An Emerging Role for isomiRs and the microRNA Epitranscriptome in Neovascularization
Abstract
1. Introduction
2. Neovascularization—Angiogenesis and Arteriogenesis
3. MicroRNAs
4. IsomiRs and the microRNA Epitranscriptome
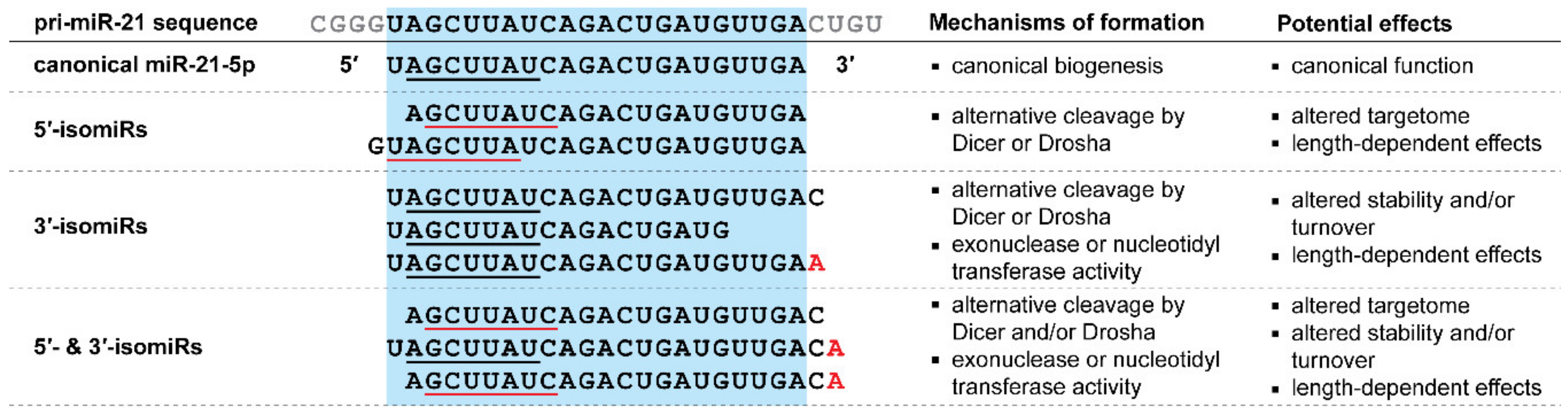
5. IsomiRs
5.1. IsomiRs
5.2. IsomiRs in Neovascularization Associated Cells and Processes
6. Adenosine-to-Inosine Editing
6.1. Adenosine-to-Inosine Editing
6.2. MicroRNA A-to-I Editing in Neovascularization
7. N6-Adenosine Methylation
7.1. N6-Adenosine Methylation
7.2. Importance of m6A in the Cardio-Vasculature and in Vasoactive MicroRNAs
8. Other Modifications in the microRNA Epitranscriptome
8.1. Pseudouridine (Ψ)
8.2. 2′-O-Methylnucleosides
8.3. N1-Methyladenosine (m1A)
8.4. N5-Methylcytosine (m5C)
8.5. N2-Methylguanosine (m2G)
9. Dynamic Regulation of the Epitranscriptome
10. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Timmis, A.; Townsend, N.; Gale, C.; Grobbee, R.; Maniadakis, N.; Flather, M.; Wilkins, E.; Wright, L.; Vos, R.; Bax, J.; et al. European Society of Cardiology: Cardiovascular Disease Statistics 2017. Eur Heart J. 2018, 39, 508–579. [Google Scholar] [CrossRef] [PubMed]
- Kaptoge, S.; Pennells, L.; De Bacquer, D.; Cooney, M.T.; Kavousi, M.; Stevens, G.; Riley, L.M.; Savin, S.; Khan, T.; Altay, S.; et al. World Health Organization cardiovascular disease risk charts: Revised models to estimate risk in 21 global regions. Lancet Glob. Health 2019, 7, e1332–e1345. [Google Scholar] [CrossRef]
- Dormandy, J.; Heeck, L.; Vig, S. Acute limb ischemia. Semin. Vasc. Surg. 1999, 12, 148–153. [Google Scholar] [PubMed]
- Powell, R.J.; Comerota, A.J.; Berceli, S.A.; Guzman, R.; Henry, T.D.; Tzeng, E.; Velazquez, O.; Marston, W.A.; Bartel, R.L.; Longcore, A.; et al. Interim analysis results from the RESTORE-CLI, a randomized, double-blind multicenter phase II trial comparing expanded autologous bone marrow-derived tissue repair cells and placebo in patients with critical limb ischemia. J. Vasc. Surg. 2011, 54, 1032–1041. [Google Scholar] [CrossRef] [PubMed]
- Van Oostrom, M.C.; van Oostrom, O.; Quax, P.H.; Verhaar, M.C.; Hoefer, I.E. Insights into mechanisms behind arteriogenesis: What does the future hold? J. Leukoc. Biol. 2008, 84, 1379–1391. [Google Scholar] [CrossRef] [PubMed]
- Raval, Z.; Losordo, D.W. Cell Therapy of Peripheral Arterial Disease: From Experimental Findings to Clinical Trials. Circ. Res. 2013, 112, 1288–1302. [Google Scholar] [CrossRef]
- Weber, C. MicroRNAs: From basic mechanisms to clinical application in cardiovascular medicine. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 168–169. [Google Scholar] [CrossRef][Green Version]
- Welten, S.M.; Goossens, E.A.; Quax, P.H.; Nossent, A.Y. The multifactorial nature of microRNAs in vascular remodelling. Cardiovasc. Res. 2016, 110, 6–22. [Google Scholar] [CrossRef]
- Welten, S.M.; Bastiaansen, A.J.; de Jong, R.C.; de Vries, M.R.; Peters, E.H.; Boonstra, M.; Sheikh, S.P.; La, M.N.; Kandimalla, E.R.; Quax, P.H.; et al. Inhibition of 14q32 MicroRNAs miR-329, miR-487b, miR-494 and miR-495 Increases Neovascularization and Blood Flow Recovery after Ischemia. Circ. Res. 2014, 115, 696–708. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Heil, M.; Schaper, W. Pathophysiology of collateral development. Coron. Artery Dis. 2004, 15, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Hoefer, I.E.; van Royen, N.; Rectenwald, J.E.; Deindl, E.; Hua, J.; Jost, M.; Grundmann, S.; Voskuil, M.; Ozaki, C.K.; Piek, J.J.; et al. Arteriogenesis proceeds via ICAM-1/Mac-1- mediated mechanisms. Circ. Res. 2004, 94, 1179–1185. [Google Scholar] [CrossRef] [PubMed]
- Scholz, D.; Ito, W.; Fleming, I.; Deindl, E.; Sauer, A.; Wiesnet, M.; Busse, R.; Schaper, J.; Schaper, W. Ultrastructure and molecular histology of rabbit hind-limb collateral artery growth (arteriogenesis). Virchows Arch. 2000, 436, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Hoefer, I.E.; van Royen, N.; Rectenwald, J.E.; Bray, E.J.; Abouhamze, Z.; Moldawer, L.L.; Voskuil, M.; Piek, J.J.; Buschmann, I.R.; Ozaki, C.K. Direct evidence for tumor necrosis factor-alpha signaling in arteriogenesis. Circulation 2002, 105, 1639–1641. [Google Scholar] [CrossRef] [PubMed]
- Kosaki, K.; Ando, J.; Korenaga, R.; Kurokawa, T.; Kamiya, A. Fluid shear stress increases the production of granulocyte-macrophage colony-stimulating factor by endothelial cells via mRNA stabilization. Circ. Res. 1998, 82, 794–802. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, C.E.; Hoefer, I.E.; Meder, B.; Roth, H.; van Royen, N.; Breit, S.M.; Jost, M.M.; Aharinejad, S.; Hartmann, S.; Buschmann, I.R. Arteriogenesis depends on circulating monocytes and macrophage accumulation and is severely depressed in op/op mice. J. Leukoc. Biol. 2006, 80, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Stabile, E.; Burnett, M.S.; Watkins, C.; Kinnaird, T.; Bachis, A.; la Sala, A.; Miller, J.M.; Shou, M.; Epstein, S.E.; Fuchs, S. Impaired arteriogenic response to acute hindlimb ischemia in CD4-knockout mice. Circulation 2003, 108, 205–210. [Google Scholar] [CrossRef]
- Van Weel, V.; Toes, R.E.; Seghers, L.; Deckers, M.M.; de Vries, M.R.; Eilers, P.H.; Sipkens, J.; Schepers, A.; Eefting, D.; van Hinsbergh, V.W.; et al. Natural killer cells and CD4+ T-cells modulate collateral artery development. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 2310–2318. [Google Scholar] [CrossRef]
- Wolf, C.; Cai, W.J.; Vosschulte, R.; Koltai, S.; Mousavipour, D.; Scholz, D.; Afsah-Hedjri, A.; Schaper, W.; Schaper, J. Vascular remodeling and altered protein expression during growth of coronary collateral arteries. J. Mol. Cell. Cardiol. 1998, 30, 2291–2305. [Google Scholar] [CrossRef]
- Buschmann, I.; Schaper, W. Arteriogenesis Versus Angiogenesis: Two Mechanisms of Vessel Growth. News Physiol Sci. 1999, 14, 121–125. [Google Scholar] [CrossRef]
- Carmeliet, P. Mechanisms of angiogenesis and arteriogenesis. Nat. Med. 2000, 6, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, C.J.; Mitchell, J.R. Cellular infiltration of the human arterial adventitia associated with atheromatous plaques. Circulation 1962, 26, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.C.; Nakatsu, M.N.; Chou, W.; Gershon, P.D.; Hughes, C.C.W. The requirement for fibroblasts in angiogenesis: Fibroblast-derived matrix proteins are essential for endothelial cell lumen formation. Mol. Biol. Cell 2011, 22, 3791–3800. [Google Scholar] [CrossRef] [PubMed]
- Noonan, D.M.; De Lerma Barbaro, A.; Vannini, N.; Mortara, L.; Albini, A. Inflammation, inflammatory cells and angiogenesis: Decisions and indecisions. Cancer Metastasis Rev. 2008, 27, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, J.; Stark, A.; Russell, R.B.; Cohen, S.M. Principles of microRNA-target recognition. PLoS Biol. 2005, 3, e85. [Google Scholar] [CrossRef]
- Lewis, B.P.; Shih, I.H.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of mammalian microRNA targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef]
- Lee, Y.; Jeon, K.; Lee, J.T.; Kim, S.; Kim, V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002, 21, 4663–4670. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Kobayashi, H.; Tomari, Y. RISC assembly: Coordination between small RNAs and Argonaute proteins. Biochim. Biophys. Acta 2016, 1859, 71–81. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Plessmann, U.; Harlander, S.; Daiss, J.L.; Eichner, N.; Lehmann, G.; Schall, K.; Urlaub, H.; Meister, G. A Compendium of RNA-Binding Proteins that Regulate MicroRNA Biogenesis. Mol. Cell 2017, 66, 270. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Liang, G.; Egger, G.; Friedman, J.M.; Chuang, J.C.; Coetzee, G.A.; Jones, P.A. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 2006, 9, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Vrba, L.; Munoz-Rodriguez, J.L.; Stampfer, M.R.; Futscher, B.W. miRNA gene promoters are frequent targets of aberrant DNA methylation in human breast cancer. PLoS ONE 2013, 8, e54398. [Google Scholar] [CrossRef] [PubMed]
- Downie Ruiz Velasco, A.; Welten, S.M.J.; Goossens, E.A.C.; Quax, P.H.A.; Rappsilber, J.; Michlewski, G.; Nossent, A.Y. Posttranscriptional Regulation of 14q32 MicroRNAs by the CIRBP and HADHB during Vascular Regeneration after Ischemia. Mol. Ther. Nucleic Acids 2019, 14, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Kuehbacher, A.; Urbich, C.; Zeiher, A.M.; Dimmeler, S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ. Res. 2007, 101, 59–68. [Google Scholar] [CrossRef]
- Suarez, Y.; Fernandez-Hernando, C.; Pober, J.S.; Sessa, W.C. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ. Res. 2007, 100, 1164–1173. [Google Scholar] [CrossRef]
- Suarez, Y.; Fernandez-Hernando, C.; Yu, J.; Gerber, S.A.; Harrison, K.D.; Pober, J.S.; Iruela-Arispe, M.L.; Merkenschlager, M.; Sessa, W.C. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 14082–14087. [Google Scholar] [CrossRef]
- Kir, D.; Schnettler, E.; Modi, S.; Ramakrishnan, S. Regulation of angiogenesis by microRNAs in cardiovascular diseases. Angiogenesis 2018, 21, 699–710. [Google Scholar] [CrossRef]
- Lin, X.; Zhan, J.K.; Wang, Y.J.; Tan, P.; Chen, Y.Y.; Deng, H.Q.; Liu, Y.S. Function, Role, and Clinical Application of MicroRNAs in Vascular Aging. BioMed Res. Int. 2016, 2016, 6021394. [Google Scholar] [CrossRef]
- Sun, L.L.; Li, W.D.; Lei, F.R.; Li, X.Q. The regulatory role of microRNAs in angiogenesis-related diseases. J. Cell Mol. Med. 2018, 22, 4568–4587. [Google Scholar] [CrossRef]
- Lucas, T.; Bonauer, A.; Dimmeler, S. RNA Therapeutics in Cardiovascular Disease. Circ. Res. 2018, 123, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Bonauer, A.; Carmona, G.; Iwasaki, M.; Mione, M.; Koyanagi, M.; Fischer, A.; Burchfield, J.; Fox, H.; Doebele, C.; Ohtani, K.; et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science 2009, 324, 1710–1713. [Google Scholar] [CrossRef] [PubMed]
- Hinkel, R.; Penzkofer, D.; Zuhlke, S.; Fischer, A.; Husada, W.; Xu, Q.F.; Baloch, E.; van Rooij, E.; Zeiher, A.M.; Kupatt, C.; et al. Inhibition of microRNA-92a protects against ischemia/reperfusion injury in a large-animal model. Circulation 2013, 128, 1066–1075. [Google Scholar] [CrossRef] [PubMed]
- Bellera, N.; Barba, I.; Rodriguez-Sinovas, A.; Ferret, E.; Asin, M.A.; Gonzalez-Alujas, M.T.; Perez-Rodon, J.; Esteves, M.; Fonseca, C.; Toran, N.; et al. Single intracoronary injection of encapsulated antagomir-92a promotes angiogenesis and prevents adverse infarct remodeling. J. Am. Heart Assoc. 2014, 3, e000946. [Google Scholar] [CrossRef] [PubMed]
- Hanna, J.; Hossain, G.S.; Kocerha, J. The Potential for microRNA Therapeutics and Clinical Research. Front. Genet. 2019, 10, 478. [Google Scholar] [CrossRef] [PubMed]
- Fish, J.E.; Santoro, M.M.; Morton, S.U.; Yu, S.; Yeh, R.F.; Wythe, J.D.; Ivey, K.N.; Bruneau, B.G.; Stainier, D.Y.; Srivastava, D. miR-126 regulates angiogenic signaling and vascular integrity. Dev. Cell 2008, 15, 272–284. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Aurora, A.B.; Johnson, B.A.; Qi, X.; McAnally, J.; Hill, J.A.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell 2008, 15, 261–271. [Google Scholar] [CrossRef]
- Schober, A.; Nazari-Jahantigh, M.; Wei, Y.; Bidzhekov, K.; Gremse, F.; Grommes, J.; Megens, R.T.; Heyll, K.; Noels, H.; Hristov, M.; et al. MicroRNA-126-5p promotes endothelial proliferation and limits atherosclerosis by suppressing Dlk1. Nat. Med. 2014, 20, 368–376. [Google Scholar] [CrossRef]
- Harris, T.A.; Yamakuchi, M.; Ferlito, M.; Mendell, J.T.; Lowenstein, C.J. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc. Natl. Acad. Sci. USA 2008, 105, 1516–1521. [Google Scholar] [CrossRef]
- Van Solingen, C.; Seghers, L.; Bijkerk, R.; Duijs, J.M.; Roeten, M.K.; van Oeveren-Rietdijk, A.M.; Baelde, H.J.; Monge, M.; Vos, J.B.; de Boer, H.C.; et al. Antagomir-mediated silencing of endothelial cell specific microRNA-126 impairs ischemia-induced angiogenesis. J. Cell Mol. Med. 2009, 13, 1577–1585. [Google Scholar] [CrossRef]
- Katare, R.; Rawal, S.; Munasinghe, P.E.; Tsuchimochi, H.; Inagaki, T.; Fujii, Y.; Dixit, P.; Umetani, K.; Kangawa, K.; Shirai, M.; et al. Ghrelin Promotes Functional Angiogenesis in a Mouse Model of Critical Limb Ischemia Through Activation of Proangiogenic MicroRNAs. Endocrinology 2016, 157, 432–445. [Google Scholar] [CrossRef] [PubMed]
- Fichtlscherer, S.; De Rosa, S.; Fox, H.; Schwietz, T.; Fischer, A.; Liebetrau, C.; Weber, M.; Hamm, C.W.; Roxe, T.; Muller-Ardogan, M.; et al. Circulating microRNAs in patients with coronary artery disease. Circ. Res. 2010, 107, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ling, C.C.; Li, L.; Qin, Y.; Qi, J.; Liu, X.; You, B.; Shi, Y.; Zhang, J.; Jiang, Q.; et al. MicroRNA-10a/10b represses a novel target gene mib1 to regulate angiogenesis. Cardiovasc. Res. 2016, 110, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Hergenreider, E.; Heydt, S.; Treguer, K.; Boettger, T.; Horrevoets, A.J.; Zeiher, A.M.; Scheffer, M.P.; Frangakis, A.S.; Yin, X.; Mayr, M.; et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat. Cell Biol. 2012, 14, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Shi, C.; Manduchi, E.; Civelek, M.; Davies, P.F. MicroRNA-10a regulation of proinflammatory phenotype in athero-susceptible endothelium in vivo and in vitro. Proc. Natl. Acad Sci. USA 2010, 107, 13450–13455. [Google Scholar] [CrossRef]
- Hassel, D.; Cheng, P.; White, M.P.; Ivey, K.N.; Kroll, J.; Augustin, H.G.; Katus, H.A.; Stainier, D.Y.; Srivastava, D. MicroRNA-10 regulates the angiogenic behavior of zebrafish and human endothelial cells by promoting vascular endothelial growth factor signaling. Circ. Res. 2012, 111, 1421–1433. [Google Scholar] [CrossRef]
- Liu, L.Z.; Li, C.; Chen, Q.; Jing, Y.; Carpenter, R.; Jiang, Y.; Kung, H.F.; Lai, L.; Jiang, B.H. MiR-21 induced angiogenesis through AKT and ERK activation and HIF-1alpha expression. PLoS ONE 2011, 6, e19139. [Google Scholar] [CrossRef]
- Maegdefessel, L.; Azuma, J.; Toh, R.; Deng, A.; Merk, D.R.; Raiesdana, A.; Leeper, N.J.; Raaz, U.; Schoelmerich, A.M.; McConnell, M.V.; et al. MicroRNA-21 blocks abdominal aortic aneurysm development and nicotine-augmented expansion. Sci. Transl. Med. 2012, 4, 122ra122. [Google Scholar] [CrossRef]
- Ji, R.; Cheng, Y.; Yue, J.; Yang, J.; Liu, X.; Chen, H.; Dean, D.B.; Zhang, C. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ. Res. 2007, 100, 1579–1588. [Google Scholar] [CrossRef]
- Thum, T.; Gross, C.; Fiedler, J.; Fischer, T.; Kissler, S.; Bussen, M.; Galuppo, P.; Just, S.; Rottbauer, W.; Frantz, S.; et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 2008, 456, 980–984. [Google Scholar] [CrossRef]
- Kaluza, D.; Kroll, J.; Gesierich, S.; Manavski, Y.; Boeckel, J.N.; Doebele, C.; Zelent, A.; Rossig, L.; Zeiher, A.M.; Augustin, H.G.; et al. Histone deacetylase 9 promotes angiogenesis by targeting the antiangiogenic microRNA-17-92 cluster in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Landskroner-Eiger, S.; Qiu, C.; Perrotta, P.; Siragusa, M.; Lee, M.Y.; Ulrich, V.; Luciano, A.K.; Zhuang, Z.W.; Corti, F.; Simons, M.; et al. Endothelial miR-17 approximately 92 cluster negatively regulates arteriogenesis via miRNA-19 repression of WNT signaling. Proc. Natl. Acad. Sci. USA 2015, 112, 12812–12817. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Hoernes, T.P.; Erlacher, M.D. Translating the epitranscriptome. Wiley Interdiscip. Rev. RNA 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129, 1401–1414. [Google Scholar] [CrossRef]
- Blow, M.J.; Grocock, R.J.; van Dongen, S.; Enright, A.J.; Dicks, E.; Futreal, P.A.; Wooster, R.; Stratton, M.R. RNA editing of human microRNAs. Genome Biol. 2006, 7, R27. [Google Scholar] [CrossRef]
- Cloonan, N.; Wani, S.; Xu, Q.; Gu, J.; Lea, K.; Heater, S.; Barbacioru, C.; Steptoe, A.L.; Martin, H.C.; Nourbakhsh, E.; et al. MicroRNAs and their isomiRs function cooperatively to target common biological pathways. Genome Biol. 2011, 12, R126. [Google Scholar] [CrossRef]
- Neilsen, C.T.; Goodall, G.J.; Bracken, C.P. IsomiRs—The overlooked repertoire in the dynamic microRNAome. Trends Genet. 2012, 28, 544–549. [Google Scholar] [CrossRef]
- Llorens, F.; Banez-Coronel, M.; Pantano, L.; del Rio, J.A.; Ferrer, I.; Estivill, X.; Marti, E. A highly expressed miR-101 isomiR is a functional silencing small RNA. BMC Genom. 2013, 14, 104. [Google Scholar] [CrossRef]
- Loher, P.; Londin, E.R.; Rigoutsos, I. IsomiR expression profiles in human lymphoblastoid cell lines exhibit population and gender dependencies. Oncotarget 2014, 5, 8790–8802. [Google Scholar] [CrossRef]
- McCall, M.N.; Kim, M.S.; Adil, M.; Patil, A.H.; Lu, Y.; Mitchell, C.J.; Leal-Rojas, P.; Xu, J.; Kumar, M.; Dawson, V.L.; et al. Toward the human cellular microRNAome. Genome Res. 2017, 27, 1769–1781. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.C.; Chan, E.; Molnar, A.; Sarkar, R.; Alexieva, D.; Isa, I.M.; Robinson, S.; Zhang, S.; Ellis, P.; Langford, C.F.; et al. 5′ isomiR variation is of functional and evolutionary importance. Nucleic Acids Res. 2014, 42, 9424–9435. [Google Scholar] [CrossRef] [PubMed]
- Bofill-De Ros, X.; Yang, A.; Gu, S. IsomiRs: Expanding the miRNA repression toolbox beyond the seed. Biochim. Biophys. Acta Gene Regul. Mech. 2019. [Google Scholar] [CrossRef] [PubMed]
- Telonis, A.G.; Loher, P.; Jing, Y.; Londin, E.; Rigoutsos, I. Beyond the one-locus-one-miRNA paradigm: microRNA isoforms enable deeper insights into breast cancer heterogeneity. Nucleic Acids Res. 2015, 43, 9158–9175. [Google Scholar] [CrossRef] [PubMed]
- Burroughs, A.M.; Ando, Y.; de Hoon, M.J.; Tomaru, Y.; Nishibu, T.; Ukekawa, R.; Funakoshi, T.; Kurokawa, T.; Suzuki, H.; Hayashizaki, Y.; et al. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res. 2010, 20, 1398–1410. [Google Scholar] [CrossRef] [PubMed]
- Wyman, S.K.; Knouf, E.C.; Parkin, R.K.; Fritz, B.R.; Lin, D.W.; Dennis, L.M.; Krouse, M.A.; Webster, P.J.; Tewari, M. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Res. 2011, 21, 1450–1461. [Google Scholar] [CrossRef]
- Lee, L.W.; Zhang, S.; Etheridge, A.; Ma, L.; Martin, D.; Galas, D.; Wang, K. Complexity of the microRNA repertoire revealed by next-generation sequencing. RNA 2010, 16, 2170–2180. [Google Scholar] [CrossRef]
- Newman, M.A.; Mani, V.; Hammond, S.M. Deep sequencing of microRNA precursors reveals extensive 3′ end modification. RNA 2011, 17, 1795–1803. [Google Scholar] [CrossRef]
- Mercey, O.; Popa, A.; Cavard, A.; Paquet, A.; Chevalier, B.; Pons, N.; Magnone, V.; Zangari, J.; Brest, P.; Zaragosi, L.E.; et al. Characterizing isomiR variants within the microRNA-34/449 family. FEBS Lett. 2017, 591, 693–705. [Google Scholar] [CrossRef]
- Karali, M.; Persico, M.; Mutarelli, M.; Carissimo, A.; Pizzo, M.; Singh Marwah, V.; Ambrosio, C.; Pinelli, M.; Carrella, D.; Ferrari, S.; et al. High-resolution analysis of the human retina miRNome reveals isomiR variations and novel microRNAs. Nucleic Acids Res. 2016, 44, 1525–1540. [Google Scholar] [CrossRef]
- Manzano, M.; Forte, E.; Raja, A.N.; Schipma, M.J.; Gottwein, E. Divergent target recognition by coexpressed 5′-isomiRs of miR-142-3p and selective viral mimicry. RNA 2015, 21, 1606–1620. [Google Scholar] [CrossRef] [PubMed]
- Marzi, M.J.; Ghini, F.; Cerruti, B.; de Pretis, S.; Bonetti, P.; Giacomelli, C.; Gorski, M.M.; Kress, T.; Pelizzola, M.; Muller, H.; et al. Degradation dynamics of microRNAs revealed by a novel pulse-chase approach. Genome Res. 2016, 26, 554–565. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Liu, J.; Elfenbein, S.J.; Ma, Y.; Zhong, M.; Qiu, C.; Ding, Y.; Lu, J. Characterization of the mammalian miRNA turnover landscape. Nucleic Acids Res. 2015, 43, 2326–2341. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Vazquez, C.; Enright, A.J.; Rodriguez-Galan, A.; Perez-Garcia, A.; Collier, P.; Jones, M.R.; Benes, V.; Mizgerd, J.P.; Mittelbrunn, M.; Ramiro, A.R.; et al. 3′ Uridylation controls mature microRNA turnover during CD4 T-cell activation. RNA 2017, 23, 882–891. [Google Scholar] [CrossRef] [PubMed]
- Katoh, T.; Hojo, H.; Suzuki, T. Destabilization of microRNAs in human cells by 3′ deadenylation mediated by PARN and CUGBP1. Nucleic Acids Res. 2015, 43, 7521–7534. [Google Scholar] [CrossRef]
- Katoh, T.; Sakaguchi, Y.; Miyauchi, K.; Suzuki, T.; Kashiwabara, S.; Baba, T.; Suzuki, T. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 2009, 23, 433–438. [Google Scholar] [CrossRef]
- Yamane, D.; Selitsky, S.R.; Shimakami, T.; Li, Y.; Zhou, M.; Honda, M.; Sethupathy, P.; Lemon, S.M. Differential hepatitis C virus RNA target site selection and host factor activities of naturally occurring miR-122 3 variants. Nucleic Acids Res. 2017, 45, 4743–4755. [Google Scholar] [CrossRef][Green Version]
- Yu, F.; Pillman, K.A.; Neilsen, C.T.; Toubia, J.; Lawrence, D.M.; Tsykin, A.; Gantier, M.P.; Callen, D.F.; Goodall, G.J.; Bracken, C.P. Naturally existing isoforms of miR-222 have distinct functions. Nucleic Acids Res. 2017, 45, 11371–11385. [Google Scholar] [CrossRef]
- Voellenkle, C.; Rooij, J.; Guffanti, A.; Brini, E.; Fasanaro, P.; Isaia, E.; Croft, L.; David, M.; Capogrossi, M.C.; Moles, A.; et al. Deep-sequencing of endothelial cells exposed to hypoxia reveals the complexity of known and novel microRNAs. RNA 2012, 18, 472–484. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Orekhov, A.N.; Bobryshev, Y.V. The role of miR-126 in embryonic angiogenesis, adult vascular homeostasis, and vascular repair and its alterations in atherosclerotic disease. J. Mol. Cell. Cardiol. 2016, 97, 47–55. [Google Scholar] [CrossRef]
- Guduric-Fuchs, J.; O’Connor, A.; Cullen, A.; Harwood, L.; Medina, R.J.; O’Neill, C.L.; Stitt, A.W.; Curtis, T.M.; Simpson, D.A. Deep sequencing reveals predominant expression of miR-21 amongst the small non-coding RNAs in retinal microvascular endothelial cells. J. Cell. Biochem. 2012, 113, 2098–2111. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Valverde, S.L.; Taft, R.J.; Mattick, J.S. Dynamic isomiR regulation in Drosophila development. RNA 2010, 16, 1881–1888. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, S.H.; Fledderus, J.O.; Bot, P.T.; Moerland, P.D.; Hoefer, I.E.; Baan, J., Jr.; Henriques, J.P.; van der Schaaf, R.J.; Vis, M.M.; Horrevoets, A.J.; et al. Interferon-beta signaling is enhanced in patients with insufficient coronary collateral artery development and inhibits arteriogenesis in mice. Circ. Res. 2008, 102, 1286–1294. [Google Scholar] [CrossRef] [PubMed]
- Yildirim, C.; Nieuwenhuis, S.; Teunissen, P.F.; Horrevoets, A.J.; van Royen, N.; van der Pouw Kraan, T.C. Interferon-Beta, a Decisive Factor in Angiogenesis and Arteriogenesis. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 2015, 35, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, S.H.; Bot, P.T.; Fledderus, J.O.; van der Laan, A.M.; Volger, O.L.; Laufs, U.; Bohm, M.; de Vries, C.J.; Horrevoets, A.J.; Piek, J.J.; et al. Blocking interferon-beta stimulates vascular smooth muscle cell proliferation and arteriogenesis. J. Biol. Chem. 2010, 285, 34677–34685. [Google Scholar] [CrossRef] [PubMed]
- Nejad, C.; Pillman, K.A.; Siddle, K.J.; Pepin, G.; Anko, M.L.; McCoy, C.E.; Beilharz, T.H.; Quintana-Murci, L.; Goodall, G.J.; Bracken, C.P.; et al. miR-222 isoforms are differentially regulated by type-I interferon. RNA 2018, 24, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Poliseno, L.; Tuccoli, A.; Mariani, L.; Evangelista, M.; Citti, L.; Woods, K.; Mercatanti, A.; Hammond, S.; Rainaldi, G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood 2006, 108, 3068–3071. [Google Scholar] [CrossRef]
- Dentelli, P.; Rosso, A.; Orso, F.; Olgasi, C.; Taverna, D.; Brizzi, M.F. microRNA-222 controls neovascularization by regulating signal transducer and activator of transcription 5A expression. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 1562–1568. [Google Scholar] [CrossRef]
- Van der Kwast, R.; Woudenberg, T.; Quax, P.H.A.; Nossent, A.Y. MicroRNA-411 and Its 5′-IsomiR Have Distinct Targets and Functions and Are Differentially Regulated in the Vasculature under Ischemia. Mol. Ther. J. Am. Soc. Gene Ther. 2019. [Google Scholar] [CrossRef]
- Wagner, R.W.; Smith, J.E.; Cooperman, B.S.; Nishikura, K. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc. Natl. Acad. Sci. USA 1989, 86, 2647–2651. [Google Scholar] [CrossRef]
- Gommans, W.M. A-to-I editing of microRNAs: Regulating the regulators? Semin. Cell Dev. Biol. 2012, 23, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Kume, H.; Hino, K.; Galipon, J.; Ui-Tei, K. A-to-I editing in the miRNA seed region regulates target mRNA selection and silencing efficiency. Nucleic Acids Res. 2014, 42, 10050–10060. [Google Scholar] [CrossRef] [PubMed]
- Savva, Y.A.; Rieder, L.E.; Reenan, R.A. The ADAR protein family. Genome Biol. 2012, 13, 252. [Google Scholar] [CrossRef] [PubMed]
- Nishikura, K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2016, 17, 83–96. [Google Scholar] [CrossRef]
- Mallela, A.; Nishikura, K. A-to-I editing of protein coding and noncoding RNAs. Crit. Rev. Biochem. Mol. Biol. 2012, 47, 493–501. [Google Scholar] [CrossRef]
- Nigita, G.; Veneziano, D.; Ferro, A. A-to-I RNA Editing: Current Knowledge Sources and Computational Approaches with Special Emphasis on Non-Coding RNA Molecules. Front. Bioeng. Biotechnol. 2015, 3. [Google Scholar] [CrossRef]
- Wang, Q.; Khillan, J.; Gadue, P.; Nishikura, K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science 2000, 290, 1765–1768. [Google Scholar] [CrossRef]
- Higuchi, M.; Maas, S.; Single, F.N.; Hartner, J.; Rozov, A.; Burnashev, N.; Feldmeyer, D.; Sprengel, R.; Seeburg, P.H. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 2000, 406, 78–81. [Google Scholar] [CrossRef]
- Brusa, R.; Zimmermann, F.; Koh, D.S.; Feldmeyer, D.; Gass, P.; Seeburg, P.H.; Sprengel, R. Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science 1995, 270, 1677–1680. [Google Scholar] [CrossRef]
- Hong, H.; Lin, J.; Chen, L. Regulatory factors governing adenosine-to-inosine (A-to-I) RNA editing. Biosci Rep. 2015, 35. [Google Scholar] [CrossRef]
- Lai, F.; Chen, C.X.; Lee, V.M.; Nishikura, K. Dramatic increase of the RNA editing for glutamate receptor subunits during terminal differentiation of clonal human neurons. J. Neurochem. 1997, 69, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Emeson, R.B.; Samuel, C.E. Serotonin-2C receptor pre-mRNA editing in rat brain and in vitro by splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase ADAR1. J. Biol. Chem. 1999, 274, 18351–18358. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Chendrimada, T.P.; Wang, Q.; Higuchi, M.; Seeburg, P.H.; Shiekhattar, R.; Nishikura, K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006, 13, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, Y.; Zinshteyn, B.; Chendrimada, T.P.; Shiekhattar, R.; Nishikura, K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007, 8, 763–769. [Google Scholar] [CrossRef]
- Kawahara, Y.; Megraw, M.; Kreider, E.; Iizasa, H.; Valente, L.; Hatzigeorgiou, A.G.; Nishikura, K. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 2008, 36, 5270–5280. [Google Scholar] [CrossRef]
- Kawahara, Y.; Zinshteyn, B.; Sethupathy, P.; Iizasa, H.; Hatzigeorgiou, A.G.; Nishikura, K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science 2007, 315, 1137–1140. [Google Scholar] [CrossRef]
- Li, L.; Song, Y.; Shi, X.; Liu, J.; Xiong, S.; Chen, W.; Fu, Q.; Huang, Z.; Gu, N.; Zhang, R. The landscape of miRNA editing in animals and its impact on miRNA biogenesis and targeting. Genome Res. 2018, 28, 132–143. [Google Scholar] [CrossRef]
- Vitsios, D.M.; Davis, M.P.; van Dongen, S.; Enright, A.J. Large-scale analysis of microRNA expression, epi-transcriptomic features and biogenesis. Nucleic Acids Res. 2016. [Google Scholar] [CrossRef]
- Wang, Y.; Liang, H. When MicroRNAs Meet RNA Editing in Cancer: A Nucleotide Change Can Make a Difference. Bioessays News Rev. Mol. Cell. Dev. Biol. 2018, 40. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, X.; Yu, S.; Jeong, K.J.; Zhou, Z.; Han, L.; Tsang, Y.H.; Li, J.; Chen, H.; Mangala, L.S.; et al. Systematic characterization of A-to-I RNA editing hotspots in microRNAs across human cancers. Genome Res. 2017, 27, 1112–1125. [Google Scholar] [CrossRef]
- Cho, C.J.; Myung, S.J.; Chang, S. ADAR1 and MicroRNA.; A Hidden Crosstalk in Cancer. Int. J. Mol. Sci. 2017, 18, 799. [Google Scholar] [CrossRef] [PubMed]
- Shoshan, E.; Mobley, A.K.; Braeuer, R.R.; Kamiya, T.; Huang, L.; Vasquez, M.E.; Salameh, A.; Lee, H.J.; Kim, S.J.; Ivan, C.; et al. Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma growth and metastasis. Nat. Cell Biol. 2015, 17, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Van der Kwast, R.V.C.T.; van Ingen, E.; Parma, L.; Peters, H.A.B.; Quax, P.H.A.; Nossent, A.Y. Adenosine-to-Inosine Editing of MicroRNA-487b Alters Target Gene Selection After Ischemia and Promotes Neovascularization. Circ. Res. 2018, 122, 444–456. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef]
- Wang, P.; Doxtader, K.A.; Nam, Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol. Cell 2016, 63, 306–317. [Google Scholar] [CrossRef]
- Yang, Y.; Hsu, P.J.; Chen, Y.S.; Yang, Y.G. Dynamic transcriptomic m(6)A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef]
- Schwartz, S.; Mumbach, M.R.; Jovanovic, M.; Wang, T.; Maciag, K.; Bushkin, G.G.; Mertins, P.; Ter-Ovanesyan, D.; Habib, N.; Cacchiarelli, D.; et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014, 8, 284–296. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vagbo, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Nachtergaele, S.; He, C. The emerging biology of RNA post-transcriptional modifications. RNA Biol. 2017, 14, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Yoon, K.J.; Ringeling, F.R.; Vissers, C.; Jacob, F.; Pokrass, M.; Jimenez-Cyrus, D.; Su, Y.; Kim, N.S.; Zhu, Y.; Zheng, L.; et al. Temporal Control of Mammalian Cortical Neurogenesis by m(6)A Methylation. Cell 2017, 171, 877–889. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Y.; Toth, J.I.; Petroski, M.D.; Zhang, Z.; Zhao, J.C. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014, 16, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, K.; Umetani, M.; Minami, T.; Okayama, H.; Takada, S.; Yamamoto, M.; Aburatani, H.; Reid, P.C.; Housman, D.E.; Hamakubo, T.; et al. Wilms’ tumor 1-associating protein regulates G2/M transition through stabilization of cyclin A2 mRNA. Proc. Natl. Acad. Sci. USA 2006, 103, 17278–17283. [Google Scholar] [CrossRef]
- Liu, N.; Zhou, K.I.; Parisien, M.; Dai, Q.; Diatchenko, L.; Pan, T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017, 45, 6051–6063. [Google Scholar] [CrossRef]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L.; et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018, 20, 285–295. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Alarcon, C.R.; Goodarzi, H.; Lee, H.; Liu, X.; Tavazoie, S.; Tavazoie, S.F. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell 2015, 162, 1299–1308. [Google Scholar] [CrossRef] [PubMed]
- Alarcon, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Dai, Q.; Fong, R.; Saikia, M.; Stephenson, D.; Yu, Y.T.; Pan, T.; Piccirilli, J.A. Identification of recognition residues for ligation-based detection and quantitation of pseudouridine and N6-methyladenosine. Nucleic Acids Res. 2007, 35, 6322–6329. [Google Scholar] [CrossRef]
- Konno, M.; Koseki, J.; Asai, A.; Yamagata, A.; Shimamura, T.; Motooka, D.; Okuzaki, D.; Kawamoto, K.; Mizushima, T.; Eguchi, H.; et al. Distinct methylation levels of mature microRNAs in gastrointestinal cancers. Nat. Commun. 2019, 10, 3888. [Google Scholar] [CrossRef]
- Muller, S.; Glass, M.; Singh, A.K.; Haase, J.; Bley, N.; Fuchs, T.; Lederer, M.; Dahl, A.; Huang, H.; Chen, J.; et al. IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- and miRNA-dependent manner. Nucleic Acids Res. 2019, 47, 375–390. [Google Scholar] [CrossRef]
- Dorn, L.E.; Lasman, L.; Chen, J.; Xu, X.; Hund, T.J.; Medvedovic, M.; Hanna, J.H.; van Berlo, J.H.; Accornero, F. The N(6)-Methyladenosine mRNA Methylase METTL3 Controls Cardiac Homeostasis and Hypertrophy. Circulation 2019, 139, 533–545. [Google Scholar] [CrossRef]
- Mathiyalagan, P.; Adamiak, M.; Mayourian, J.; Sassi, Y.; Liang, Y.; Agarwal, N.; Jha, D.; Zhang, S.; Kohlbrenner, E.; Chepurko, E.; et al. FTO-Dependent N(6)-Methyladenosine Regulates Cardiac Function During Remodeling and Repair. Circulation 2019, 139, 518–532. [Google Scholar] [CrossRef]
- Ma, J.Z.; Yang, F.; Zhou, C.C.; Liu, F.; Yuan, J.H.; Wang, F.; Wang, T.T.; Xu, Q.G.; Zhou, W.P.; Sun, S.H. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6)-methyladenosine-dependent primary MicroRNA processing. Hepatology 2017, 65, 529–543. [Google Scholar] [CrossRef]
- Han, J.; Wang, J.Z.; Yang, X.; Yu, H.; Zhou, R.; Lu, H.C.; Yuan, W.B.; Lu, J.C.; Zhou, Z.J.; Lu, Q.; et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol. Cancer 2019, 18, 110. [Google Scholar] [CrossRef]
- Cech, T.R.; Steitz, J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Heo, I.; Kim, V.N. Modifications of small RNAs and their associated proteins. Cell 2010, 143, 703–709. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Cozen, A.E.; Liu, Y.; Chen, Q.; Lowe, T.M. Small RNA Modifications: Integral to Function and Disease. Trends Mol. Med. 2016, 22, 1025–1034. [Google Scholar] [CrossRef] [PubMed]
- Lan, M.D.; Xiong, J.; You, X.J.; Weng, X.C.; Zhou, X.; Yuan, B.F.; Feng, Y.Q. Existence of Diverse Modifications in Small-RNA Species Composed of 16-28 Nucleotides. Chem. Eur. J. 2018, 24, 9949–9956. [Google Scholar] [CrossRef] [PubMed]
- Charette, M.; Gray, M.W. Pseudouridine in RNA: What, where, how, and why. Iubmb Life 2000, 49, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Karijolich, J.; Yi, C.; Yu, Y.T. Transcriptome-wide dynamics of RNA pseudouridylation. Nat. Rev. Mol. Cell Biol. 2015, 16, 581–585. [Google Scholar] [CrossRef]
- Schwartz, S.; Bernstein, D.A.; Mumbach, M.R.; Jovanovic, M.; Herbst, R.H.; Leon-Ricardo, B.X.; Engreitz, J.M.; Guttman, M.; Satija, R.; Lander, E.S.; et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 2014, 159, 148–162. [Google Scholar] [CrossRef]
- Arnez, J.G.; Steitz, T.A. Crystal structure of unmodified tRNA(Gln) complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry 1994, 33, 7560–7567. [Google Scholar] [CrossRef]
- Cohn, W.E. Pseudouridine, a carbon-carbon linked ribonucleoside in ribonucleic acids: Isolation, structure, and chemical characteristics. J. Biol. Chem. 1960, 235, 1488–1498. [Google Scholar]
- Carlile, T.M.; Rojas-Duran, M.F.; Zinshteyn, B.; Shin, H.; Bartoli, K.M.; Gilbert, W.V. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 2014, 515, 143–146. [Google Scholar] [CrossRef]
- Ayadi, L.; Galvanin, A.; Pichot, F.; Marchand, V.; Motorin, Y. RNA ribose methylation (2′-O-methylation): Occurrence, biosynthesis and biological functions. Biochim. Biophys. Acta Gene Regul. Mech. 2018. [Google Scholar] [CrossRef] [PubMed]
- Dimitrova, D.G.; Teysset, L.; Carre, C. RNA 2′-O-Methylation (Nm) Modification in Human Diseases. Genes 2019, 10, 117. [Google Scholar] [CrossRef] [PubMed]
- Bachellerie, J.P.; Cavaille, J.; Huttenhofer, A. The expanding snoRNA world. Biochimie 2002, 84, 775–790. [Google Scholar] [CrossRef]
- Cavaille, J.; Buiting, K.; Kiefmann, M.; Lalande, M.; Brannan, C.I.; Horsthemke, B.; Bachellerie, J.P.; Brosius, J.; Huttenhofer, A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. USA 2000, 97, 14311–14316. [Google Scholar] [CrossRef]
- Vitali, P.; Basyuk, E.; Le, M.E.; Bertrand, E.; Muscatelli, F.; Cavaille, J.; Huttenhofer, A. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J. Cell Biol. 2005, 169, 745–753. [Google Scholar] [CrossRef]
- Yi-Brunozzi, H.Y.; Easterwood, L.M.; Kamilar, G.M.; Beal, P.A. Synthetic substrate analogs for the RNA-editing adenosine deaminase ADAR-2. Nucleic Acids Res. 1999, 27, 2912–2917. [Google Scholar] [CrossRef]
- Mizrahi, R.A.; Phelps, K.J.; Ching, A.Y.; Beal, P.A. Nucleoside analog studies indicate mechanistic differences between RNA-editing adenosine deaminases. Nucleic Acids Res. 2012, 40, 9825–9835. [Google Scholar] [CrossRef]
- Inoue, H.; Hayase, Y.; Imura, A.; Iwai, S.; Miura, K.; Ohtsuka, E. Synthesis and hybridization studies on two complementary nona(2′-O-methyl)ribonucleotides. Nucleic Acids Res. 1987, 15, 6131–6148. [Google Scholar] [CrossRef]
- Majlessi, M.; Nelson, N.C.; Becker, M.M. Advantages of 2′-O-methyl oligoribonucleotide probes for detecting RNA targets. Nucleic Acids Res. 1998, 26, 2224–2229. [Google Scholar] [CrossRef]
- Tsourkas, A.; Behlke, M.A.; Bao, G. Hybridization of 2′-O-methyl and 2′-deoxy molecular beacons to RNA and DNA targets. Nucleic Acids Res. 2002, 30, 5168–5174. [Google Scholar] [CrossRef]
- Yu, B.; Yang, Z.; Li, J.; Minakhina, S.; Yang, M.; Padgett, R.W.; Steward, R.; Chen, X. Methylation as a crucial step in plant microRNA biogenesis. Science 2005, 307, 932–935. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Nachtergaele, S.; Moshitch-Moshkovitz, S.; Peer, E.; Kol, N.; Ben-Haim, M.S.; Dai, Q.; Di Segni, A.; Salmon-Divon, M.; Clark, W.C.; et al. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature 2016, 530, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xiong, X.; Wang, K.; Wang, L.; Shu, X.; Ma, S.; Yi, C. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat. Chem. Biol. 2016, 12, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Jia, G. Reversible RNA Modification N(1)-methyladenosine (m(1)A) in mRNA and tRNA. Genom. Proteom. Bioinform. 2018, 16, 155–161. [Google Scholar] [CrossRef]
- Liu, F.; Clark, W.; Luo, G.; Wang, X.; Fu, Y.; Wei, J.; Wang, X.; Hao, Z.; Dai, Q.; Zheng, G.; et al. ALKBH1-Mediated tRNA Demethylation Regulates Translation. Cell 2016, 167, 816–828. [Google Scholar] [CrossRef]
- Zhou, H.; Kimsey, I.J.; Nikolova, E.N.; Sathyamoorthy, B.; Grazioli, G.; McSally, J.; Bai, T.; Wunderlich, C.H.; Kreutz, C.; Andricioaei, I.; et al. m(1)A and m(1)G disrupt A-RNA structure through the intrinsic instability of Hoogsteen base pairs. Nat. Struct. Mol. Biol. 2016, 23, 803–810. [Google Scholar] [CrossRef]
- Squires, J.E.; Patel, H.R.; Nousch, M.; Sibbritt, T.; Humphreys, D.T.; Parker, B.J.; Suter, C.M.; Preiss, T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012, 40, 5023–5033. [Google Scholar] [CrossRef]
- Van Haute, L.; Dietmann, S.; Kremer, L.; Hussain, S.; Pearce, S.F.; Powell, C.A.; Rorbach, J.; Lantaff, R.; Blanco, S.; Sauer, S.; et al. Deficient methylation and formylation of mt-tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3. Nat. Commun. 2016, 7, 12039. [Google Scholar] [CrossRef]
- Nakano, S.; Suzuki, T.; Kawarada, L.; Iwata, H.; Asano, K.; Suzuki, T. NSUN3 methylase initiates 5-formylcytidine biogenesis in human mitochondrial tRNA(Met). Nat. Chem. Biol. 2016, 12, 546–551. [Google Scholar] [CrossRef]
- Goll, M.G.; Kirpekar, F.; Maggert, K.A.; Yoder, J.A.; Hsieh, C.L.; Zhang, X.; Golic, K.G.; Jacobsen, S.E.; Bestor, T.H. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 2006, 311, 395–398. [Google Scholar] [CrossRef]
- Trixl, L.; Lusser, A. The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark. Wiley Interdiscip. Rev. RNA 2019, 10, e1510. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Sajini, A.A.; Blanco, S.; Dietmann, S.; Lombard, P.; Sugimoto, Y.; Paramor, M.; Gleeson, J.G.; Odom, D.T.; Ule, J.; et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013, 4, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Khoddami, V.; Cairns, B.R. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat. Biotechnol. 2013, 31, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Jurkowski, T.P.; Meusburger, M.; Phalke, S.; Helm, M.; Nellen, W.; Reuter, G.; Jeltsch, A. Human DNMT2 methylates tRNA(Asp) molecules using a DNA methyltransferase-like catalytic mechanism. RNA 2008, 14, 1663–1670. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.; Pollex, T.; Hanna, K.; Tuorto, F.; Meusburger, M.; Helm, M.; Lyko, F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010, 24, 1590–1595. [Google Scholar] [CrossRef]
- Aguilo, F.; Li, S.; Balasubramaniyan, N.; Sancho, A.; Benko, S.; Zhang, F.; Vashisht, A.; Rengasamy, M.; Andino, B.; Chen, C.H.; et al. Deposition of 5-Methylcytosine on Enhancer RNAs Enables the Coactivator Function of PGC-1alpha. Cell Rep. 2016, 14, 479–492. [Google Scholar] [CrossRef]
- Sergiev, P.V.; Bogdanov, A.A.; Dontsova, O.A. Ribosomal RNA guanine-(N2)-methyltransferases and their targets. Nucleic Acids Res. 2007, 35, 2295–2301. [Google Scholar] [CrossRef]
- Chen, W.; Song, X.; Lv, H.; Lin, H. iRNA-m2G: Identifying N(2)-methylguanosine Sites Based on Sequence-Derived Information. Mol. Ther. Nucleic Acids 2019, 18, 253–258. [Google Scholar] [CrossRef]
- Bavi, R.S.; Kamble, A.D.; Kumbhar, N.M.; Kumbhar, B.V.; Sonawane, K.D. Conformational preferences of modified nucleoside N(2)-methylguanosine (m(2)G) and its derivative N(2), N(2)-dimethylguanosine (m(2)(2)G) occur at 26th position (hinge region) in tRNA. Cell Biochem. Biophys. 2011, 61, 507–521. [Google Scholar] [CrossRef]
- Bavi, R.S.; Sambhare, S.B.; Sonawane, K.D. MD simulation studies to investigate iso-energetic conformational behaviour of modified nucleosides m(2)G and m(2) 2G present in tRNA. Comput. Struct. Biotechnol. J. 2013, 5, e201302015. [Google Scholar] [CrossRef]
- Pinto, Y.; Buchumenski, I.; Levanon, E.Y.; Eisenberg, E. Human cancer tissues exhibit reduced A-to-I editing of miRNAs coupled with elevated editing of their targets. Nucleic Acids Res. 2018, 46, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M.; Pandolfi, P.P. The Epitranscriptome of Noncoding RNAs in Cancer. Cancer Discov. 2017, 7, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.B.; Jaffrey, S.R. 5′ UTR m(6)A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Laurent, B.; Hsu, C.H.; Nachtergaele, S.; Lu, Z.; Sheng, W.; Xu, C.; Chen, H.; Ouyang, J.; Wang, S.; et al. RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature 2017, 543, 573–576. [Google Scholar] [CrossRef]
- Zhou, J.; Wan, J.; Gao, X.; Zhang, X.; Jaffrey, S.R.; Qian, S.B. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 2015, 526, 591–594. [Google Scholar] [CrossRef]
- Blanco, S.; Bandiera, R.; Popis, M.; Hussain, S.; Lombard, P.; Aleksic, J.; Sajini, A.; Tanna, H.; Cortes-Garrido, R.; Gkatza, N.; et al. Stem cell function and stress response are controlled by protein synthesis. Nature 2016, 534, 335–340. [Google Scholar] [CrossRef]
- Blanco, S.; Dietmann, S.; Flores, J.V.; Hussain, S.; Kutter, C.; Humphreys, P.; Lukk, M.; Lombard, P.; Treps, L.; Popis, M.; et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014, 33, 2020–2039. [Google Scholar] [CrossRef]
- Krogh, N.; Jansson, M.D.; Hafner, S.J.; Tehler, D.; Birkedal, U.; Christensen-Dalsgaard, M.; Lund, A.H.; Nielsen, H. Profiling of 2′-O-Me in human rRNA reveals a subset of fractionally modified positions and provides evidence for ribosome heterogeneity. Nucleic Acids Res. 2016, 44, 7884–7895. [Google Scholar] [CrossRef]
- Marcel, V.; Ghayad, S.E.; Belin, S.; Therizols, G.; Morel, A.P.; Solano-Gonzalez, E.; Vendrell, J.A.; Hacot, S.; Mertani, H.C.; Albaret, M.A.; et al. p53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell 2013, 24, 318–330. [Google Scholar] [CrossRef]
- Gatsiou, A.; Stellos, K. Dawn of Epitranscriptomic Medicine. Circ. Genom. Precis. Med. 2018, 11, e001927. [Google Scholar] [CrossRef]
- Stellos, K.; Gatsiou, A.; Stamatelopoulos, K.; Perisic Matic, L.; John, D.; Lunella, F.F.; Jae, N.; Rossbach, O.; Amrhein, C.; Sigala, F.; et al. Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation. Nat. Med. 2016, 22, 1140–1150. [Google Scholar] [CrossRef] [PubMed]
- Veliz, E.A.; Easterwood, L.M.; Beal, P.A. Substrate analogues for an RNA-editing adenosine deaminase: Mechanistic investigation and inhibitor design. J. Am. Chem. Soc. 2003, 125, 10867–10876. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.F.; Yang, Q.; Liu, C.X.; Wu, M.; Chen, L.L.; Yang, L. N(6)-Methyladenosines Modulate A-to-I RNA Editing. Mol. Cell 2018, 69, 126–135.e126. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Telonis, A.G.; Magee, R.; Loher, P.; Chervoneva, I.; Londin, E.; Rigoutsos, I. Knowledge about the presence or absence of miRNA isoforms (isomiRs) can successfully discriminate amongst 32 TCGA cancer types. Nucleic Acids Res. 2017, 45, 2973–2985. [Google Scholar] [CrossRef]
- Schaefer, M.; Kapoor, U.; Jantsch, M.F. Understanding RNA modifications: The promises and technological bottlenecks of the ‘epitranscriptome’. Open Biol. 2017, 7, 170077. [Google Scholar] [CrossRef]
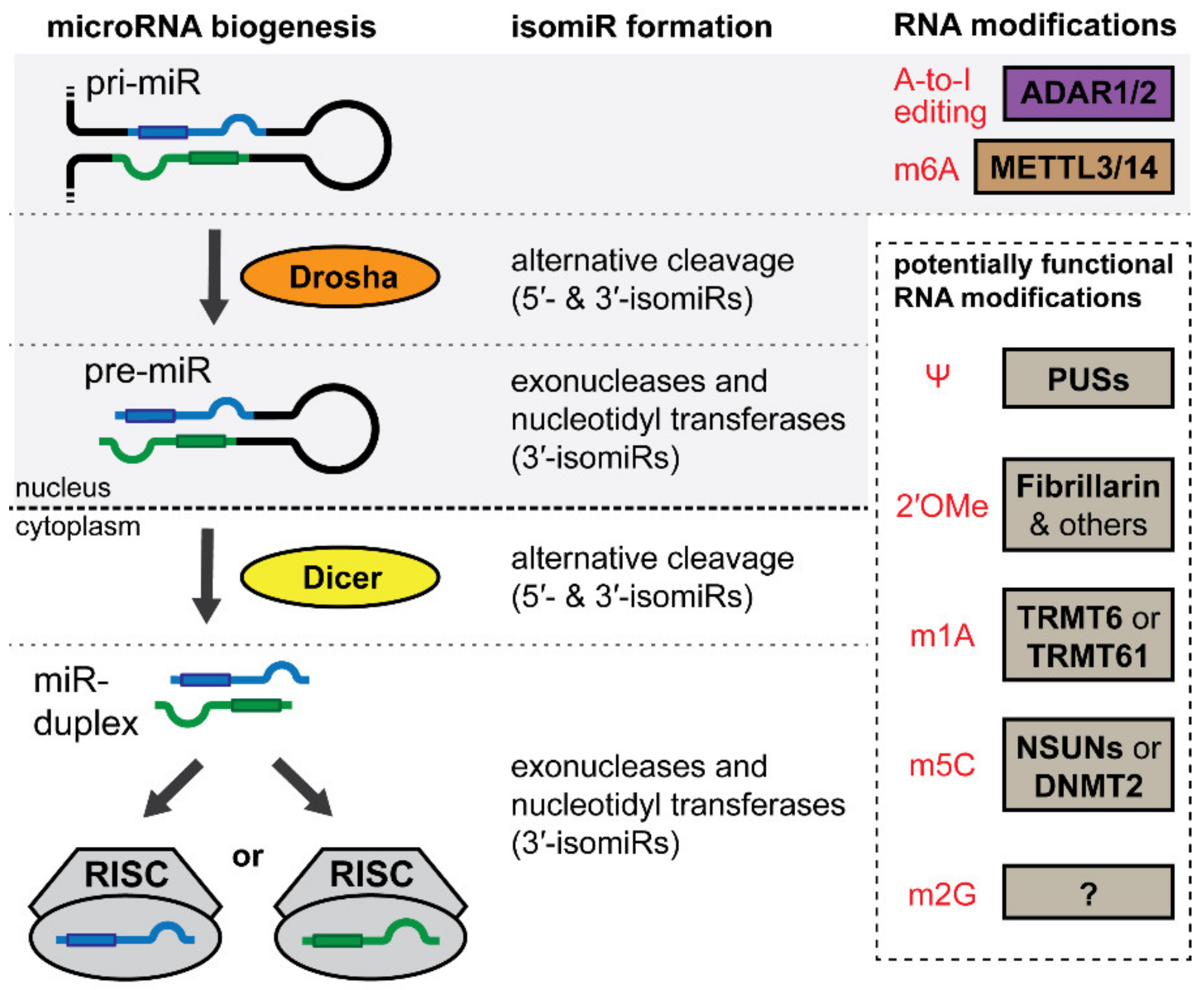
| Topic | Key Findings | References |
|---|---|---|
| Prevalence of isomiRs |
| [71,72,89,91] |
| [74] | |
| Potential functional effect of isomiRs |
| [67,79,80,81,99] |
| [82,83,84,85,86] | |
| [87,88] | |
| Abundant vasoactive microRNAs with isomiRs |
| [74,89,91] |
| [74] | |
| [89,91] | |
| [88,96] | |
| [99] | |
| Regulation of isomiRs |
| [75,76,78,92] |
| [89] | |
| [99] |
| Nucleotide Modification | Abbreviation | Writers | Erasers | Potential Effects on microRNAs |
|---|---|---|---|---|
| Adenosine-to-inosine editing | A-to-I editing | ADAR1 or ADAR2 | - |
|
| N6-methyl-adenosine | m6A | METTL3/14 | ALKBH5 FTO |
|
| Pseudouridine | Ψ | PUSs | - |
|
| 2′-O-methyl-nucleosides | 2′OMe | Methyl-transferases | - |
|
| N1-methyl-adenosine | m1A | TRMT6 & 61 | ALKBH3 |
|
| N5-methyl-cytosine | m5C | NSUNs DNMT2 | - |
|
| N2-methyl-guanosine | m2G | unclear | - |
|
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
van der Kwast, R.V.C.T.; Quax, P.H.A.; Nossent, A.Y. An Emerging Role for isomiRs and the microRNA Epitranscriptome in Neovascularization. Cells 2020, 9, 61. https://doi.org/10.3390/cells9010061
van der Kwast RVCT, Quax PHA, Nossent AY. An Emerging Role for isomiRs and the microRNA Epitranscriptome in Neovascularization. Cells. 2020; 9(1):61. https://doi.org/10.3390/cells9010061
Chicago/Turabian Stylevan der Kwast, Reginald V.C.T., Paul H.A. Quax, and A. Yaël Nossent. 2020. "An Emerging Role for isomiRs and the microRNA Epitranscriptome in Neovascularization" Cells 9, no. 1: 61. https://doi.org/10.3390/cells9010061
APA Stylevan der Kwast, R. V. C. T., Quax, P. H. A., & Nossent, A. Y. (2020). An Emerging Role for isomiRs and the microRNA Epitranscriptome in Neovascularization. Cells, 9(1), 61. https://doi.org/10.3390/cells9010061






