Increased Lamin B1 Levels Promote Cell Migration by Altering Perinuclear Actin Organization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Datasets
2.2. Cell Culture
2.3. Plasmids
2.4. Motility Assays
2.5. Immunostaining
2.6. Western Blot Analysis
3. Results
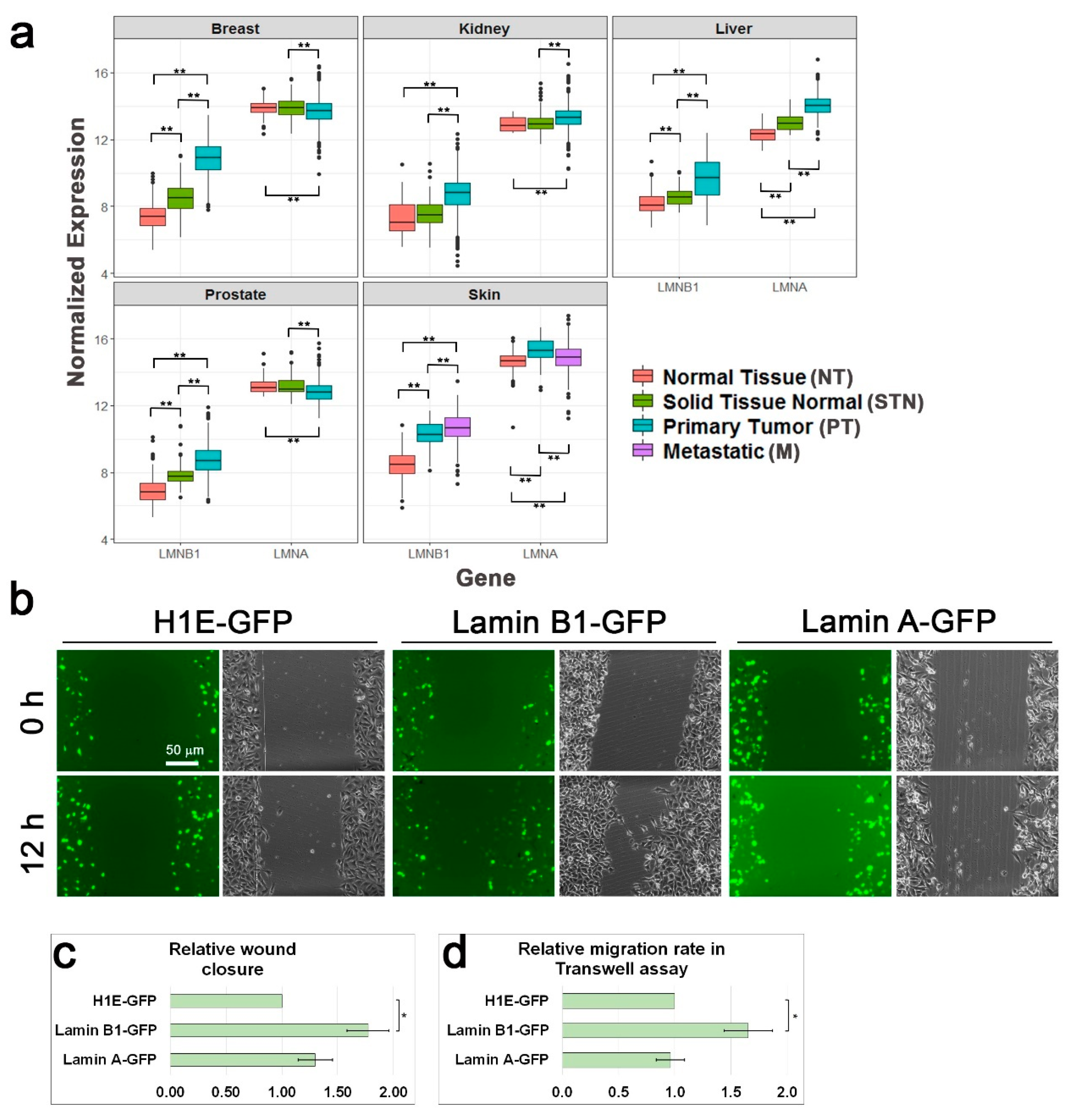
3.1. Lamin B1 Over-Expression Elevates the Migration Rate of Melanoma Cells
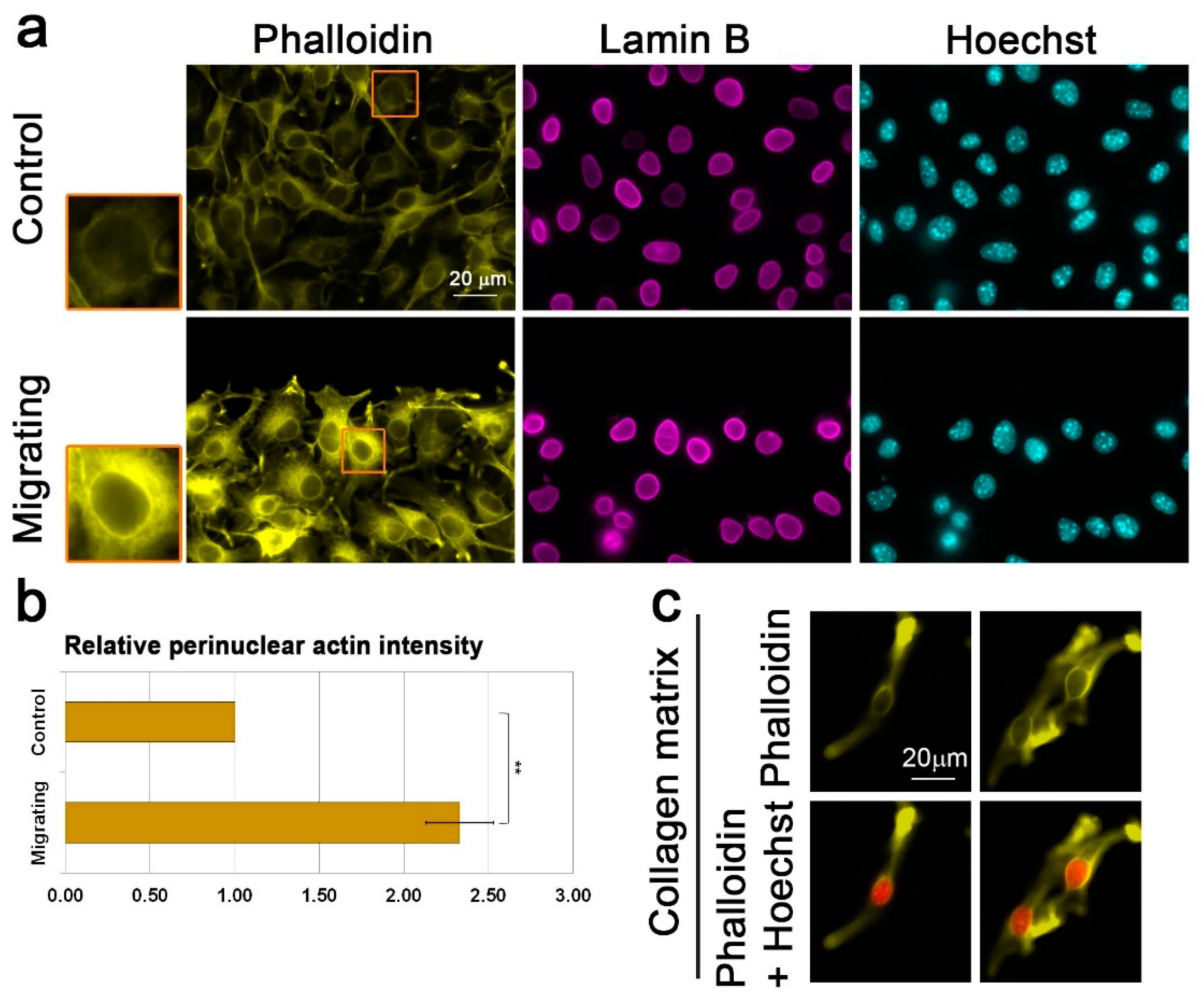
3.2. Migration Signals Induce the Formation of a Perinuclear Actin Rim
3.3. Lamin B1 Over-Expression Interferes with Perinuclear Actin Rim Formation
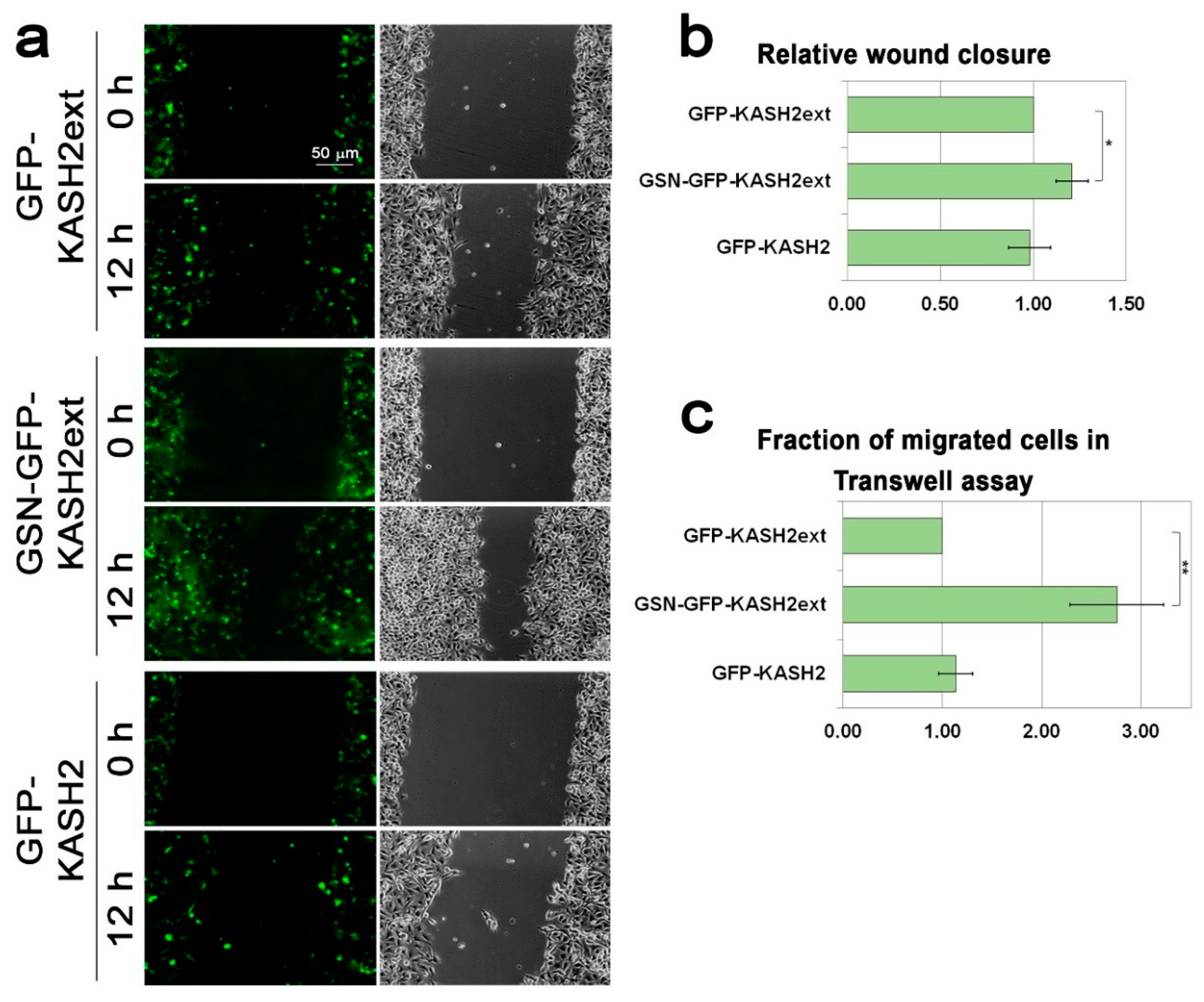
3.4. Interference with Perinuclear Actin Filaments Increases the Cellular Migration Rate
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hetzer, M.W. The nuclear envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000539. [Google Scholar] [CrossRef] [PubMed]
- Wilson, K.L.; Berk, J.M. The nuclear envelope at a glance. J. Cell Sci. 2010, 123, 1973–1978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, W.; Worman, H.J.; Gundersen, G.G. Accessorizing and anchoring the LINC complex for multifunctionality. J. Cell Biol. 2015, 208, 11–22. [Google Scholar] [CrossRef] [Green Version]
- Luxton, G.G.; Starr, D.A. KASHing up with the nucleus: Novel functional roles of KASH proteins at the cytoplasmic surface of the nucleus. Curr. Opin. Cell Biol. 2014, 28, 69–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beck, M.; Hurt, E. The nuclear pore complex: Understanding its function through structural insight. Nat. Rev. Mol. Cell Biol. 2017, 18, 73–89. [Google Scholar] [CrossRef]
- Gruenbaum, Y.; Foisner, R. Lamins: Nuclear Intermediate Filament Proteins with Fundamental Functions in Nuclear Mechanics and Genome Regulation. Ann. Rev. Biochem. 2015, 84, 131–164. [Google Scholar] [CrossRef]
- Ho, C.Y.; Lammerding, J. Lamins at a glance. J. Cell Sci. 2012, 125, 2087–2093. [Google Scholar] [CrossRef] [Green Version]
- de Leeuw, R.; Gruenbaum, Y.; Medalia, O. Nuclear Lamins: Thin Filaments with Major Functions. Trends Cell Biol. 2018, 28, 34–45. [Google Scholar] [CrossRef]
- Davidson, P.M.; Lammerding, J. Broken nuclei—Lamins, nuclear mechanics, and disease. Trends Cell Biol. 2014, 24, 247–256. [Google Scholar] [CrossRef] [Green Version]
- Osmanagic-Myers, S.; Dechat, T.; Foisner, R. Lamins at the crossroads of mechanosignaling. Genes. Dev. 2015, 29, 225–237. [Google Scholar] [CrossRef] [Green Version]
- Shevelyov, Y.Y.; Ulianov, S.V. The Nuclear Lamina as an Organizer of Chromosome Architecture. Cells 2019, 8, 136. [Google Scholar] [CrossRef] [Green Version]
- Coffinier, C.; Chang, S.Y.; Nobumori, C.; Tu, Y.; Farber, E.A.; Toth, J.I.; Fong, L.G.; Young, S.G. Abnormal development of the cerebral cortex and cerebellum in the setting of lamin B2 deficiency. Proc. Natl. Acad. Sci. USA 2010, 107, 5076–5081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coffinier, C.; Jung, H.-J.; Nobumori, C.; Chang, S.; Tu, Y.; Barnes, R.H.; Yoshinaga, Y.; de Jong, P.J.; Vergnes, L.; Reue, K.; et al. Deficiencies in lamin B1 and lamin B2 cause neurodevelopmental defects and distinct nuclear shape abnormalities in neurons. Mol. Biol. Cell 2011, 22, 4683–4693. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.-J.; Nobumori, C.; Goulbourne, C.N.; Tu, Y.; Lee, J.M.; Tatar, A.; Wu, D.; Yoshinaga, Y.; de Jong, P.J.; Coffinier, C.; et al. Farnesylation of lamin B1 is important for retention of nuclear chromatin during neuronal migration. Proc. Natl. Acad. Sci. USA 2013, 110, E1923–E1932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, J.R.; Zheng, X.; Zheng, Y. Lamin-B1 contributes to the proper timing of epicardial cell migration and function during embryonic heart development. Mol. Biol. Cell 2016, 27, 3956–3963. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Vong, J.S.-L.; Asafova, A.; Garvalov, B.K.; Caputo, L.; Cordero, J.; Singh, A.; Boettger, T.; Günther, S.; Fink, L.; et al. Lamin B1 loss promotes lung cancer development and metastasis by epigenetic derepression of RET. J. Exp. Med. 2019, 216, 1377–1395. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Du, Y.; Kong, X.; Li, Z.; Jia, Z.; Cui, J.; Gao, J.; Wang, G.; Xie, K. Lamin B1 Is a Novel Therapeutic Target of Betulinic Acid in Pancreatic Cancer. Clin. Cancer Res. 2013, 19, 4651–4661. [Google Scholar] [CrossRef] [Green Version]
- Rowat, A.C.; Jaalouk, D.E.; Zwerger, M.; Ung, W.L.; Eydelnant, I.A.; Olins, D.E.; Olins, A.L.; Herrmann, H.; Weitz, D.A.; Lammerding, J. Nuclear Envelope Composition Determines the Ability of Neutrophil-type Cells to Passage through Micron-scale Constrictions. J. Biol. Chem. 2013, 288, 8610–8618. [Google Scholar] [CrossRef] [Green Version]
- Harada, T.; Swift, J.; Irianto, J.; Shin, J.-W.; Spinler, K.R.; Athirasala, A.; Diegmiller, R.; Dingal, P.C.D.P.; Ivanovska, I.L.; Discher, D.E. Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J. Cell Biol. 2014, 204, 669–682. [Google Scholar] [CrossRef] [Green Version]
- Yadav, S.K.; Feigelson, S.W.; Roncato, F.; Antman-Passig, M.; Shefi, O.; Lammerding, J.; Alon, R. Frontline Science: Elevated nuclear lamin A is permissive for granulocyte transendothelial migration but not for motility through collagen I barriers. J. Leukoc Biol. 2018, 104, 239–251. [Google Scholar] [CrossRef]
- Krause, M.; Yang, F.W.; te Lindert, M.; Isermann, P.; Schepens, J.; Maas, R.J.A.; Venkataraman, C.; Lammerding, J.; Madzvamuse, A.; Hendriks, W.; et al. Cell migration through three-dimensional confining pores: Speed accelerations by deformation and recoil of the nucleus. Phil. Trans. R Soc. B 2019, 374, 20180225. [Google Scholar] [CrossRef] [PubMed]
- Evangelisti, C.; Paganelli, F.; Giuntini, G.; Mattioli, E.; Cappellini, A.; Ramazzotti, G.; Faenza, I.; Maltarello, M.; Martelli, A.; Scotlandi, K.; et al. Lamin A and Prelamin A Counteract Migration of Osteosarcoma Cells. Cells 2020, 9, 774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reis-Sobreiro, M.; Chen, J.-F.; Novitskaya, T.; You, S.; Morley, S.; Steadman, K.; Gill, N.K.; Eskaros, A.; Rotinen, M.; Chu, C.-Y.; et al. Emerin Deregulation Links Nuclear Shape Instability to Metastatic Potential. Cancer Res. 2018, 78, 6086–6097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorland, Y.L.; Cornelissen, A.S.; Kuijk, C.; Tol, S.; Hoogenboezem, M.; van Buul, J.D.; Nolte, M.A.; Voermans, C.; Huveneers, S. Nuclear shape, protrusive behaviour and in vivo retention of human bone marrow mesenchymal stromal cells is controlled by Lamin-A/C expression. Sci. Rep. 2019, 9, 14401. [Google Scholar] [CrossRef]
- Goldberg, M.W.; Huttenlauch, I.; Hutchison, C.J.; Stick, R. Filaments made from A- and B-type lamins differ in structure and organization. J. Cell Sci. 2008, 121, 215–225. [Google Scholar] [CrossRef] [Green Version]
- Lammerding, J.; Fong, L.G.; Ji, J.Y.; Reue, K.; Stewart, C.L.; Young, S.G.; Lee, R.T. Lamins A and C but not lamin B1 regulate nuclear mechanics. J. Biol. Chem. 2006, 281, 25768–25780. [Google Scholar] [CrossRef] [Green Version]
- Stephens, A.D.; Banigan, E.J.; Adam, S.A.; Goldman, R.D.; Marko, J.F. Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol. Biol. Cell 2017, 28, 1984–1996. [Google Scholar] [CrossRef]
- Swift, J.; Ivanovska, I.L.; Buxboim, A.; Harada, T.; Dingal, P.C.D.P.; Pinter, J.; Pajerowski, J.D.; Spinler, K.R.; Shin, J.-W.; Tewari, M.; et al. Nuclear Lamin-A Scales with Tissue Stiffness and Enhances Matrix-Directed Differentiation. Science 2013, 341, 1240104. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Tamashunas, A.C.; Agrawal, A.; Torbati, M.; Katiyar, A.; Dickinson, R.B.; Lammerding, J.; Lele, T.P. Local, transient tensile stress on the nuclear membrane causes membrane rupture. Mol. Biol. Cell 2019, 30, 899–906. [Google Scholar] [CrossRef]
- Alcorta-Sevillano, N.; Macías, I.; Rodríguez, C.I.; Infante, A. Crucial Role of Lamin A/C in the Migration and Differentiation of MSCs in Bone. Cells 2020, 9, 1330. [Google Scholar] [CrossRef]
- Kim, D.-H.; Cho, S.; Wirtz, D. Tight coupling between nucleus and cell migration through the perinuclear actin cap. J. Cell Sci. 2014, 127, 2528–2541. [Google Scholar] [CrossRef] [Green Version]
- Maninova, M.; Caslavsky, J.; Vomastek, T. The assembly and function of perinuclear actin cap in migrating cells. Protoplasma 2017, 254, 1207–1218. [Google Scholar] [CrossRef] [PubMed]
- Luxton, G.W.G.; Gomes, E.R.; Folker, E.S.; Vintinner, E.; Gundersen, G.G. Linear Arrays of Nuclear Envelope Proteins Harness Retrograde Actin Flow for Nuclear Movement. Science 2010, 329, 956–959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osorio, D.S.; Gomes, E.R. Connecting the Nucleus to the Cytoskeleton for Nuclear Positioning and Cell Migration. In Cancer Biology and the Nuclear Envelope; Schirmer, E.C., de las Heras, J.I., Eds.; Springer: New York, NY, USA, 2014; Volume 773, pp. 505–520. ISBN 978-1-4899-8031-1. [Google Scholar]
- Thiam, H.-R.; Vargas, P.; Carpi, N.; Crespo, C.L.; Raab, M.; Terriac, E.; King, M.C.; Jacobelli, J.; Alberts, A.S.; Stradal, T.; et al. Perinuclear Arp2/3-driven actin polymerization enables nuclear deformation to facilitate cell migration through complex environments. Nat. Commun. 2016, 7, 10997. [Google Scholar] [CrossRef]
- Goldman, M.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. The UCSC Xena platform for public and private cancer genomics data visualization and interpretation. bioRxiv 2018. [Google Scholar] [CrossRef] [Green Version]
- Maizels, Y.; Elbaz, A.; Hernandez-Vicens, R.; Sandrusy, O.; Rosenberg, A.; Gerlitz, G. Increased chromatin plasticity supports enhanced metastatic potential of mouse melanoma cells. Exp. Cell Res. 2017, 357, 282–290. [Google Scholar] [CrossRef]
- Dahl, K.N.; Scaffidi, P.; Islam, M.F.; Yodh, A.G.; Wilson, K.L.; Misteli, T. Distinct structural and mechanical properties of the nuclear lamina in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA 2006, 103, 10271–10276. [Google Scholar] [CrossRef] [Green Version]
- Stewart-Hutchinson, P.J.; Hale, C.M.; Wirtz, D.; Hodzic, D. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp. Cell Res. 2008, 314, 1892–1905. [Google Scholar] [CrossRef] [Green Version]
- Gerlitz, G.; Livnat, I.; Ziv, C.; Yarden, O.; Bustin, M.; Reiner, O. Migration cues induce chromatin alterations. Traffic 2007, 8, 1521–1529. [Google Scholar] [CrossRef]
- Segal, T.; Salmon-Divon, M.; Gerlitz, G. The Heterochromatin Landscape in Migrating Cells and the Importance of H3K27me3 for Associated Transcriptome Alterations. Cells 2018, 7. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, X.; Li, Q.; Mogilner, A.; Bershadsky, A.D.; Shivashankar, G.V. Mechanical stimulation induces formin-dependent assembly of a perinuclear actin rim. Proc. Natl. Acad. Sci. USA 2015, 112, E2595–E2601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, X.; Kawauchi, K.; Shivashankar, G.V.; Bershadsky, A.D. Novel localization of formin mDia2: Importin -mediated delivery to and retention at the cytoplasmic side of the nuclear envelope. Biol. Open 2015, 4, 1569–1575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wales, P.; Schuberth, C.E.; Aufschnaiter, R.; Fels, J.; García-Aguilar, I.; Janning, A.; Dlugos, C.P.; Schäfer-Herte, M.; Klingner, C.; Wälte, M.; et al. Calcium-mediated actin reset (CaAR) mediates acute cell adaptations. eLife 2016, 5. [Google Scholar] [CrossRef]
- Shiu, J.-Y.; Aires, L.; Lin, Z.; Vogel, V. Nanopillar force measurements reveal actin-cap-mediated YAP mechanotransduction. Nat. Cell Biol. 2018, 20, 262–271. [Google Scholar] [CrossRef]
- Khatau, S.B.; Bloom, R.J.; Bajpai, S.; Razafsky, D.; Zang, S.; Giri, A.; Wu, P.-H.; Marchand, J.; Celedon, A.; Hale, C.M.; et al. The distinct roles of the nucleus and nucleus-cytoskeleton connections in three-dimensional cell migration. Sci. Rep. 2012, 2, 488. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.-K.; Louhghalam, A.; Lee, G.; Schafer, B.W.; Wirtz, D.; Kim, D.-H. Nuclear lamin A/C harnesses the perinuclear apical actin cables to protect nuclear morphology. Nat. Commun. 2017, 8, 2123. [Google Scholar] [CrossRef]
- Hieda, M. Signal Transduction across the Nuclear Envelope: Role of the LINC Complex in Bidirectional Signaling. Cells 2019, 8, 124. [Google Scholar] [CrossRef] [Green Version]
- Lv, X.; Liu, L.; Cheng, C.; Yu, B.; Xiong, L.; Hu, K.; Tang, J.; Zeng, L.; Sang, Y. SUN2 exerts tumor suppressor functions by suppressing the Warburg effect in lung cancer. Sci. Rep. 2015, 5, 17940. [Google Scholar] [CrossRef]
- Matsumoto, A.; Hieda, M.; Yokoyama, Y.; Nishioka, Y.; Yoshidome, K.; Tsujimoto, M.; Matsuura, N. Global loss of a nuclear lamina component, lamin A/C, and LINC complex components SUN1, SUN2, and nesprin-2 in breast cancer. Cancer Med. 2015, 4, 1547–1557. [Google Scholar] [CrossRef]
- Yajun, C.; Chen, Y.; Xiaosa, L.; Xiao, W.; Jia, C.; Zhong, W.; Bin, X. Loss of Sun2 promotes the progression of prostate cancer by regulating fatty acid oxidation. Oncotarget 2017, 8, 89620–89630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cartwright, S.; Karakesisoglou, I. Nesprins in health and disease. Semin. Cell Dev. Biol. 2014, 29, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Sur-Erdem, I.; Hussain, M.S.; Asif, M.; Pınarbası, N.; Aksu, A.C.; Noegel, A.A. Nesprin-1 impact on tumorigenic cell phenotypes. Mol. Biol. Rep. 2020, 47, 921–934. [Google Scholar] [CrossRef] [PubMed]
- Marmé, A.; Zimmermann, H.-P.; Moldenhauer, G.; Schorpp-Kistner, M.; Müller, C.; Keberlein, O.; Giersch, A.; Kretschmer, J.; Seib, B.; Spiess, E.; et al. Loss of Drop1 expression already at early tumor stages in a wide range of human carcinomas. Int. J. Cancer 2008, 123, 2048–2056. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fracchia, A.; Asraf, T.; Salmon-Divon, M.; Gerlitz, G. Increased Lamin B1 Levels Promote Cell Migration by Altering Perinuclear Actin Organization. Cells 2020, 9, 2161. https://doi.org/10.3390/cells9102161
Fracchia A, Asraf T, Salmon-Divon M, Gerlitz G. Increased Lamin B1 Levels Promote Cell Migration by Altering Perinuclear Actin Organization. Cells. 2020; 9(10):2161. https://doi.org/10.3390/cells9102161
Chicago/Turabian StyleFracchia, Andrea, Tal Asraf, Mali Salmon-Divon, and Gabi Gerlitz. 2020. "Increased Lamin B1 Levels Promote Cell Migration by Altering Perinuclear Actin Organization" Cells 9, no. 10: 2161. https://doi.org/10.3390/cells9102161
APA StyleFracchia, A., Asraf, T., Salmon-Divon, M., & Gerlitz, G. (2020). Increased Lamin B1 Levels Promote Cell Migration by Altering Perinuclear Actin Organization. Cells, 9(10), 2161. https://doi.org/10.3390/cells9102161




