Zinc Finger Transcription Factor MZF1—A Specific Regulator of Cancer Invasion
Abstract
1. Transcription Factors as Drug Targets in Cancer
2. What Is MZF1?
2.1. MZF1 Is a Sequence-Specific, Oncogenic Transcription Factor Involved in Myeloid Differentiation
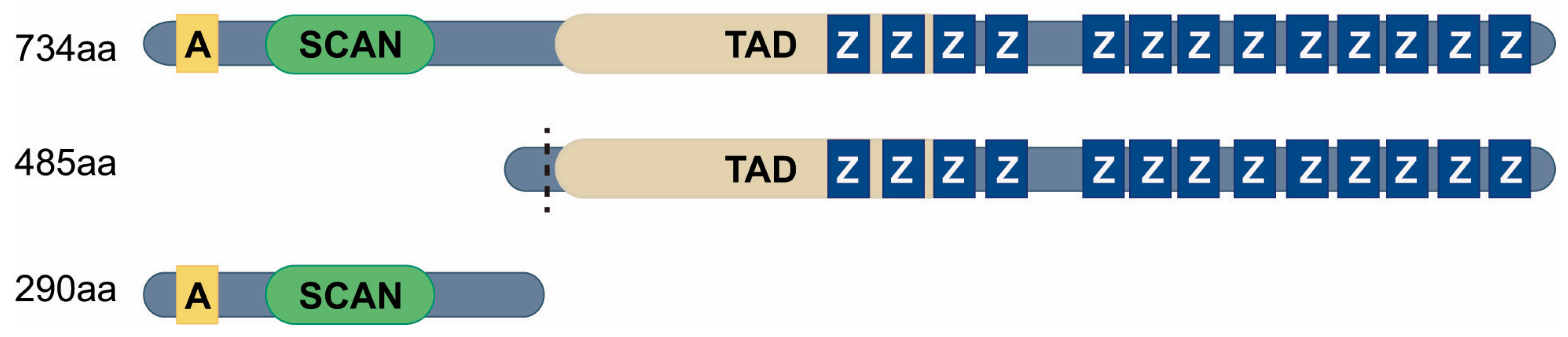
2.2. MZF1 Transcript Variants and Functional Domains
3. MZF1 and Cancer
3.1. MZF1 and Hematological Malignancies
3.2. MZF1 Acts as an Oncogene in Solid Cancers
3.2.1. MZF1 in Breast Cancer
3.2.2. MZF1 in Cervical and Colorectal Cancers
3.2.3. MZF1 in Liver and Lung Cancer
3.2.4. MZF1 in Prostate Cancer
3.2.5. MZF1 in Other Type of Cancers
3.2.6. Many MZF1 Target Genes Have a Role in Cancer
4. How Does MZF1 Function?
4.1. Regulation of MZF1 Expression
4.2. Regulation of the Transcriptional Activity of MZF1
4.2.1. Interaction with Other Transcription Factors
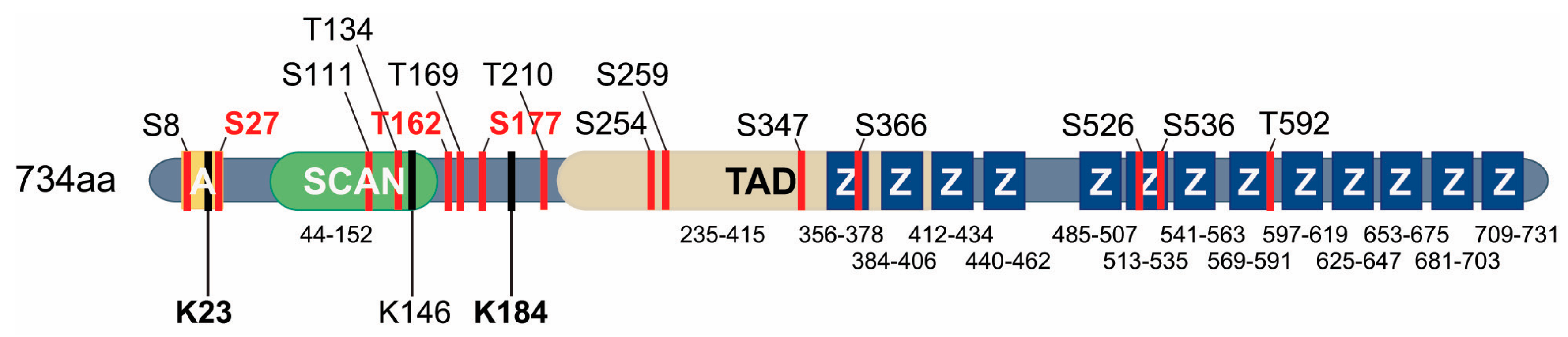
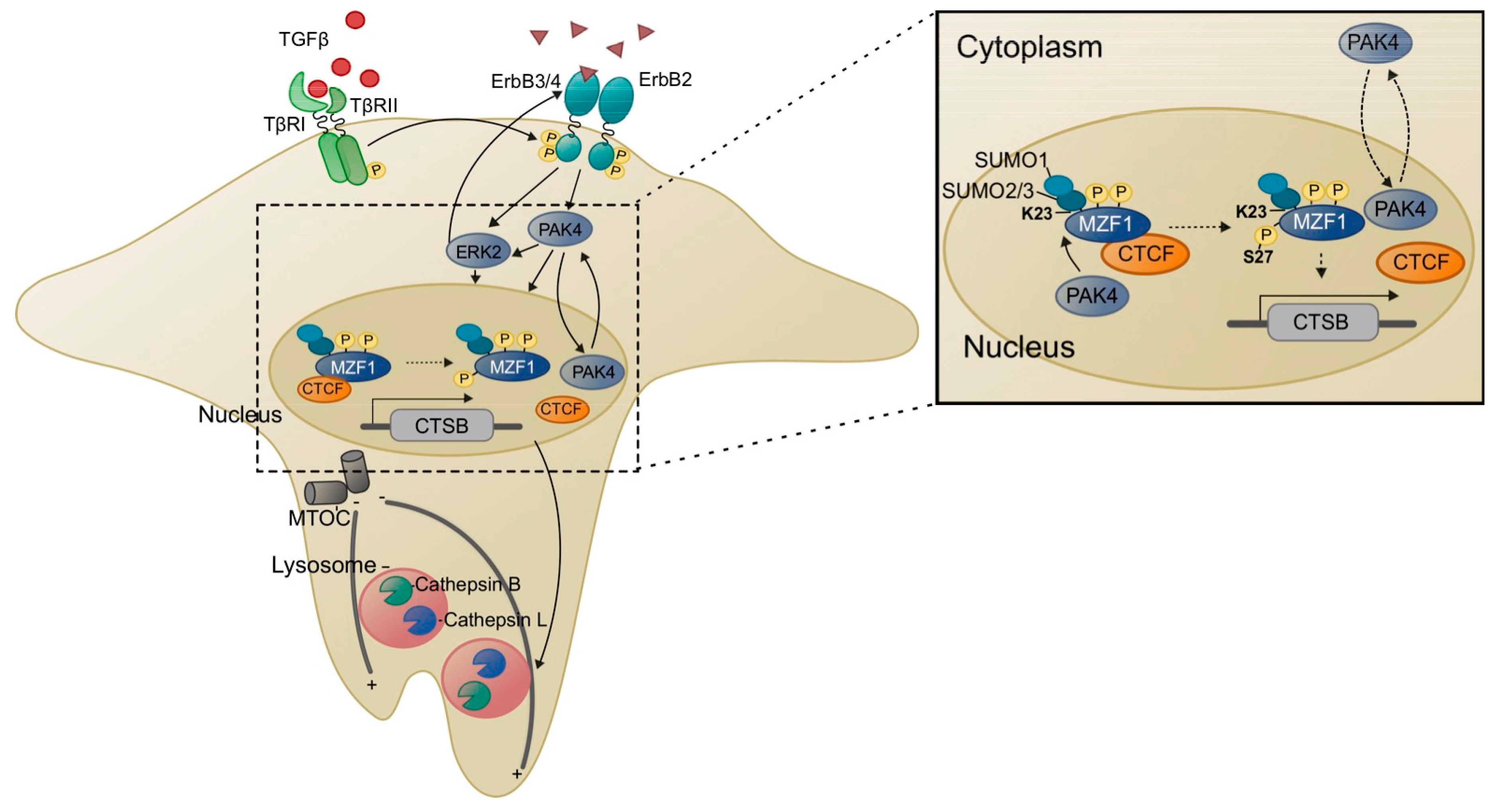
4.2.2. SUMOylation of MZF1
4.2.3. Phosphorylation of MZF1
4.2.4. Mutations Creating New MZF1 Binding Sites in the Genome
5. How Does MZF1 Promote Cancer Invasion and Metastasis?
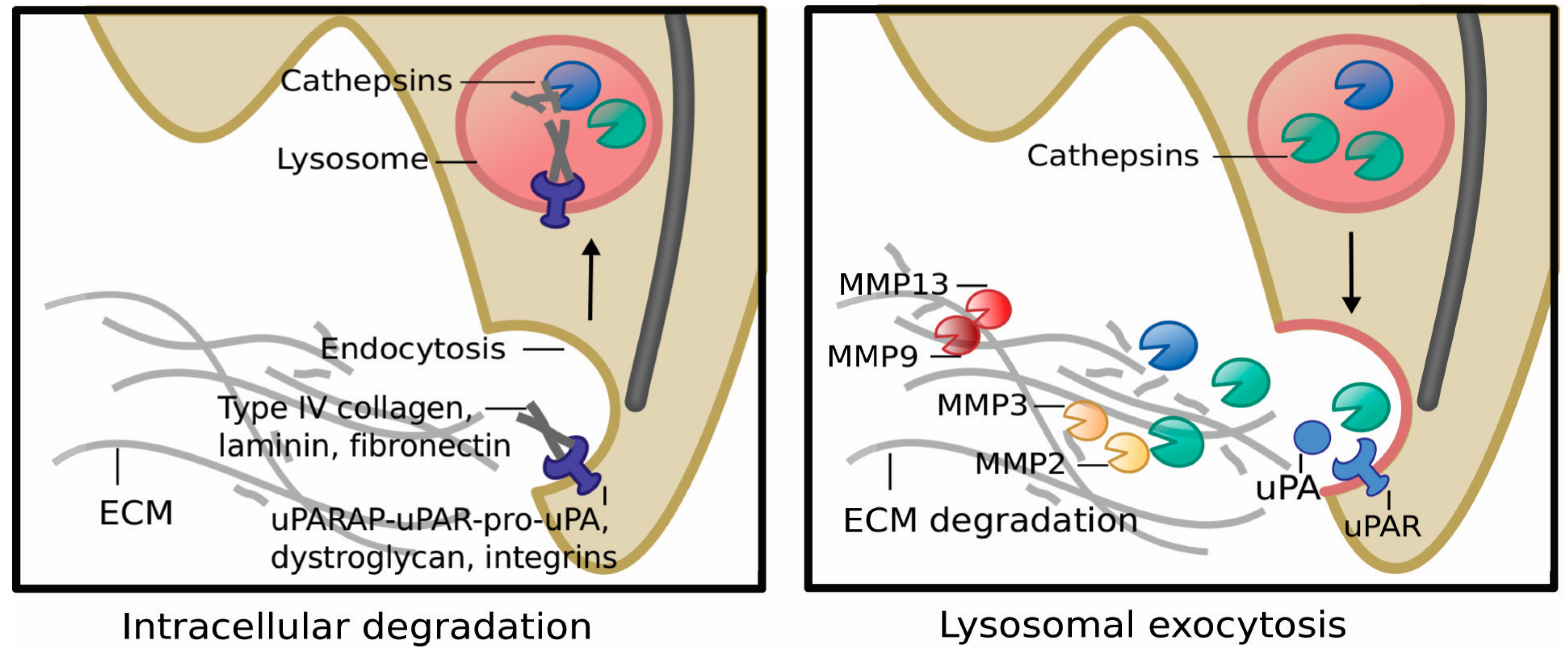
5.1. Lysosomes, MZF1 and Invasion
5.2. MZF1 and EMT
6. Conclusions and Future Directions
Funding
Conflicts of Interest
References
- Gandhi, A.K.; Kang, J.; Havens, C.G.; Conklin, T.; Ning, Y.; Wu, L.; Ito, T.; Ando, H.; Waldman, M.F.; Thakurta, A.; et al. Immunomodulatory agents lenalidomide and pomalidomide co-stimulate T cells by inducing degradation of T cell repressors Ikaros and Aiolos via modulation of the E3 ubiquitin ligase complex CRL4(CRBN.). Br. J. Haematol. 2014, 164, 811–821. [Google Scholar] [CrossRef]
- Licht, J.D.; Shortt, J.; Johnstone, R. From anecdote to targeted therapy: The curious case of thalidomide in multiple myeloma. Cancer Cell 2014, 25, 9–11. [Google Scholar] [CrossRef]
- Peterson, F.C.; Hayes, P.L.; Waltner, J.K.; Heisner, A.K.; Jensen, D.R.; Sander, T.L.; Volkman, B.F. Structure of the SCAN domain from the tumor suppressor protein MZF1. J. Mol. Biol. 2006, 363, 137–147. [Google Scholar] [CrossRef][Green Version]
- Hromas, R.; Collins, S.J.; Hickstein, D.; Raskind, W.; Deaven, L.L.; O’Hara, P.; Hagen, F.S.; Kaushansky, K. A retinoic acid-responsive human zinc finger gene, MZF-1, preferentially expressed in myeloid cells. J. Biol. Chem. 1991, 266, 14183–14187. [Google Scholar]
- Morris, J.F.; Rauscher, F.J., 3rd; Davis, B.; Klemsz, M.; Xu, D.; Tenen, D.; Hromas, R. The myeloid zinc finger gene, MZF-1, regulates the CD34 promoter in vitro. Blood 1995, 86, 3640–3647. [Google Scholar]
- Perrotti, D.; Melotti, P.; Skorski, T.; Casella, I.; Peschle, C.; Calabretta, B. Overexpression of the zinc finger protein MZF1 inhibits hematopoietic development from embryonic stem cells: Correlation with negative regulation of CD34 and c-myb promoter activity. Mol. Cell Biol. 1995, 15, 6075–6087. [Google Scholar] [CrossRef]
- Rafn, B.; Nielsen, C.F.; Andersen, S.H.; Szyniarowski, P.; Corcelle-Termeau, E.; Valo, E.; Fehrenbacher, N.; Olsen, C.J.; Daugaard, M.; Egebjerg, C.; et al. ErbB2-driven breast cancer cell invasion depends on a complex signaling network activating myeloid zinc finger-1-dependent cathepsin B expression. Mol. Cell 2012, 45, 764–776. [Google Scholar] [CrossRef] [PubMed]
- Mudduluru, G.; Vajkoczy, P.; Allgayer, H. Myeloid zinc finger 1 induces migration, invasion, and in vivo metastasis through Axl gene expression in solid cancer. Mol. Cancer Res. 2010, 8, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, Y.H.; Wu, T.T.; Tsai, J.H.; Huang, C.Y.; Hsieh, Y.S.; Liu, J.Y. PKCalpha expression regulated by Elk-1 and MZF-1 in human HCC cells. Biochem. Biophys. Res. Commun. 2006, 339, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, Y.H.; Wu, T.T.; Huang, C.Y.; Hsieh, Y.S.; Liu, J.Y. Suppression of tumorigenicity of human hepatocellular carcinoma cells by antisense oligonucleotide MZF-1. Chin. J. Physiol. 2007, 50, 9–15. [Google Scholar] [PubMed]
- Yue, C.H.; Chiu, Y.W.; Tung, J.N.; Tzang, B.S.; Shiu, J.J.; Huang, W.H.; Liu, J.Y.; Hwang, J.M. Expression of protein kinase C alpha and the MZF-1 and Elk-1 transcription factors in human breast cancer cells. Chin. J. Physiol. 2012, 55, 31–36. [Google Scholar] [CrossRef]
- Yue, C.H.; Huang, C.Y.; Tsai, J.H.; Hsu, C.W.; Hsieh, Y.H.; Lin, H.; Liu, J.Y. MZF-1/Elk-1 Complex Binds to Protein Kinase Calpha Promoter and Is Involved in Hepatocellular Carcinoma. PLoS ONE 2015, 10, e0127420. [Google Scholar] [CrossRef]
- Tsai, L.H.; Wu, J.Y.; Cheng, Y.W.; Chen, C.Y.; Sheu, G.T.; Wu, T.C.; Lee, H. The MZF1/c-MYC axis mediates lung adenocarcinoma progression caused by wild-type lkb1 loss. Oncogene 2015, 34, 1641–1649. [Google Scholar] [CrossRef] [PubMed]
- Weber, C.E.; Kothari, A.N.; Wai, P.Y.; Li, N.Y.; Driver, J.; Zapf, M.A.; Franzen, C.A.; Gupta, G.N.; Osipo, C.; Zlobin, A.; et al. Osteopontin mediates an MZF1-TGF-beta1-dependent transformation of mesenchymal stem cells into cancer-associated fibroblasts in breast cancer. Oncogene 2015, 34, 4821–4833. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Wang, J.; Wang, G.; Jin, Y.; Luo, X.; Xia, X.; Gong, J.; Hu, J. p55PIK transcriptionally activated by MZF1 promotes colorectal cancer cell proliferation. Biomed. Res. Int. 2013, 2013, 868131. [Google Scholar] [CrossRef] [PubMed]
- Murai, K.; Murakami, H.; Nagata, S. A novel form of the myeloid-specific zinc finger protein (MZF-2). Genes Cells 1997, 2, 581–591. [Google Scholar] [CrossRef] [PubMed]
- Peterson, M.J.; Morris, J.F. Human myeloid zinc finger gene MZF produces multiple transcripts and encodes a SCAN box protein. Gene 2000, 254, 105–118. [Google Scholar] [CrossRef]
- Murai, K.; Murakami, H.; Nagata, S. Myeloid-specific transcriptional activation by murine myeloid zinc-finger protein 2. Proc. Natl. Acad. Sci. USA 1998, 95, 3461–3466. [Google Scholar] [CrossRef]
- Sander, T.L.; Stringer, K.F.; Maki, J.L.; Szauter, P.; Stone, J.R.; Collins, T. The SCAN domain defines a large family of zinc finger transcription factors. Gene 2003, 310, 29–38. [Google Scholar] [CrossRef]
- Edelstein, L.C.; Collins, T. The SCAN domain family of zinc finger transcription factors. Gene 2005, 359, 1–17. [Google Scholar] [CrossRef]
- Ogawa, H.; Murayama, A.; Nagata, S.; Fukunaga, R. Regulation of myeloid zinc finger protein 2A transactivation activity through phosphorylation by mitogen-activated protein kinases. J. Biol. Chem. 2003, 278, 2921–2927. [Google Scholar] [CrossRef] [PubMed]
- Brix, D.M.; Tvingsholm, S.A.; Hansen, M.B.; Clemmensen, K.B.; Ohman, T.; Siino, V.; Lambrughi, M.; Hansen, K.; Puustinen, P.; Gromova, I.; et al. Release of transcriptional repression via ErbB2-induced, SUMO-directed phosphorylation of myeloid zinc finger-1 serine 27 activates lysosome redistribution and invasion. Oncogene 2019, 38, 3170–3184. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.J.; Hsu, L.S.; Yue, C.H.; Lin, H.; Chiu, Y.W.; Lin, Y.Y.; Huang, C.Y.; Hung, M.C.; Liu, J.Y. MZF-1/Elk-1 interaction domain as therapeutic target for protein kinase Calpha-based triple-negative breast cancer cells. Oncotarget 2016, 7, 59845–59859. [Google Scholar] [CrossRef] [PubMed]
- Yue, C.H.; Liu, J.Y.; Chi, C.S.; Hu, C.W.; Tan, K.T.; Huang, F.M.; Pan, Y.R.; Lin, K.I.; Lee, C.J. Myeloid Zinc Finger 1 (MZF1) Maintains the Mesenchymal Phenotype by Down-regulating IGF1R/p38 MAPK/ERalpha Signaling Pathway in High-level MZF1-expressing TNBC cells. Anticancer Res. 2019, 39, 4149–4164. [Google Scholar] [CrossRef]
- Pengue, G.; Calabro, V.; Bartoli, P.C.; Pagliuca, A.; Lania, L. Repression of transcriptional activity at a distance by the evolutionarily conserved KRAB domain present in a subfamily of zinc finger proteins. Nucleic Acids Res. 1994, 22, 2908–2914. [Google Scholar] [CrossRef]
- Sander, T.L.; Haas, A.L.; Peterson, M.J.; Morris, J.F. Identification of a novel SCAN box-related protein that interacts with MZF1B. The leucine-rich SCAN box mediates hetero- and homoprotein associations. J. Biol. Chem. 2000, 275, 12857–12867. [Google Scholar] [CrossRef]
- Williams, A.J.; Khachigian, L.M.; Shows, T.; Collins, T. Isolation and characterization of a novel zinc-finger protein with transcription repressor activity. J. Biol. Chem. 1995, 270, 22143–22152. [Google Scholar] [CrossRef]
- Williams, A.J.; Blacklow, S.C.; Collins, T. The zinc finger-associated SCAN box is a conserved oligomerization domain. Mol. Cell Biol. 1999, 19, 8526–8535. [Google Scholar] [CrossRef]
- Morris, J.F.; Hromas, R.; Rauscher, F.J., 3rd. Characterization of the DNA-binding properties of the myeloid zinc finger protein MZF1: Two independent DNA-binding domains recognize two DNA consensus sequences with a common G-rich core. Mol. Cell Biol. 1994, 14, 1786–1795. [Google Scholar] [CrossRef]
- Robertson, K.A.; Hill, D.P.; Kelley, M.R.; Tritt, R.; Crum, B.; Van Epps, S.; Srour, E.; Rice, S.; Hromas, R. The myeloid zinc finger gene (MZF-1) delays retinoic acid-induced apoptosis and differentiation in myeloid leukemia cells. Leukemia 1998, 12, 690–698. [Google Scholar] [CrossRef]
- Bavisotto, L.; Kaushansky, K.; Lin, N.; Hromas, R. Antisense oligonucleotides from the stage-specific myeloid zinc finger gene MZF-1 inhibit granulopoiesis in vitro. J. Exp. Med. 1991, 174, 1097–1101. [Google Scholar] [CrossRef] [PubMed]
- Gaboli, M.; Kotsi, P.A.; Gurrieri, C.; Cattoretti, G.; Ronchetti, S.; Cordon-Cardo, C.; Broxmeyer, H.E.; Hromas, R.; Pandolfi, P.P. Mzf1 controls cell proliferation and tumorigenesis. Genes Dev. 2001, 15, 1625–1630. [Google Scholar] [CrossRef] [PubMed]
- Hromas, R.; Morris, J.; Cornetta, K.; Berebitsky, D.; Davidson, A.; Sha, M.; Sledge, G.; Rauscher, F., 3rd. Aberrant expression of the myeloid zinc finger gene, MZF-1, is oncogenic. Cancer Res. 1995, 55, 3610–3614. [Google Scholar]
- Brix, D.M.; Clemmensen, K.K.; Kallunki, T. When Good Turns Bad: Regulation of Invasion and Metastasis by ErbB2 Receptor Tyrosine Kinase. Cells 2014, 3, 53–78. [Google Scholar] [CrossRef]
- Mason, S.D.; Joyce, J.A. Proteolytic networks in cancer. Trends Cell Biol. 2011, 21, 228–237. [Google Scholar] [CrossRef]
- Kallunki, T.; Olsen, O.D.; Jaattela, M. Cancer-associated lysosomal changes: Friends or foes? Oncogene 2013, 32, 1995–2004. [Google Scholar] [CrossRef]
- Hamalisto, S.; Jaattela, M. Lysosomes in cancer-living on the edge (of the cell). Curr. Opin. Cell Biol. 2016, 39, 69–76. [Google Scholar] [CrossRef]
- Imamura, T.; Hikita, A.; Inoue, Y. The roles of TGF-beta signaling in carcinogenesis and breast cancer metastasis. Breast Cancer 2012, 19, 118–124. [Google Scholar] [CrossRef]
- Gupta, P.; Srivastava, S.K. HER2 mediated de novo production of TGFbeta leads to SNAIL driven epithelial-to-mesenchymal transition and metastasis of breast cancer. Mol. Oncol. 2014, 8, 1532–1547. [Google Scholar] [CrossRef]
- Pradeep, C.R.; Zeisel, A.; Kostler, W.J.; Lauriola, M.; Jacob-Hirsch, J.; Haibe-Kains, B.; Amariglio, N.; Ben-Chetrit, N.; Emde, A.; Solomonov, I.; et al. Modeling invasive breast cancer: Growth factors propel progression of HER2-positive premalignant lesions. Oncogene 2012, 31, 3569–3583. [Google Scholar] [CrossRef]
- Seton-Rogers, S.E.; Lu, Y.; Hines, L.M.; Koundinya, M.; LaBaer, J.; Muthuswamy, S.K.; Brugge, J.S. Cooperation of the ErbB2 receptor and transforming growth factor beta in induction of migration and invasion in mammary epithelial cells. Proc. Natl. Acad. Sci. USA 2004, 101, 1257–1262. [Google Scholar] [CrossRef] [PubMed]
- Ueda, Y.; Wang, S.; Dumont, N.; Yi, J.Y.; Koh, Y.; Arteaga, C.L. Overexpression of HER2 (erbB2) in human breast epithelial cells unmasks transforming growth factor beta-induced cell motility. J. Biol. Chem. 2004, 279, 24505–24513. [Google Scholar] [CrossRef] [PubMed]
- Le Mee, S.; Fromigue, O.; Marie, P.J. Sp1/Sp3 and the myeloid zinc finger gene MZF1 regulate the human N-cadherin promoter in osteoblasts. Exp. Cell Res. 2005, 302, 129–142. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Greger, J.; Shi, H.; Liu, Y.; Greshock, J.; Annan, R.; Halsey, W.; Sathe, G.M.; Martin, A.M.; Gilmer, T.M. Novel mechanism of lapatinib resistance in HER2-positive breast tumor cells: Activation of AXL. Cancer Res. 2009, 69, 6871–6878. [Google Scholar] [CrossRef]
- Asiedu, M.K.; Beauchamp-Perez, F.D.; Ingle, J.N.; Behrens, M.D.; Radisky, D.C.; Knutson, K.L. AXL induces epithelial-to-mesenchymal transition and regulates the function of breast cancer stem cells. Oncogene 2014, 33, 1316–1324. [Google Scholar] [CrossRef]
- Paccez, J.D.; Vogelsang, M.; Parker, M.I.; Zerbini, L.F. The receptor tyrosine kinase Axl in cancer: Biological functions and therapeutic implications. Int. J. Cancer 2014, 134, 1024–1033. [Google Scholar] [CrossRef]
- Arteaga, C.L.; Engelman, J.A. ERBB receptors: From oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell 2014, 25, 282–303. [Google Scholar] [CrossRef]
- Chen, P.M.; Cheng, Y.W.; Wang, Y.C.; Wu, T.C.; Chen, C.Y.; Lee, H. Up-regulation of FOXM1 by E6 oncoprotein through the MZF1/NKX2-1 axis is required for human papillomavirus-associated tumorigenesis. Neoplasia 2014, 16, 961–971. [Google Scholar] [CrossRef]
- Tsai, S.J.; Hwang, J.M.; Hsieh, S.C.; Ying, T.H.; Hsieh, Y.H. Overexpression of myeloid zinc finger 1 suppresses matrix metalloproteinase-2 expression and reduces invasiveness of SiHa human cervical cancer cells. Biochem. Biophys. Res. Commun. 2012, 425, 462–467. [Google Scholar] [CrossRef]
- Tsai, L.H.; Chen, P.M.; Cheng, Y.W.; Chen, C.Y.; Sheu, G.T.; Wu, T.C.; Lee, H. LKB1 loss by alteration of the NKX2-1/p53 pathway promotes tumor malignancy and predicts poor survival and relapse in lung adenocarcinomas. Oncogene 2014, 33, 3851–3860. [Google Scholar] [CrossRef][Green Version]
- Eguchi, T.; Prince, T.L.; Tran, M.T.; Sogawa, C.; Lang, B.J.; Calderwood, S.K. MZF1 and SCAND1 Reciprocally Regulate CDC37 Gene Expression in Prostate Cancer. Cancers 2019, 11, 792. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, Z.; Yang, K.; Du, J.; Xu, Y.; Liu, S. Myeloid zinc-finger 1 (MZF-1) suppresses prostate tumor growth through enforcing ferroportin-conducted iron egress. Oncogene 2015, 34, 3839–3847. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lei, Q.; Yu, Z.; Xu, G.; Tang, H.; Wang, W.; Wang, Z.; Li, G.; Wu, M. MiR-101 reverses the hypomethylation of the LMO3 promoter in glioma cells. Oncotarget 2015, 6, 7930–7943. [Google Scholar] [CrossRef]
- Sonabend, A.M.; Lesniak, M.S. Oligodendrogliomas: Clinical significance of 1p and 19q chromosomal deletions. Expert. Rev. Neurother. 2005, 5, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Min, L.; Xu, C.; Shao, L.; Guo, S.; Cheng, R.; Xing, J.; Zhu, S.; Zhang, S. Construction of disease-specific transcriptional regulatory networks identifies co-activation of four gene in esophageal squamous cell carcinoma. Oncol. Rep. 2017, 38, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Jiao, W.; Mei, H.; Song, H.; Li, D.; Xiang, X.; Chen, Y.; Yang, F.; Li, H.; Huang, K.; et al. miRNA-337-3p inhibits gastric cancer progression through repressing myeloid zinc finger 1-facilitated expression of matrix metalloproteinase 14. Oncotarget 2016, 7, 40314–40328. [Google Scholar] [CrossRef] [PubMed]
- Li, G.Q.; He, Q.; Yang, L.; Wang, S.B.; Yu, D.D.; He, Y.Q.; Hu, J.; Pan, Y.M.; Wu, Y. Clinical Significance of Myeloid Zinc Finger 1 Expression in the Progression of Gastric Tumourigenesis. Cell Physiol. Biochem. 2017, 44, 1242–1250. [Google Scholar] [CrossRef]
- Eguchi, T.; Prince, T.; Wegiel, B.; Calderwood, S.K. Role and Regulation of Myeloid Zinc Finger Protein 1 in Cancer. J. Cell Biochem. 2015, 116, 2146–2154. [Google Scholar] [CrossRef]
- Moeenrezakhanlou, A.; Shephard, L.; Lam, L.; Reiner, N.E. Myeloid cell differentiation in response to calcitriol for expression CD11b and CD14 is regulated by myeloid zinc finger-1 protein downstream of phosphatidylinositol 3-kinase. J. Leukoc. Biol. 2008, 84, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Ko, H.; Kim, S.; Yang, K.; Kim, K. Phosphorylation-dependent stabilization of MZF1 upregulates N-cadherin expression during protein kinase CK2-mediated epithelial-mesenchymal transition. Oncogenesis 2018, 7, 27. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Han, L.; Zhou, C.; Wei, W.; Chen, X.; Yi, H.; Wu, X.; Bai, X.; Guo, S.; Yu, Y.; et al. TGF-beta1-induced CK17 enhances cancer stem cell-like properties rather than EMT in promoting cervical cancer metastasis via the ERK1/2-MZF1 signaling pathway. FEBS J. 2017, 284, 3000–3017. [Google Scholar] [CrossRef] [PubMed]
- Piszczatowski, R.T.; Rafferty, B.J.; Rozado, A.; Parziale, J.V.; Lents, N.H. Myeloid Zinc Finger 1 (MZF-1) Regulates Expression of the CCN2/CTGF and CCN3/NOV Genes in the Hematopoietic Compartment. J. Cell Physiol. 2015, 230, 2634–2639. [Google Scholar] [CrossRef] [PubMed]
- Piszczatowski, R.T.; Rafferty, B.J.; Rozado, A.; Tobak, S.; Lents, N.H. The glyceraldehyde 3-phosphate dehydrogenase gene (GAPDH) is regulated by myeloid zinc finger 1 (MZF-1) and is induced by calcitriol. Biochem. Biophys. Res. Commun. 2014, 451, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Sheikh, T.; Sen, E. SIRT6 regulated nucleosomal occupancy affects Hexokinase 2 expression. Exp. Cell Res. 2017, 357, 98–106. [Google Scholar] [CrossRef]
- Vishwamitra, D.; Curry, C.V.; Alkan, S.; Song, Y.H.; Gallick, G.E.; Kaseb, A.O.; Shi, P.; Amin, H.M. The transcription factors Ik-1 and MZF1 downregulate IGF-IR expression in NPM-ALK(+) T-cell lymphoma. Mol. Cancer 2015, 14, 53. [Google Scholar] [CrossRef]
- Zhang, S.; Shi, W.; Ramsay, E.S.; Bliskovsky, V.; Eiden, A.M.; Connors, D.; Steinsaltz, M.; DuBois, W.; Mock, B.A. The transcription factor MZF1 differentially regulates murine Mtor promoter variants linked to tumor susceptibility. J. Biol. Chem. 2019, 294, 16756–16764. [Google Scholar] [CrossRef]
- Lin, S.; Wang, X.; Pan, Y.; Tian, R.; Lin, B.; Jiang, G.; Chen, K.; He, Y.; Zhang, L.; Zhai, W.; et al. Transcription Factor Myeloid Zinc-Finger 1 Suppresses Human Gastric Carcinogenesis by Interacting with Metallothionein 2A. Clin. Cancer Res. 2019, 25, 1050–1062. [Google Scholar] [CrossRef]
- Doppler, S.A.; Werner, A.; Barz, M.; Lahm, H.; Deutsch, M.A.; Dressen, M.; Schiemann, M.; Voss, B.; Gregoire, S.; Kuppusamy, R.; et al. Myeloid zinc finger 1 (Mzf1) differentially modulates murine cardiogenesis by interacting with an Nkx2.5 cardiac enhancer. PLoS ONE 2014, 9, e113775. [Google Scholar] [CrossRef]
- Chen, H.; Zuo, Q.; Wang, Y.; Song, J.; Yang, H.; Zhang, Y.; Li, B. Inducing goat pluripotent stem cells with four transcription factor mRNAs that activate endogenous promoters. BMC Biotechnol. 2017, 17, 11. [Google Scholar] [CrossRef]
- Jia, N.; Wang, J.; Li, Q.; Tao, X.; Chang, K.; Hua, K.; Yu, Y.; Wong, K.K.; Feng, W. DNA methylation promotes paired box 2 expression via myeloid zinc finger 1 in endometrial cancer. Oncotarget 2016, 7, 84785–84797. [Google Scholar] [CrossRef]
- Lee, Y.K.; Park, U.H.; Kim, E.J.; Hwang, J.T.; Jeong, J.C.; Um, S.J. Tumor antigen PRAME is up-regulated by MZF1 in cooperation with DNA hypomethylation in melanoma cells. Cancer Lett. 2017, 403, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, S.S.; Lee, H.S.; Hong, S.; Rajasekaran, N.; Wang, L.H.; Choi, J.S.; Shin, Y.K. Upregulation of SMAD4 by MZF1 inhibits migration of human gastric cancer cells. Int. J. Oncol. 2017, 50, 272–282. [Google Scholar] [CrossRef] [PubMed]
- Driver, J.; Weber, C.E.; Callaci, J.J.; Kothari, A.N.; Zapf, M.A.; Roper, P.M.; Borys, D.; Franzen, C.A.; Gupta, G.N.; Wai, P.Y.; et al. Alcohol inhibits osteopontin-dependent transforming growth factor-beta1 expression in human mesenchymal stem cells. J. Biol. Chem. 2015, 290, 9959–9973. [Google Scholar] [CrossRef] [PubMed]
- Horinaka, M.; Yoshida, T.; Tomosugi, M.; Yasuda, S.; Sowa, Y.; Sakai, T. Myeloid zinc finger 1 mediates sulindac sulfide-induced upregulation of death receptor 5 of human colon cancer cells. Sci. Rep. 2014, 4, 6000. [Google Scholar] [CrossRef]
- Verma, N.K.; Gadi, A.; Maurizi, G.; Roy, U.B.; Mansukhani, A.; Basilico, C. Myeloid Zinc Finger 1 and GA Binding Protein Co-Operate with Sox2 in Regulating the Expression of Yes-Associated Protein 1 in Cancer Cells. Stem Cells 2017, 35, 2340–2350. [Google Scholar] [CrossRef] [PubMed]
- Hromas, R.; Davis, B.; Rauscher, F.J., 3rd; Klemsz, M.; Tenen, D.; Hoffman, S.; Xu, D.; Morris, J.F. Hematopoietic transcriptional regulation by the myeloid zinc finger gene, MZF-1. Curr. Top. Microbiol. Immunol. 1996, 211, 159–164. [Google Scholar] [CrossRef]
- Tvingsholm, S.A.; Hansen, M.B.; Clemmensen, K.K.B.; Brix, D.M.; Rafn, B.; Frankel, L.B.; Louhimo, R.; Moreira, J.; Hautaniemi, S.; Gromova, I.; et al. Let-7 microRNA controls invasion-promoting lysosomal changes via the oncogenic transcription factor myeloid zinc finger-1. Oncogenesis 2018, 7, 14. [Google Scholar] [CrossRef]
- Nana-Sinkam, S.P.; Croce, C.M. MicroRNAs as therapeutic targets in cancer. Transl. Res. 2011, 157, 216–225. [Google Scholar] [CrossRef]
- Yu, F.; Yao, H.; Zhu, P.; Zhang, X.; Pan, Q.; Gong, C.; Huang, Y.; Hu, X.; Su, F.; Lieberman, J.; et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007, 131, 1109–1123. [Google Scholar] [CrossRef]
- Noll, L.; Peterson, F.C.; Hayes, P.L.; Volkman, B.F.; Sander, T. Heterodimer formation of the myeloid zinc finger 1 SCAN domain and association with promyelocytic leukemia nuclear bodies. Leuk. Res. 2008, 32, 1582–1592. [Google Scholar] [CrossRef]
- Nygaard, M.; Terkelsen, T.; Vidas Olsen, A.; Sora, V.; Salamanca Viloria, J.; Rizza, F.; Bergstrand-Poulsen, S.; Di Marco, M.; Vistesen, M.; Tiberti, M.; et al. The Mutational Landscape of the Oncogenic MZF1 SCAN Domain in Cancer. Front. Mol. Biosci. 2016, 3, 78. [Google Scholar] [CrossRef] [PubMed]
- Raman, N.; Nayak, A.; Muller, S. The SUMO system: A master organizer of nuclear protein assemblies. Chromosoma 2013, 122, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Hay, R.T. SUMO: A history of modification. Mol. Cell 2005, 18, 1–12. [Google Scholar] [CrossRef]
- Zhong, S.; Salomoni, P.; Pandolfi, P.P. The transcriptional role of PML and the nuclear body. Nat. Cell Biol. 2000, 2, E85–E90. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, R.; Pandolfi, P.P. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 2007, 8, 1006–1016. [Google Scholar] [CrossRef] [PubMed]
- Lallemand-Breitenbach, V.; de The, H. PML nuclear bodies: From architecture to function. Curr. Opin. Cell Biol. 2018, 52, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Mertins, P.; Mani, D.R.; Ruggles, K.V.; Gillette, M.A.; Clauser, K.R.; Wang, P.; Wang, X.; Qiao, J.W.; Cao, S.; Petralia, F.; et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 2016, 534, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Melton, C.; Reuter, J.A.; Spacek, D.V.; Snyder, M. Recurrent somatic mutations in regulatory regions of human cancer genomes. Nat. Genet. 2015, 47, 710–716. [Google Scholar] [CrossRef]
- Tian, E.; Borset, M.; Sawyer, J.R.; Brede, G.; Vatsveen, T.K.; Hov, H.; Waage, A.; Barlogie, B.; Shaughnessy, J.D., Jr.; Epstein, J.; et al. Allelic mutations in noncoding genomic sequences construct novel transcription factor binding sites that promote gene overexpression. Genes Chromosomes Cancer 2015, 54, 692–701. [Google Scholar] [CrossRef]
- Rafn, B.; Kallunki, T. A way to invade: A story of ErbB2 and lysosomes. Cell Cycle 2012, 11, 2415–2416. [Google Scholar] [CrossRef][Green Version]
- Brix, D.M.; Rafn, B.; Bundgaard Clemmensen, K.; Andersen, S.H.; Ambartsumian, N.; Jaattela, M.; Kallunki, T. Screening and identification of small molecule inhibitors of ErbB2-induced invasion. Mol. Oncol. 2014, 8, 1703–1718. [Google Scholar] [CrossRef] [PubMed]
- Olson, O.C.; Joyce, J.A. Cysteine cathepsin proteases: Regulators of cancer progression and therapeutic response. Nat. Rev. Cancer 2015, 15, 712–729. [Google Scholar] [CrossRef] [PubMed]
- Bawa-Khalfe, T.; Yeh, E.T. SUMO Losing Balance: SUMO Proteases Disrupt SUMO Homeostasis to Facilitate Cancer Development and Progression. Genes Cancer 2010, 1, 748–752. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.I.; Baek, S.H. SUMOylation code in cancer development and metastasis. Mol. Cells 2006, 22, 247–253. [Google Scholar]
- Yang, Y.; Xia, Z.; Wang, X.; Zhao, X.; Sheng, Z.; Ye, Y.; He, G.; Zhou, L.; Zhu, H.; Xu, N.; et al. Small-Molecule Inhibitors Targeting Protein SUMOylation as Novel Anticancer Compounds. Mol. Pharmacol. 2018, 94, 885–894. [Google Scholar] [CrossRef]
- P21-Activated Kinase 4 (PAK4) Inhibitors as Potential Cancer Therapy. Available online: https://pubs.acs.org/doi/pdf/10.1021/ml500445c (accessed on 13 January 2020).
- This Is the First Study Using Escalating Doses of PF-03758309, an Oral Compound, in Patients with Advanced Solid Tumors. Available online: https://clinicaltrials.gov/ct2/show/NCT00932126 (accessed on 13 January 2020).
- PAK4 and NAMPT in Patients with Solid Malignancies or NHL (PANAMA) (PANAMA). Available online: https://clinicaltrials.gov/ct2/show/NCT02702492 (accessed on 13 January 2020).
| Gene | Method | Reference | Function | EMT |
|---|---|---|---|---|
| AXL | ChIP, Luciferase | [8] | Activator | yes |
| CD14 | EMSA,luciferase | [59] | Activator | yes |
| CD34 | EMSA, Acetyltransferase activity | [6] | Activator | |
| CDC37 | ChIP, Luciferase | [51] | Activator | |
| CDH2 (N-Cadherin) | EMSA | [43] | Activator | yes |
| CDH2 (N-Cadherin) | ChIP, Luciferase | [60] | Activator | |
| CK17 | Luciferase, qPCR | [61] | Activator | |
| CTGF | ChIP | [62] | Activator | yes |
| CTSB | ChIP, Luciferase | [7] | Activator | |
| GAPDH | ChIP | [63] | Activator | yes |
| HK2 | ChIP | [64] | Suppressor | |
| IGFIR | ChIP, Luciferase | [65] | Suppressor | yes |
| ITGAM (CD11b) | EMSA, luciferase | [59] | Activator | |
| LMO3 | ChIP | [53] | Activator | |
| MMP2 | ChIP, Luciferase | [49] | Suppressor | yes |
| Mtor | ChIP, EMSA, Luciferase | [66] | Suppressor | yes |
| MYB (c-myb) | EMSA, Acetyltransferase activity | [6] | Activator | |
| MYC | ChIP, Luciferase | [13] | Activator | |
| NFKB1A | ChIP | [67] | Activator | yes |
| NKX2-1 | ChIP, Luciferase | [48] | Activator | yes |
| NKX2-5 | ChIP, Luciferase | [68] | Activator | yes |
| NOV | ChIP | [61] | Activator | |
| OOCT4 | Luciferase | [69] | Activator | yes |
| PAX2 | Luciferase | [70] | Suppressor | yes |
| PIK3R3 (p55PIK) | ChIP, Luciferase | [15] | Activator | yes |
| PRAME | ChIP | [71] | Activator | yes |
| PRKCA (PKC alpha) | ChIP, Luciferase | [12] | Activator | |
| SLC40A1 (FPN) | ChIP, Luciferase | [52] | Activator | |
| SMAD4 | ChIP, EMSA, Luciferase | [72] | Activator | |
| TGFB1 | ChIP, Luciferase | [73] | Activator | yes |
| TNFRSF10B (DR5) | Luciferase | [74] | Activator | |
| YAP1 | ChIP, EMSA, Luciferase | [75] | Activator | yes |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brix, D.M.; Bundgaard Clemmensen, K.K.; Kallunki, T. Zinc Finger Transcription Factor MZF1—A Specific Regulator of Cancer Invasion. Cells 2020, 9, 223. https://doi.org/10.3390/cells9010223
Brix DM, Bundgaard Clemmensen KK, Kallunki T. Zinc Finger Transcription Factor MZF1—A Specific Regulator of Cancer Invasion. Cells. 2020; 9(1):223. https://doi.org/10.3390/cells9010223
Chicago/Turabian StyleBrix, Ditte Marie, Knut Kristoffer Bundgaard Clemmensen, and Tuula Kallunki. 2020. "Zinc Finger Transcription Factor MZF1—A Specific Regulator of Cancer Invasion" Cells 9, no. 1: 223. https://doi.org/10.3390/cells9010223
APA StyleBrix, D. M., Bundgaard Clemmensen, K. K., & Kallunki, T. (2020). Zinc Finger Transcription Factor MZF1—A Specific Regulator of Cancer Invasion. Cells, 9(1), 223. https://doi.org/10.3390/cells9010223





