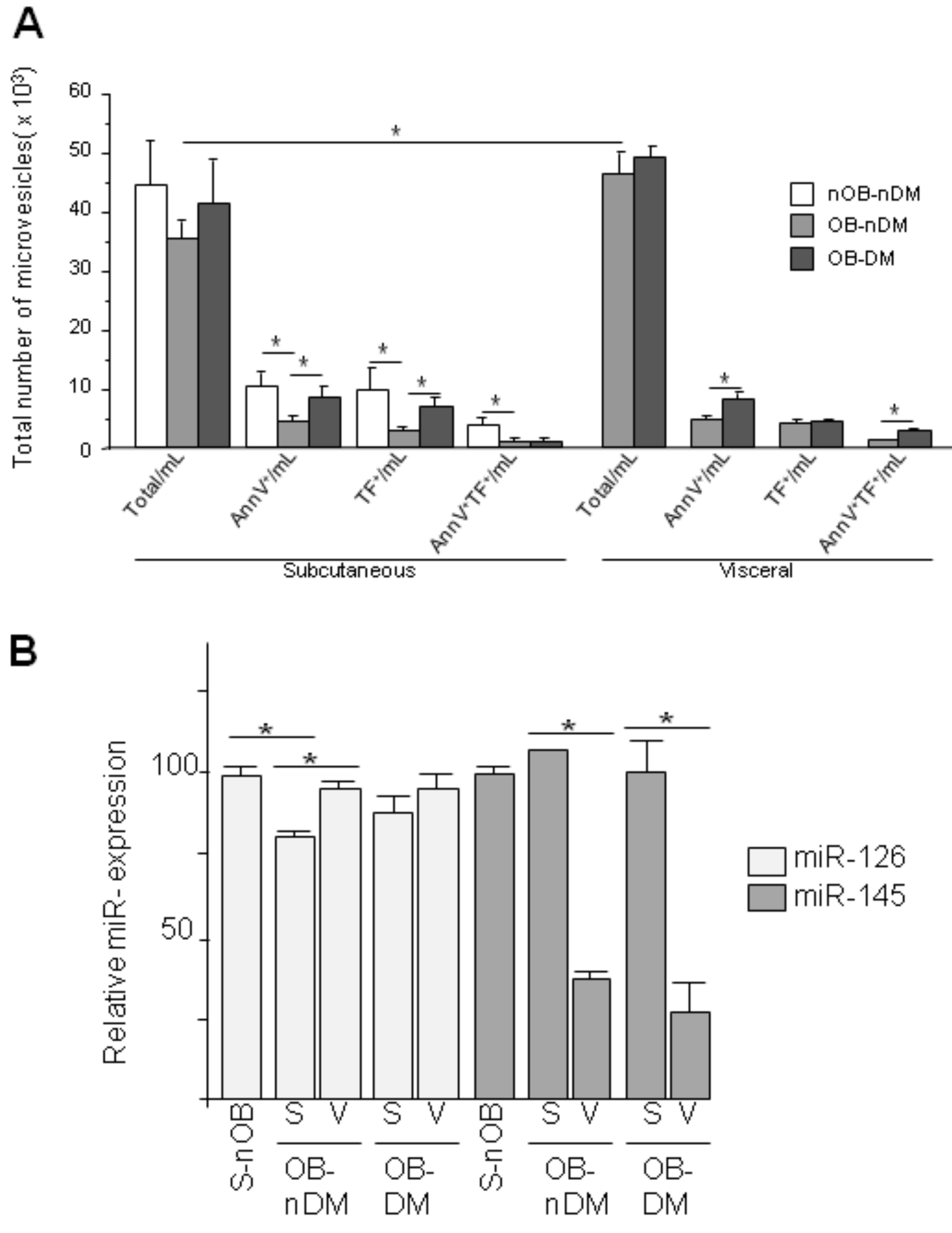
3.3. Angiogenic Related Gene Expression in Subcutaneous or Visceral Derived ASCs from Diabetic/No Diabetic Patients with Obesity
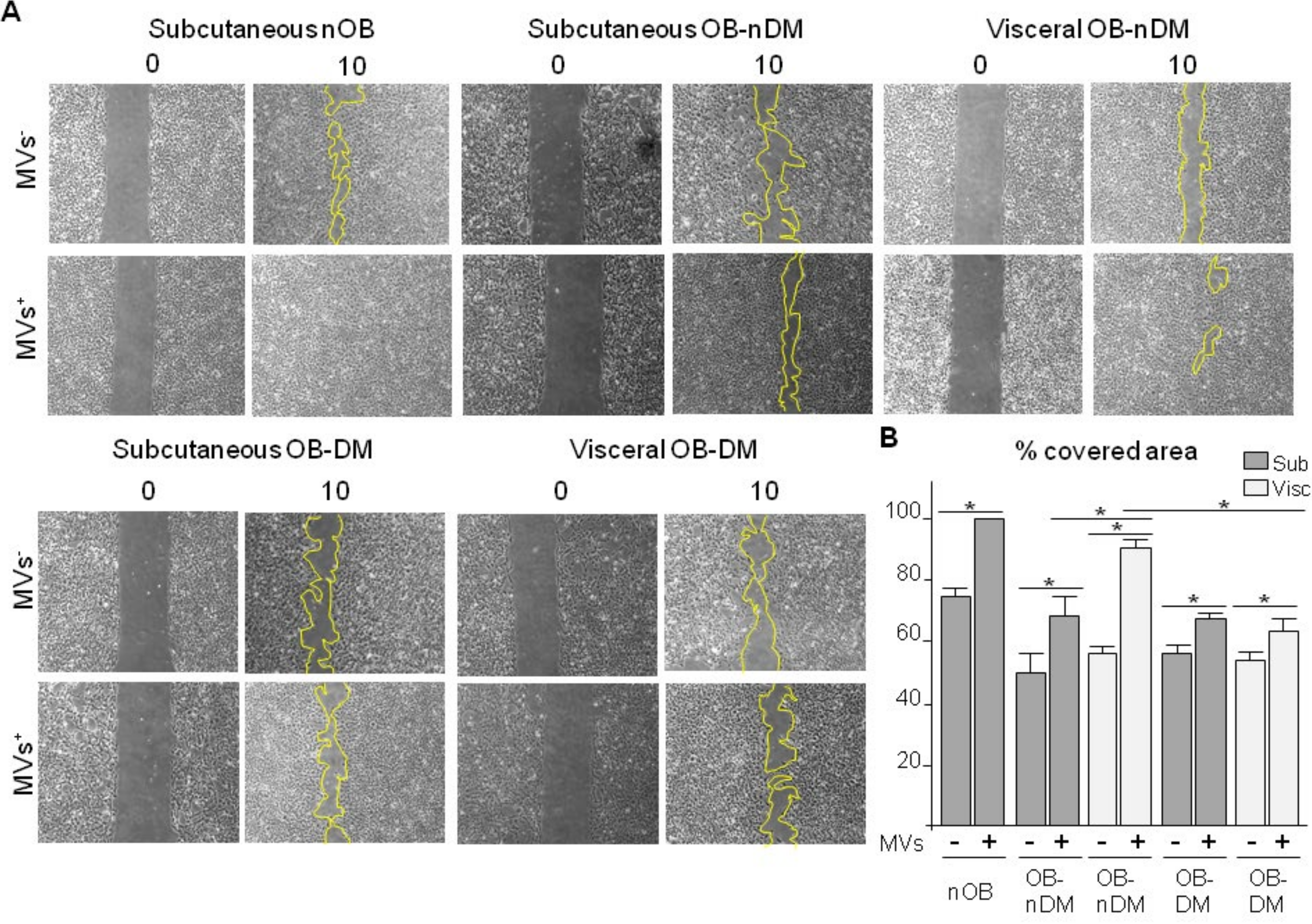
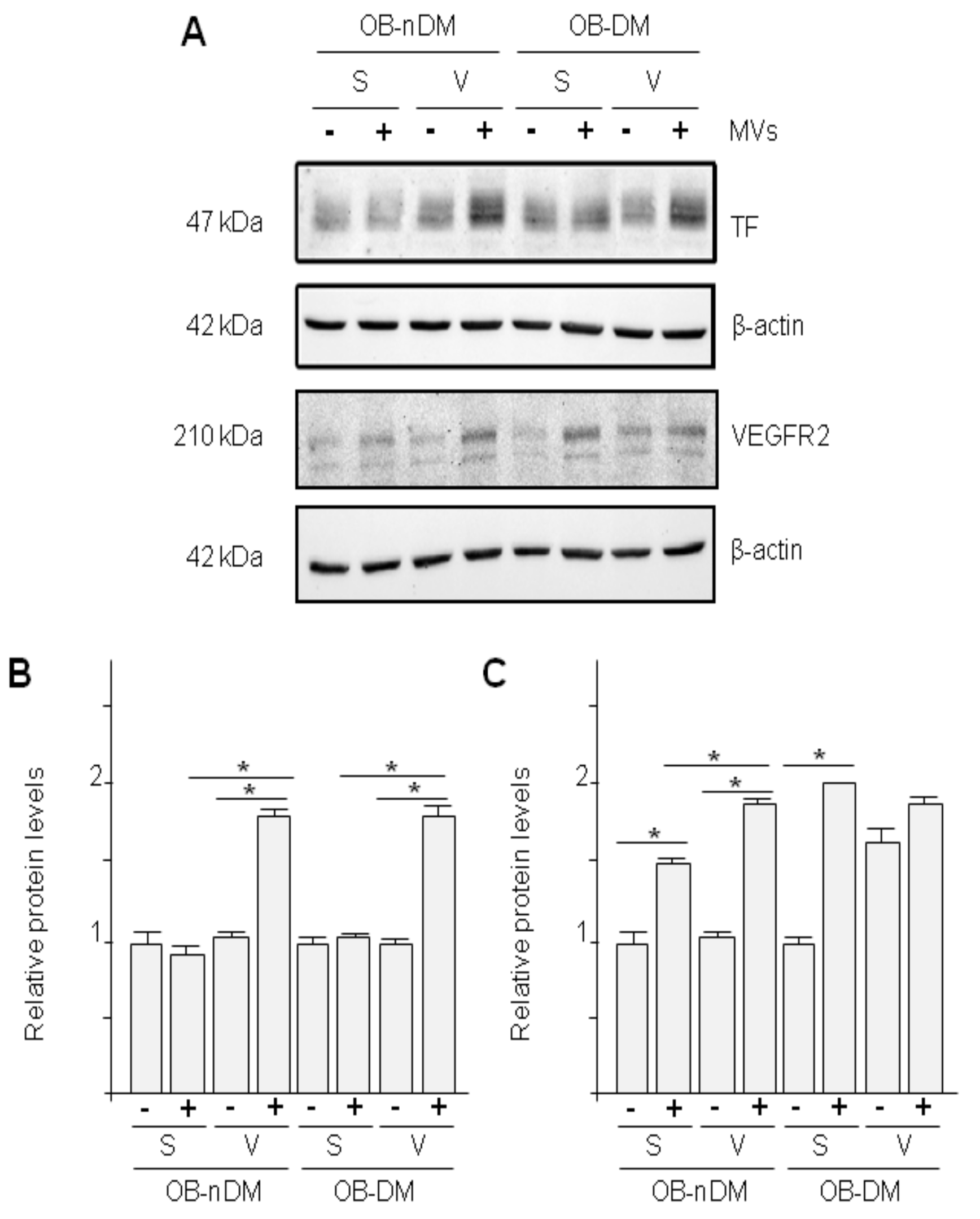
3.4. Subcutaneous and Visceral ASCs Derived Microvesicles and Effects on Wound Repair
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ASCs | Adipose derived stem cells |
| AT | Adipose tissue |
| CVRFs | Cardiovascular risk factors |
| DLP | Dyslipidemia |
| DM | Diabetes mellitus |
| HT | Hypertension |
| MVs | Microvesicles |
| OB | Obesity |
| S- | Subcutaneous |
| SAT | Subcutaneous adipose tissue |
| SAT-ASCs | Adipose derived stem cells obtained from subcutaneous adipose tissue |
| S-MVs | Microvesicles released by adipose derived stem cells obtained from subcutaneous adipose tissue |
| V- | Visceral |
| VAT | Visceral adipose tissue |
| VAT-ASCs | Adipose derived stem cells obtained from visceral adipose tissue |
| V-MVs | Microvesicles released by adipose derived stem cells obtained from visceral adipose tissue |
References
- Ailhaud, G.; Grimaldi, P.; Negrel, R. Cellular and molecular aspects of adipose tissue development. Annu. Rev. Nutr. 1992, 12, 207–233. [Google Scholar] [CrossRef]
- Smitka, K.; Maresova, D. Adipose Tissue as an Endocrine Organ: An Update on Pro-inflammatory and Anti-inflammatory Microenvironment. Prague Med. Rep. 2015, 116, 87–111. [Google Scholar] [CrossRef]
- Ley, C.; Lees, J.B.; Stevenson, J.C. Sex- and menopause-associated changes in body-fat distribution. Am. J. Clin. Nutr. 1992, 5, 950–954. [Google Scholar] [CrossRef]
- Karpe, F.; Pinnick, K.E. Biology of upper-body and lower-body adipose tissue—Link to whole-body phenotypes. Nat. Rev. Endocrinol. 2015, 11, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Ledoux, S.; Queguiner, I.; Msika, S.; Calderari, S.; Rufat, P.; Gasc, J.M.; Corvol, P.; Larger, E. Angiogenesis associated with visceral and subcutaneous adipose tissue in severe human obesity. Diabetes 2008, 57, 3247–3257. [Google Scholar] [CrossRef] [PubMed]
- Silverman, K.J.; Lund, D.P.; Zetter, B.R.; Lainey, L.L.; Shahood, J.A.; Freiman, D.G.; Folkman, J.; Barger, A.C. Angiogenic activity of adipose tissue. Biochem. Biophys. Res. Commun. 1988, 153, 347–352. [Google Scholar] [CrossRef]
- Fantuzzi, G.; Mazzone, T. Adipose tissue and atherosclerosis: Exploring the connection. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 996–1003. [Google Scholar] [CrossRef] [PubMed]
- Larsson, B.; Svärdsudd, K.; Welin, L.; Wilhelmsen, L.; Björntorp, P.; Tibblin, G. Abdominal adipose tissue distribution, obesity, and risk of cardiovascular disease and death: 13 year follow up of participants in the study of men born in 1913. Br. Med. J. Clin. Res. Ed. 1984, 288, 1401–1404. [Google Scholar] [CrossRef] [PubMed]
- Okura, T.; Nakata, Y.; Yamabuki, K.; Tanaka, K. Regional body composition changes exhibit opposing effects on coronary heart disease risk factors. Arterioscler. Thromb. Vasc. Biol. 2004, 24, 923–929. [Google Scholar] [CrossRef]
- Kranendonk, M.E.; van Herwaarden, J.A.; Stupkova, T.; de Jager, W.; Vink, K.; Moll, F.L.; Kalkhoven, E.; Visseren, F.L.J. Inflammatory characteristics of distinct abdominal adipose tissue depots relate differently to metabolic risk factors for cardiovascular disease: Distinct fat depots and vascular risk factors. Atherosclerosis 2015, 239, 419–427. [Google Scholar] [CrossRef]
- Gil, A.; Olza, J.; Gil-Campos, M.; Gomez-Llorente, C.; Aguilera, C.M. Is adipose tissue metabolically different at different sites? Int. J. Pediatr. Obes. 2011, 6 (Suppl. S1), 13–20. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.M. Subcutaneous and visceral adipose tissue: Structural and functional differences. Obes. Rev. 2010, 11, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Esteve Rafols, M. Adipose tissue: Cell heterogeneity and functional diversity. Endocrinol. Nutr. 2014, 61, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Zuk, P.A.; Zhu, M.; Ashjian, P.; De Ugarte, D.A.; Huang, J.I.; Mizuno, H.; Alfonso, Z.C.; Fraser, J.K.; Benhaim, P.; Hedrick, M.H. Human adipose tissue is a source of multipotent stem cells. Mol. Biol. Cell 2002, 13, 4279–4295. [Google Scholar] [CrossRef]
- Sadie-Van Gijsen, H.; Crowther, N.J.; Hough, F.S.; Ferris, W.F. Depot-specific differences in the insulin response of adipose-derived stromal cells. Mol. Cell Endocrinol. 2010, 328, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, M.; Matsubara, Y.; Lin, K.; Sugimachi, K.; Furue, M. Characterization and comparison of adipose tissue-derived cells from human subcutaneous and omental adipose tissues. Cell Biochem. Funct. 2009, 27, 440–447. [Google Scholar] [CrossRef]
- Kim, B.; Lee, B.; Kim, M.K.; Gong, S.P.; Park, H.Y.; Chung, Y.Y.; Kim, H.S.; No, J.H.; Park, W.Y.; Park, A.K.; et al. Gene expression profiles of human subcutaneous and visceral adipose-derived stem cells. Cell Biochem. Funct. 2016, 34, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Gimble, J.M.; Katz, A.J.; Bunnell, B.A. Adipose-derived stem cells for regenerative medicine. Circ. Res. 2007, 100, 1249–1260. [Google Scholar] [CrossRef]
- Schaffler, A.; Buchler, C. Concise review: Adipose tissue-derived stromal cells--basic and clinical implications for novel cell-based therapies. Stem. Cells 2007, 25, 818–827. [Google Scholar] [CrossRef]
- Nakagami, H.; Maeda, K.; Morishita, R.; Iguchi, S.; Nishikawa, T.; Takami, Y.; Kikuchi, Y.; Saito, Y.; Tamai, K.; Ogihara, T.; et al. Novel autologous cell therapy in ischemic limb disease through growth factor secretion by cultured adipose tissue-derived stromal cells. Arterioscler. Thromb. Vasc. Biol. 2005, 25, 2542–2547. [Google Scholar] [CrossRef]
- Cherubino, M.; Rubin, J.P.; Miljkovic, N.; Kelmendi-Doko, A.; Marra, K.G. Adipose-derived stem cells for wound healing applications. Ann. Plast. Surg. 2011, 66, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Onate, B.; Vilahur, G.; Ferrer-Lorente, R.; Ybarra, J.; Díez-Caballero, A.; Ballesta-López, C.; Moscatiello, F.; Herrero, J.; Badimon, L. The subcutaneous adipose tissue reservoir of functionally active stem cells is reduced in obese patients. FASEB J. 2012, 26, 4327–4336. [Google Scholar] [CrossRef] [PubMed]
- Perez, L.M.; Bernal, A.; San Martín, N.; Gálvez, B.G. Obese-derived ASCs show impaired migration and angiogenesis properties. Arch. Physiol. Biochem. 2013, 119, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Camussi, G.; Deregibus, M.C.; Bruno, S.; Cantaluppi, V.; Biancone, L. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int. 2010, 78, 838–848. [Google Scholar] [CrossRef]
- Kang, T.; Jones, T.M.; Naddell, C.; Bacanamwo, M.; Calvert, J.W.; Thompson, W.E.; Bond, V.C.; Chen, Y.E.; Liu, D. Adipose-Derived Stem Cells Induce Angiogenesis via Microvesicle Transport of miRNA-31. Stem Cells Transl. Med. 2016, 5, 440–450. [Google Scholar] [CrossRef]
- Lambert, C.; Arderiu, G.; Bejar, M.T.; Crespo, J.; Baldellou, M.; Juan-Babot, O.; Badimon, L. Stem cells from human cardiac adipose tissue depots show different gene expression and functional capacities. Stem Cell Res. Ther. 2019, 10, 361. [Google Scholar] [CrossRef]
- Ferrer-Lorente, R.; Bejar, M.T.; Badimon, L. Notch signaling pathway activation in normal and hyperglycemic rats differs in the stem cells of visceral and subcutaneous adipose tissue. Stem Cells Dev. 2014, 23, 3034–3048. [Google Scholar] [CrossRef]
- Ferrer-Lorente, R.; Bejar, M.T.; Tous, M.; Vilahur, G.; Badimon, L. Systems biology approach to identify alterations in the stem cell reservoir of subcutaneous adipose tissue in a rat model of diabetes: Effects on differentiation potential and function. Diabetologia 2014, 57, 246–256. [Google Scholar] [CrossRef]
- Bejar, M.T.; Ferrer-Lorente, R.; Peña, E.; Badimon, L. Inhibition of Notch rescues the angiogenic potential impaired by cardiovascular risk factors in epicardial adipose stem cells. FASEB J. 2016, 30, 2849–2859. [Google Scholar] [CrossRef]
- Onate, B.; Vilahur, G.; Camino-López, S.; Díez-Caballero, A.; Ballesta-López, C.; Ybarra, J.; Moscatiello, F.; Herrero, J.; Badimon, L. Stem cells isolated from adipose tissue of obese patients show changes in their transcriptomic profile that indicate loss in stemcellness and increased commitment to an adipocyte-like phenotype. BMC Genom. 2013, 14, 625. [Google Scholar] [CrossRef]
- Arderiu, G.; Peña, E.; Aledo, R.; Juan-Babot, O.; Crespo, J.; Vilahur, G.; Oñate, B.; Moscatiello, F.; Badimon, L. MicroRNA-145 Regulates the Differentiation of Adipose Stem Cells Toward Microvascular Endothelial Cells and Promotes Angiogenesis. Circ. Res. 2019, 125, 74–89. [Google Scholar] [CrossRef] [PubMed]
- Fish, J.E.; Santoro, M.M.; Morton, S.U.; Yu, S.; Yeh, R.F.; Wythe, J.D.; Ivey, K.N.; Bruneau, B.G.; Stainier, D.Y.; Srivastava, D. miR-126 regulates angiogenic signaling and vascular integrity. Dev. Cell 2008, 15, 272–284. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.; Ma, K.; Zhang, C.; Fu, X. Therapeutic angiogenesis using stem cell-derived extracellular vesicles: An emerging approach for treatment of ischemic diseases. Stem Cell Res. Ther. 2019, 10, 158. [Google Scholar] [CrossRef] [PubMed]
- Olshansky, S.J.; Passaro, D.J.; Hershow, R.C.; Layden, J.; Carnes, B.A.; Brody, J.; Hayflick, L.; Butler, R.N.; Allison, D.B.; Ludwig, D.S. A potential decline in life expectancy in the United States in the 21st century. N. Engl. J. Med. 2005, 352, 1138–1145. [Google Scholar] [CrossRef] [PubMed]
- Badimon, L.; Onate, B.; Vilahur, G. Adipose-derived Mesenchymal Stem Cells and Their Reparative Potential in Ischemic Heart Disease. Rev. Esp. Cardiol. Engl. Ed. 2015, 68, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Becerra, S.P.; Notario, V. The effects of PEDF on cancer biology: Mechanisms of action and therapeutic potential. Nat. Rev. Cancer 2013, 13, 258–271. [Google Scholar] [CrossRef]
- Cai, J.; Chen, Z.; Ruan, Q.; Han, S.; Liu, L.; Qi, X.; Boye, S.L.; Hauswirth, W.W.; Grant, M.B.; Boulton, M.E. gamma-Secretase and presenilin mediate cleavage and phosphorylation of vascular endothelial growth factor receptor-1. J. Biol. Chem. 2011, 286, 42514–42523. [Google Scholar] [CrossRef]
- He, X.; Cheng, R.; Benyajati, S.; Ma, J.X. PEDF and its roles in physiological and pathological conditions: Implication in diabetic and hypoxia-induced angiogenic diseases. Clin. Sci. 2015, 128, 805–823. [Google Scholar] [CrossRef]
- Hosseinzadeh-Attar, M.J.; Mahdavi-Mazdeh, M.; Yaseri, M.; Zahed, N.S.; Alipoor, E. Comparative Assessment of Serum Adipokines Zinc-alpha2-glycoprotein and Adipose Triglyceride Lipase, and Cardiovascular Risk Factors Between Normal Weight and Obese Patients with Hemodialysis. Arch. Med. Res. 2017, 48, 459–466. [Google Scholar] [CrossRef]
- Mairal, A.; Langin, D.; Arner, P.; Hoffstedt, J. Human adipose triglyceride lipase (PNPLA2) is not regulated by obesity and exhibits low in vitro triglyceride hydrolase activity. Diabetologia 2006, 49, 1629–1636. [Google Scholar] [CrossRef]
- Steinberg, G.R.; Kemp, B.E.; Watt, M.J. Adipocyte triglyceride lipase expression in human obesity. Am. J. Physiol. Endocrinol. Metab. 2007, 293, E958–E964. [Google Scholar] [CrossRef] [PubMed]
- Schramme, A.; Mussbacher, M.; Wölkart, G.; Stessel, H.; Pail, K.; Winkler, S.; Schweiger, M.; Haemmerle, G.; Al Zoughbi, W.; Höfler, G.; et al. Endothelial dysfunction in adipose triglyceride lipase deficiency. Biochimica. Biophysica. Acta 2014, 1841, 906–917. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J. Exercise training regulates angiogenic gene expression in white adipose tissue. J. Exerc. Rehabil. 2018, 14, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Mazaki-Tovi, S.; Tarca, A.L.; Vaisbuch, E.; Kusanovic, J.P.; Than, N.G.; Chaiworapongsa, T.; Dong, Z.; Hassan, S.S.; Romero, R. Characterization of visceral and subcutaneous adipose tissue transcriptome in pregnant women with and without spontaneous labor at term: Implication of alternative splicing in the metabolic adaptations of adipose tissue to parturition. J. Perinat. Med. 2016, 44, 813–835. [Google Scholar] [CrossRef]
- Peinado, J.R.; Pardo, M.; de la Rosa, O.; Malagón, M.M. Proteomic characterization of adipose tissue constituents, a necessary step for understanding adipose tissue complexity. Proteomics 2012, 12, 607–620. [Google Scholar] [CrossRef]
- Perez-Perez, R.; Ortega-Delgado, F.J.; García-Santos, E.; López, J.A.; Camafeita, E.; Ricart, W.; Fernández-Real, J.M.; Peral, B. Differential proteomics of omental and subcutaneous adipose tissue reflects their unalike biochemical and metabolic properties. J. Proteome Res. 2009, 8, 1682–1693. [Google Scholar] [CrossRef]
- Lemoine, A.Y.; Ledoux, S.; Larger, E. Adipose tissue angiogenesis in obesity. Thromb. Haemost. 2013, 110, 661–668. [Google Scholar] [CrossRef]
- Huang, H.; Bhat, A.; Woodnutt, G.; Lappe, R. Targeting the ANGPT-TIE2 pathway in malignancy. Nat. Rev. Cancer 2010, 10, 575–585. [Google Scholar] [CrossRef]
- Linares, P.M.; Chaparro, M.; Gisbert, J.P. Angiopoietins in inflammation and their implication in the development of inflammatory bowel disease. A review. J. Crohns Colitis 2014, 8, 183–190. [Google Scholar] [CrossRef]
- Abramovich, D.; Irusta, G.; Bas, D.; Cataldi, N.I.; Parborell, F.; Tesone, M. Angiopoietins/TIE2 system and VEGF are involved in ovarian function in a DHEA rat model of polycystic ovary syndrome. Endocrinology 2012, 153, 3446–3456. [Google Scholar] [CrossRef]
- Penton, A.L.; Leonard, L.D.; Spinner, N.B. Notch signaling in human development and disease. Semin. Cell Dev. Biol. 2012, 23, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Henshall, T.L.; Keller, A.; He, L.; Johansson, B.R.; Wallgard, E.; Raschperger, E.; Mäe, M.A.; Jin, S.; Betsholtz, C.; Lendahl, U. Notch3 is necessary for blood vessel integrity in the central nervous system. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Niessen, K.; Karsan, A. Notch signaling in the developing cardiovascular system. Am. J. Physiol. Cell Physiol. 2007, 293, C1–C11. [Google Scholar] [CrossRef] [PubMed]
- Pitulescu, M.E.; Schmidt, I.; Giaimo, B.D.; Antoine, T.; Berkenfeld, F.; Ferrante, F.; Park, H.; Ehling, M.; Biljes, D.; Rocha, S.F. Dll4 and Notch signalling couples sprouting angiogenesis and artery formation. Nat. Cell Biol. 2017, 19, 915–927. [Google Scholar] [CrossRef]
- Siekmann, A.F.; Lawson, N.D. Notch signalling and the regulation of angiogenesis. Cell Adhes. Migr. 2007, 1, 104–106. [Google Scholar] [CrossRef]
- Park, S.; Sorenson, C.M.; Sheibani, N. PECAM-1 isoforms, eNOS and endoglin axis in regulation of angiogenesis. Clin. Sci. 2015, 129, 217–234. [Google Scholar] [CrossRef]
- Sun, L.; Huang, T.; Xu, W.; Sun, J.; Lv, Y.; Wang, Y. Advanced glycation end products promote VEGF expression and thus choroidal neovascularization via Cyr61-PI3K/AKT signaling pathway. Sci. Rep. 2017, 7, 14925. [Google Scholar] [CrossRef]
- Li, B.K.; Guo, K.; Li, C.Y.; Li, H.L.; Zhao, P.P.; Chen, K.; Liu, C.X. Influence of suppression of CapG gene expression by siRNA on the growth and metastasis of human prostate cancer cells. Genet. Mol. Res. 2015, 14, 15769–15778. [Google Scholar] [CrossRef]
- Silacci, P.; Mazzolai, L.; Gauci, C.; Stergiopulos, N.; Yin, H.L.; Hayoz, D. Gelsolin superfamily proteins: Key regulators of cellular functions. Cell Mol. Life Sci. 2004, 61, 2614–2623. [Google Scholar] [CrossRef]
- Turcot, V.; Bouchard, L.; Faucher, G.; Tchernof, A.; Deshaies, Y.; Pérusse, L.; Marceau, P.; Hould, F.S.; Lebel, S.; Vohl, M.C. A polymorphism of the interferon-gamma-inducible protein 30 gene is associated with hyperglycemia in severely obese individuals. Hum. Genet. 2012, 131, 57–66. [Google Scholar] [CrossRef]
- Benbow, J.H.; Thompson, K.J.; Cope, H.L.; Brandon-Warner, E.; Culberson, C.R.; Bossi, K.L.; Li, T.; Russo, M.W.; Gersin, K.S.; McKillop, I.H.; et al. Diet-Induced Obesity Enhances Progression of Hepatocellular Carcinoma through Tenascin-C/Toll-Like Receptor 4 Signaling. Am. J. Pathol. 2016, 186, 145–158. [Google Scholar] [CrossRef] [PubMed]
- Imanaka-Yoshida, K.; Yoshida, T.; Miyagawa-Tomita, S. Tenascin-C in development and disease of blood vessels. Anat. Rec. 2014, 297, 1747–1757. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Wu, Q.; Xing, X.; Wei, Y.; Shao, Z. MicroRNA-145 targets vascular endothelial growth factor and inhibits invasion and metastasis of osteosarcoma cells. Acta Biochim. Biophys. Sin. 2012, 44, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Sessa, R.; Seano, G.; di Blasio, L.; Gagliardi, P.A.; Isella, C.; Medico, E.; Cotelli, F.; Bussolino, F.; Primo. L. The miR-126 regulates angiopoietin-1 signaling and vessel maturation by targeting p85beta. Biochim. Biophys. Acta 2012, 1823, 1925–1935. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Chen, J.; Duscher, D.; Liu, Y.; Guo, G.; Kang, Y.; Xiong, H.; Zhan, P.; Wang, Y.; Wang, C.; et al. Microvesicles from human adipose stem cells promote wound healing by optimizing cellular functions via AKT and ERK signaling pathways. Stem Cell Res. Ther. 2019, 10, 47. [Google Scholar] [CrossRef]
| Variables | nOB | OB |
|---|---|---|
| N | 15 | 22 |
| Age (Years) | 40 ± 4 | 39 ± 2 |
| Sex (M/W) | 2/13 | 3/19 |
| BMI (Kg/m2) | 23.9 ± 1.1 | 43.0 ± 1.1 |
| CVRF (N) | 0 | 3 |
| Smoking (%) | 0 | 55 |
| HT (%) | 0 | 32 |
| DM (%) | 7 | 23 |
| DLP (%) | 0 | 50 |
| RBC (×106/mm3) | 3.8 ± 0.4 | 3.9 ± 0.7 |
| MCB (µm3) | 81.7 ± 4.6 | 80.3 ± 5.03 |
| RDV (%) | 14.8 ± 1 | 15.7 ± 1.8 |
| HCT (%) | 30.8 ± 2.7 | 31.05 ± 5.6 |
| PLT × 103/mm3) | 211 ± 47.4 | 182.7 ± 56.8 |
| MPV (µm3) | 9.1 ± 0.8 | 8.9 ± 1.1 |
| WBC (×103/mm3) | 4.5 ± 0.4 | 4.9 ± 0.9 |
| HGB (g/dl) | 10.7 ± 1 | 11.06 ± 2.2 |
| MCH (pg) | 28.5 ± 1.4 | 29.03 ± 2.3 |
| MCHC (g/dl) | 34.9 ± 1.2 | 36.15 ± 2.2 |
| LYMF (×103/mm3) | 1.3 ± 0.3 | 1.23 ± 0.5 |
| GRAN (×103/mm3) | 2.6 ± 0.4 | 3.2 ± 0.3 |
| MID (×103/mm3) | 0.5 ± 0.05 | 0.6 ± 0.08 |
| p Value | Ratio | |
|---|---|---|
| AGER | 0.001 | 1.75 |
| ANGPT1 | 0.134 | 1.31 |
| ANGPT2 | 0.094 | 1.39 |
| CAPG | 0.029 | 1.33 |
| CD34 | 0.041 | 1.39 |
| CD68 | <0.001 | 2.13 |
| COL18A1 | 0.417 | 0.93 |
| DLL4 | 0.001 | 0.62 |
| ENG | 0.230 | 0.93 |
| IFI30 | <0.001 | 2.55 |
| NOTCH3 | 0.005 | 1.70 |
| PECAM1 | 0.416 | 0.88 |
| PGF | 0.944 | 1.02 |
| PLIN2 | 0.002 | 0.67 |
| PNPLA2 | <0.001 | 0.62 |
| PTX3 | 0.780 | 1.00 |
| SERPINF1 | 0.939 | 1.04 |
| SPP1 | <0.001 | 4.76 |
| TEK | 0.104 | 0.83 |
| TNC | 0.183 | 1.12 |
| VEGFA | <0.001 | 0.46 |
| Gene | Ratio | All Patients N = 22 | cCRFs N = 19 | Diabetes mellitus | Dyslipidemia | Hypertension | |||
|---|---|---|---|---|---|---|---|---|---|
| OB-nDM N = 17 | OB-DM N = 5 | OB-nDLP N = 11 | OB-DLP N = 11 | OB-nHT N = 15 | OB-HT N = 7 | ||||
| AGER | 0.85 | 0.072 | 0.184 | 0.193 | 0.138 | 0.051 | 0.534 | 0.140 | 0.176 |
| ANGPT1 | 1.56 | 0.001 | 0.005 | 0.002 | 0.500 | 0.016 | 0.026 | 0.003 | 0.311 |
| ANGPT2 | 1.73 | 0.011 | 0.009 | 0.013 | 0.500 | 0.374 | 0.013 | 0.027 | 0.176 |
| CAPG | 1.17 | 0.049 | 0.045 | 0.108 | 0.225 | 0.049 | 0.373 | 0.270 | 0.066 |
| CD34 | 1.09 | 0.406 | 0.414 | 0.069 | 0.225 | 0.819 | 0.371 | 0.900 | 0.264 |
| CD68 | 1.23 | 0.013 | 0.013 | 0.028 | 0.225 | 0.182 | 0.033 | 0.054 | 0.091 |
| COL18A1 | 1.10 | 0.170 | 0.089 | 0.286 | 0.343 | 0.277 | 0.024 | 0.775 | 0.086 |
| DLL4 | 1.30 | 0.053 | 0.070 | 0.042 | 0.893 | 0.497 | 0.029 | 0.016 | 0.828 |
| ENG | 1.20 | 0.055 | 0.081 | 0.087 | 0.345 | 0.289 | 0.110 | 0.105 | 0.243 |
| IFI30 | 1.17 | 0.083 | 0.020 | 0.269 | 0.080 | 0.746 | 0.052 | 0.566 | 0.033 |
| NOTCH3 | 3.98 | <0.001 | <0.001 | <0.001 | 0.043 | 0.003 | 0.003 | <0.001 | 0.018 |
| PECAM1 | 1.52 | 0.002 | 0.006 | 0.006 | 0.225 | 0.062 | 0.016 | 0.004 | 0.176 |
| PGF | 0.83 | 0.072 | 0.091 | 0.084 | 0.686 | 0.051 | 0.594 | 0.100 | 0.866 |
| PLIN2 | 1.15 | 0.143 | 0.382 | 0.280 | 0.686 | 0.073 | 0.825 | 0.226 | 0.428 |
| PNPLA2 | 1.31 | 0.004 | 0.011 | 0.013 | 0.138 | 0.041 | 0.021 | 0.202 | 0.018 |
| PTX3 | 1.11 | 0.046 | 0.108 | 0.050 | 0.500 | 0.013 | 0.657 | 0.088 | 0.237 |
| SERPINF1 | 1.50 | <0.001 | <0.0001 | <0.001 | 0.080 | <0.001 | 0.004 | 0.002 | 0.018 |
| SPP1 | 2.90 | <0.001 | 0.003 | 0.003 | 0.225 | 0.021 | 0.016 | 0.001 | 0.237 |
| TEK | 1.24 | 0.029 | 0.031 | 0.050 | 0.500 | 0.213 | 0.086 | 0.128 | 0.145 |
| TNC | 3.03 | <0.001 | <0.001 | <0.001 | 0.080 | 0.003 | 0.008 | 0.004 | 0.018 |
| VEGFA | 0.86 | 0.076 | 0.096 | 0.376 | 0.080 | 0.024 | 0.395 | 0.005 | 0.957 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arderiu, G.; Lambert, C.; Ballesta, C.; Moscatiello, F.; Vilahur, G.; Badimon, L. Cardiovascular Risk Factors and Differential Transcriptomic Profile of the Subcutaneous and Visceral Adipose Tissue and Their Resident Stem Cells. Cells 2020, 9, 2235. https://doi.org/10.3390/cells9102235
Arderiu G, Lambert C, Ballesta C, Moscatiello F, Vilahur G, Badimon L. Cardiovascular Risk Factors and Differential Transcriptomic Profile of the Subcutaneous and Visceral Adipose Tissue and Their Resident Stem Cells. Cells. 2020; 9(10):2235. https://doi.org/10.3390/cells9102235
Chicago/Turabian StyleArderiu, Gemma, Carmen Lambert, Carlos Ballesta, Fabrizio Moscatiello, Gemma Vilahur, and Lina Badimon. 2020. "Cardiovascular Risk Factors and Differential Transcriptomic Profile of the Subcutaneous and Visceral Adipose Tissue and Their Resident Stem Cells" Cells 9, no. 10: 2235. https://doi.org/10.3390/cells9102235
APA StyleArderiu, G., Lambert, C., Ballesta, C., Moscatiello, F., Vilahur, G., & Badimon, L. (2020). Cardiovascular Risk Factors and Differential Transcriptomic Profile of the Subcutaneous and Visceral Adipose Tissue and Their Resident Stem Cells. Cells, 9(10), 2235. https://doi.org/10.3390/cells9102235






