Discovery and Preliminary Characterization of Translational Modulators that Impair the Binding of eIF6 to 60S Ribosomal Subunits
Abstract
1. Introduction
2. Materials and Methods
2.1. Screening Library and eIF6 Purification
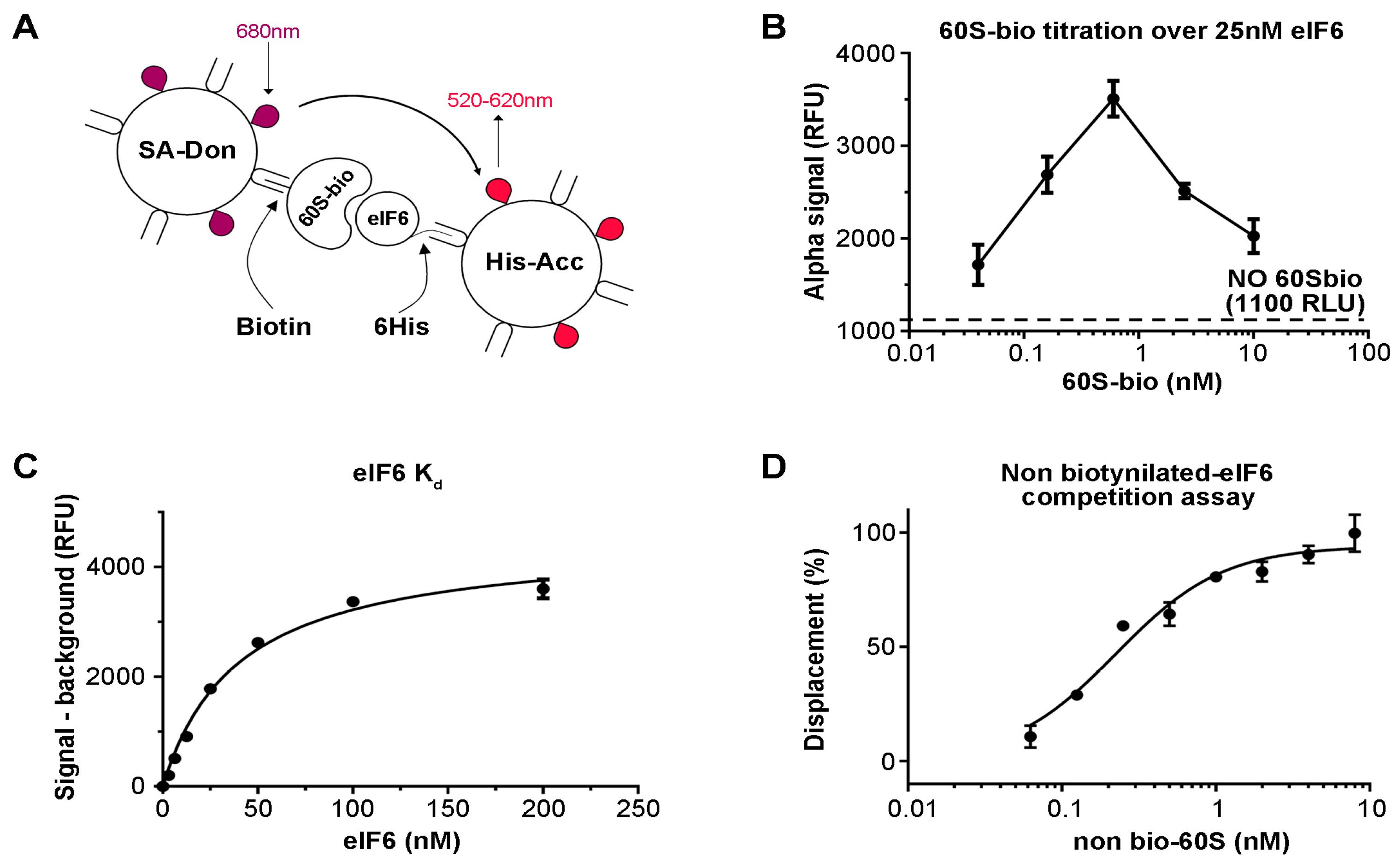
2.2. 60S–eIF6 Homogeneous Binding Assay
2.3. 60S–eIF6 Counter-Screen Assay
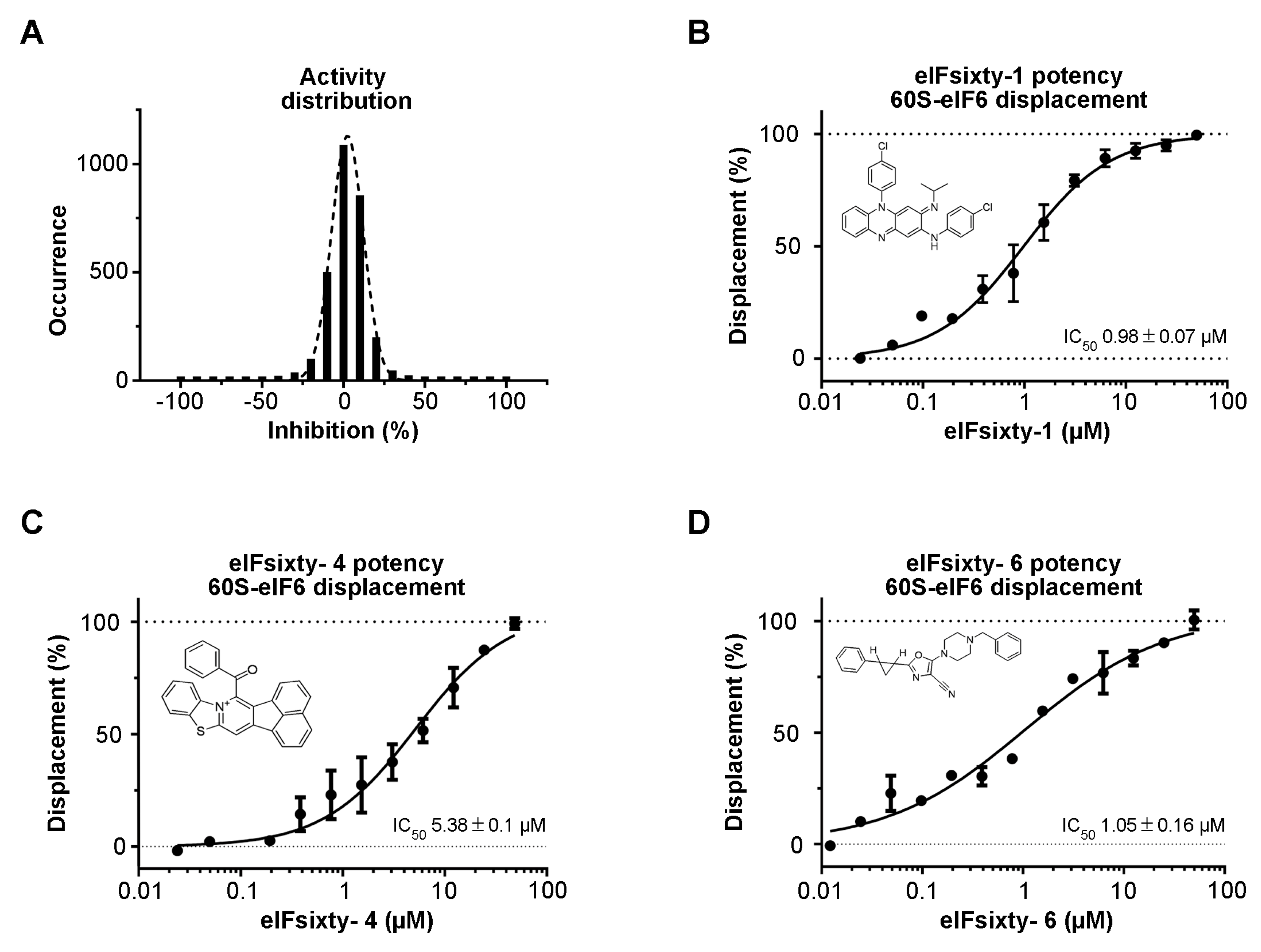
2.4. Chemistry
2.5. iRIA
2.6. Cells and Vitality Curves
2.7. Puromycin Labelling
2.8. Polysomal Profiles
2.9. Analysis of Translated mRNAs
2.10. Immunofluorescence
3. Results
3.1. eIF6-60S Homogenous Binding Assay Optimization
3.2. Hit Identification
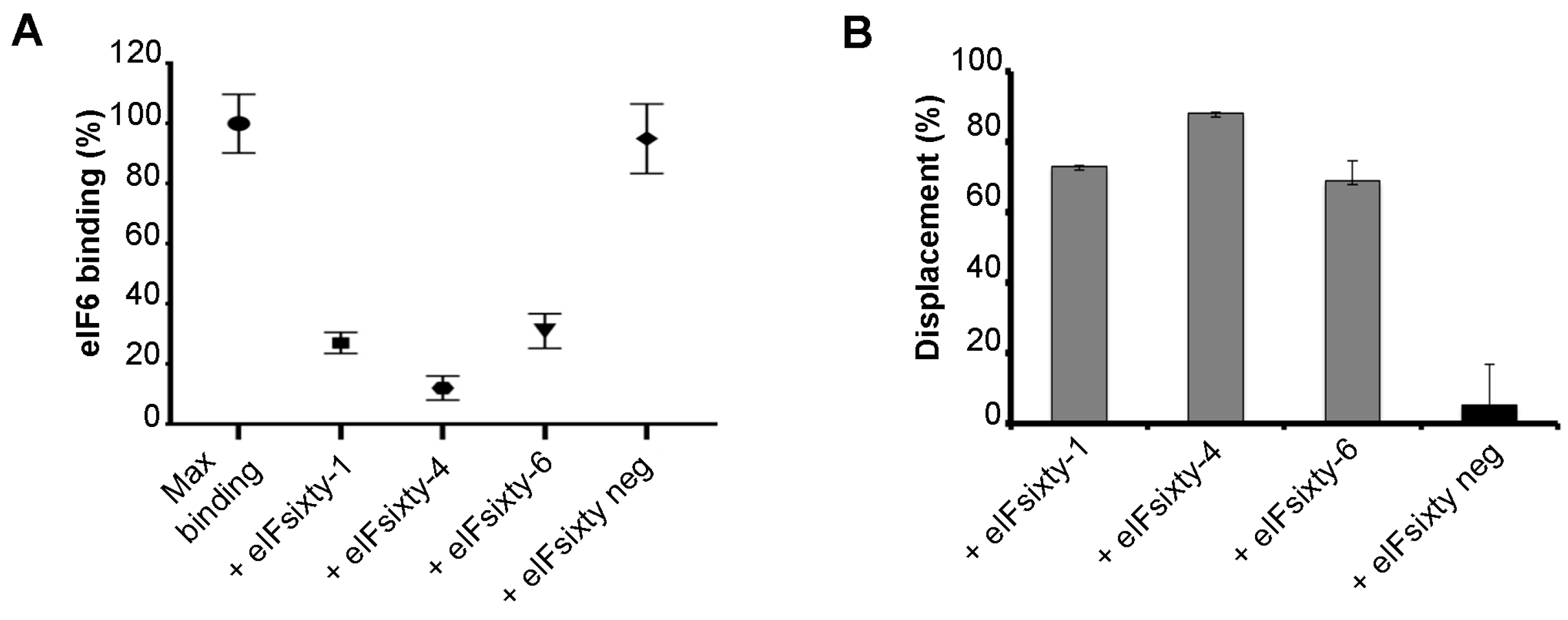
3.3. Independent ELISA Validation
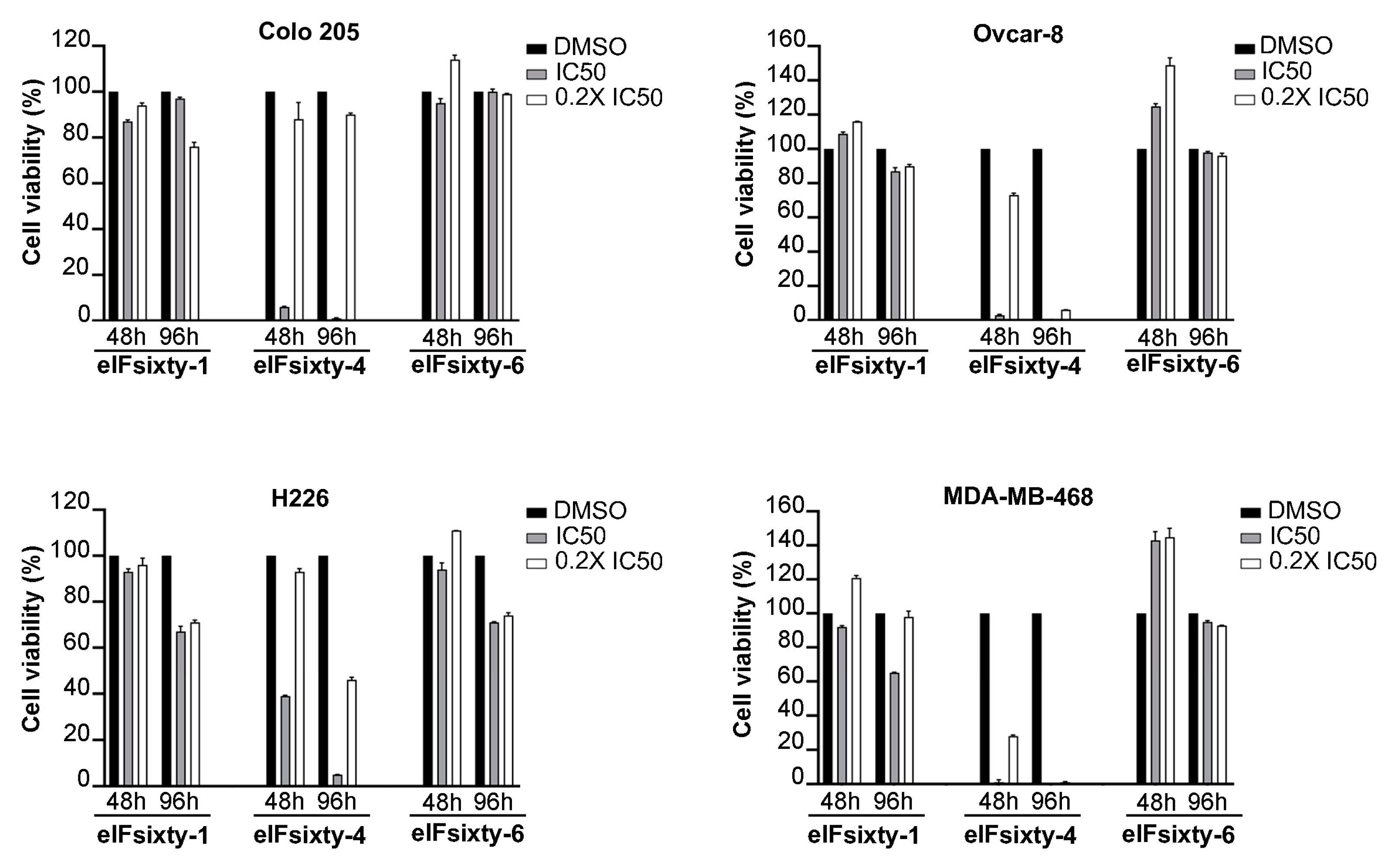
3.4. Viability Assays
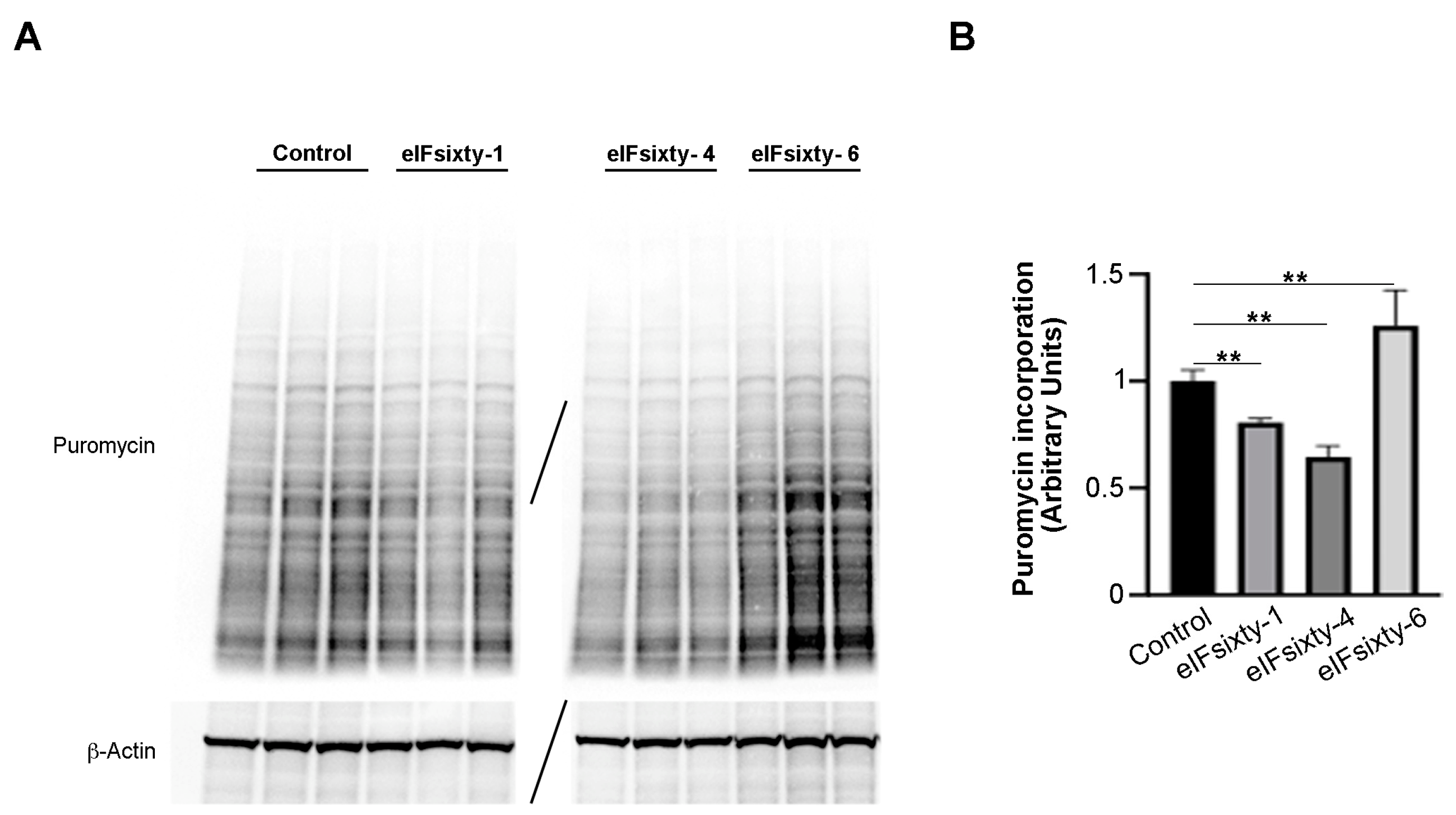
3.5. Puromycin Assays Show that eIFsixty-4 is a Strong Inhibitor of Global Translation, Whereas eIFsixty1 and eIFsixty-6 May Act As Mild Modulators
3.6. Polysomal Assays Show that eIFsixty-4 Inhibits Initiation of Translation
3.7. All Compounds Modulate the Polysomal Association of Known eIF6 Target mRNAs
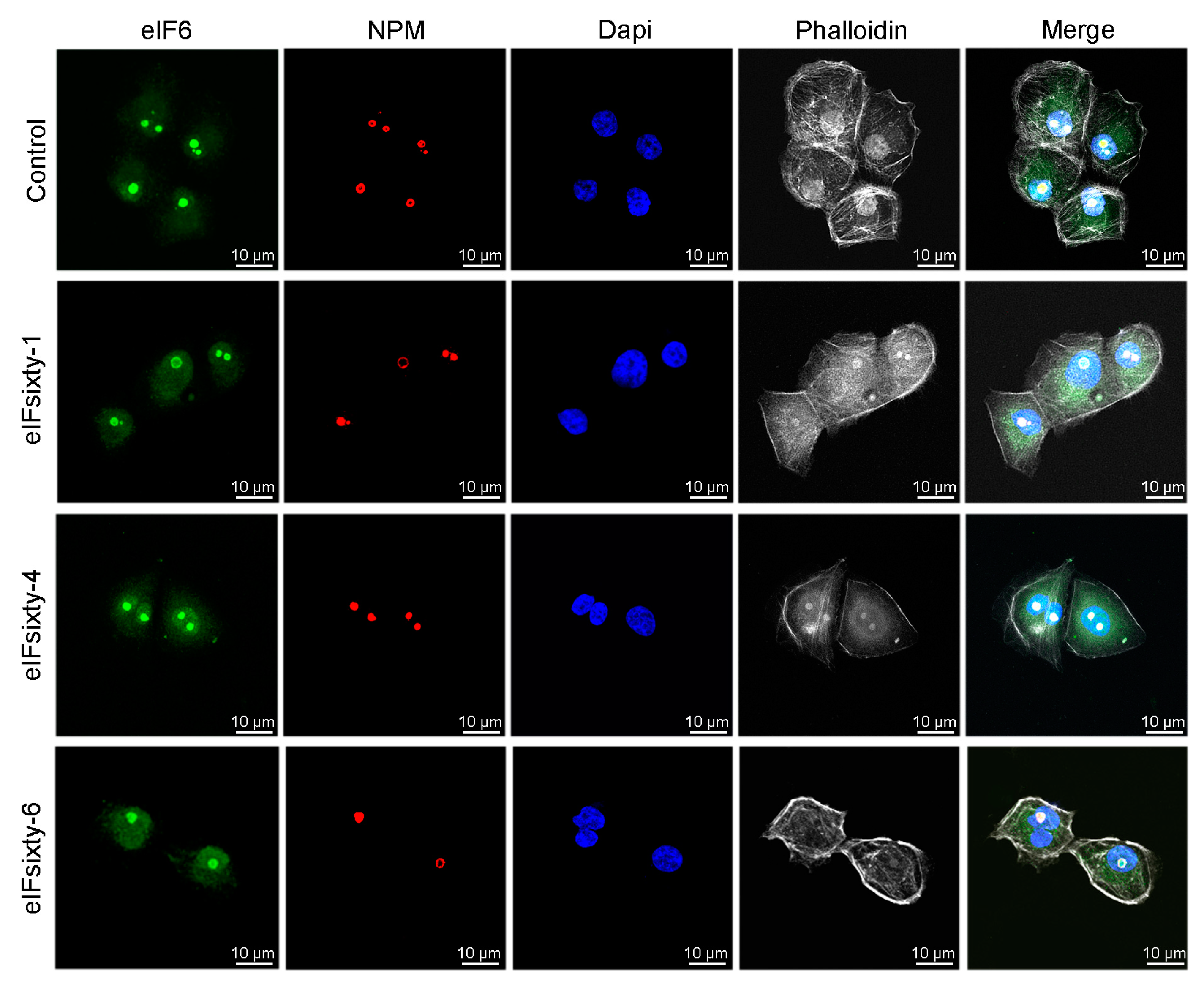
3.8. Nucleolar Structure is not Affected by eIFsixty-i Compounds
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hershey, J.W.B.; Sonenberg, N.; Mathews, M.B. Principles of Translational Control. Cold Spring Harb. Perspect. Biol. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Truitt, M.L.; Ruggero, D. New frontiers in translational control of the cancer genome. Nat. Rev. Cancer 2016, 16, 288–304. [Google Scholar] [CrossRef] [PubMed]
- Hinnebusch, A.G.; Ivanov, I.P.; Sonenberg, N. Translational control by 5’-untranslated regions of eukaryotic mRNAs. Science 2016, 352, 1413–1416. [Google Scholar] [CrossRef] [PubMed]
- Loreni, F.; Mancino, M.; Biffo, S. Translation factors and ribosomal proteins control tumor onset and progression: How? Oncogene 2014, 33, 2145–2156. [Google Scholar] [CrossRef]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 2009, 136, 731–745. [Google Scholar] [CrossRef]
- Wang, X.; Flynn, A.; Waskiewicz, A.J.; Webb, B.L.; Vries, R.G.; Baines, I.A.; Cooper, J.A.; Proud, C.G. The phosphorylation of eukaryotic initiation factor eIF4E in response to phorbol esters, cell stresses, and cytokines is mediated by distinct MAP kinase pathways. J. Biol. Chem. 1998, 273, 9373–9377. [Google Scholar] [CrossRef]
- Wendel, H.G.; Silva, R.L.; Malina, A.; Mills, J.R.; Zhu, H.; Ueda, T.; Watanabe-Fukunaga, R.; Fukunaga, R.; Teruya-Feldstein, J.; Pelletier, J.; et al. Dissecting eIF4E action in tumorigenesis. Genes Dev. 2007, 21, 3232–3237. [Google Scholar] [CrossRef]
- Bhat, M.; Robichaud, N.; Hulea, L.; Sonenberg, N.; Pelletier, J.; Topisirovic, I. Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 2015, 14, 261–278. [Google Scholar] [CrossRef]
- Hsieh, A.C.; Costa, M.; Zollo, O.; Davis, C.; Feldman, M.E.; Testa, J.R.; Meyuhas, O.; Shokat, K.M.; Ruggero, D. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell 2010, 17, 249–261. [Google Scholar] [CrossRef]
- Dowling, R.J.; Topisirovic, I.; Alain, T.; Bidinosti, M.; Fonseca, B.D.; Petroulakis, E.; Wang, X.; Larsson, O.; Selvaraj, A.; Liu, Y.; et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science 2010, 328, 1172–1176. [Google Scholar] [CrossRef]
- Di Nicolantonio, F.; Arena, S.; Tabernero, J.; Grosso, S.; Molinari, F.; Macarulla, T.; Russo, M.; Cancelliere, C.; Zecchin, D.; Mazzucchelli, L.; et al. Deregulation of the PI3K and KRAS signaling pathways in human cancer cells determines their response to everolimus. J. Clin. Investig. 2010, 120, 2858–2866. [Google Scholar] [CrossRef] [PubMed]
- Lock, R.; Ingraham, R.; Maertens, O.; Miller, A.L.; Weledji, N.; Legius, E.; Konicek, B.M.; Yan, S.C.; Graff, J.R.; Cichowski, K. Cotargeting MNK and MEK kinases induces the regression of NF1-mutant cancers. J. Clin. Investig. 2016, 126, 2181–2190. [Google Scholar] [CrossRef] [PubMed]
- Kosciuczuk, E.M.; Saleiro, D.; Kroczynska, B.; Beauchamp, E.M.; Eckerdt, F.; Blyth, G.T.; Abedin, S.M.; Giles, F.J.; Altman, J.K.; Platanias, L.C. Merestinib blocks Mnk kinase activity in acute myeloid leukemia progenitors and exhibits antileukemic effects in vitro and in vivo. Blood 2016, 128, 410–414. [Google Scholar] [CrossRef] [PubMed]
- Russell, D.W.; Spremulli, L.L. Purification and characterization of a ribosome dissociation factor (eukaryotic initiation factor 6) from wheat germ. J. Biol. Chem. 1979, 254, 8796–8800. [Google Scholar]
- Sanvito, F.; Piatti, S.; Villa, A.; Bossi, M.; Lucchini, G.; Marchisio, P.C.; Biffo, S. The beta4 integrin interactor p27(BBP/eIF6) is an essential nuclear matrix protein involved in 60S ribosomal subunit assembly. J. Cell Biol. 1999, 144, 823–837. [Google Scholar] [CrossRef]
- Gandin, V.; Miluzio, A.; Barbieri, A.M.; Beugnet, A.; Kiyokawa, H.; Marchisio, P.C.; Biffo, S. Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation. Nature 2008, 455, 684–688. [Google Scholar] [CrossRef]
- Brina, D.; Miluzio, A.; Ricciardi, S.; Clarke, K.; Davidsen, P.K.; Viero, G.; Tebaldi, T.; Offenhauser, N.; Rozman, J.; Rathkolb, B.; et al. eIF6 coordinates insulin sensitivity and lipid metabolism by coupling translation to transcription. Nat. Commun. 2015, 6, 8261. [Google Scholar] [CrossRef]
- Biffo, S.; Manfrini, N.; Ricciardi, S. Crosstalks between translation and metabolism in cancer. Curr. Opin. Genet. Dev. 2018, 48, 75–81. [Google Scholar] [CrossRef]
- Miluzio, A.; Ricciardi, S.; Manfrini, N.; Alfieri, R.; Oliveto, S.; Brina, D.; Biffo, S. Translational control by mTOR-independent routes: How eIF6 organizes metabolism. Biochem. Soc. Trans. 2016, 44, 1667–1673. [Google Scholar] [CrossRef]
- Gantenbein, N.; Bernhart, E.; Anders, I.; Golob-Schwarzl, N.; Krassnig, S.; Wodlej, C.; Brcic, L.; Lindenmann, J.; Fink-Neuboeck, N.; Gollowitsch, F.; et al. Influence of eukaryotic translation initiation factor 6 on non-small cell lung cancer development and progression. Eur. J. Cancer 2018, 101, 165–180. [Google Scholar] [CrossRef]
- Miluzio, A.; Oliveto, S.; Pesce, E.; Mutti, L.; Murer, B.; Grosso, S.; Ricciardi, S.; Brina, D.; Biffo, S. Expression and activity of eIF6 trigger malignant pleural mesothelioma growth in vivo. Oncotarget 2015, 6, 37471–37485. [Google Scholar] [CrossRef] [PubMed]
- Gatza, M.L.; Silva, G.O.; Parker, J.S.; Fan, C.; Perou, C.M. An integrated genomics approach identifies drivers of proliferation in luminal-subtype human breast cancer. Nat. Genet. 2014, 46, 1051–1059. [Google Scholar] [CrossRef] [PubMed]
- Sanvito, F.; Vivoli, F.; Gambini, S.; Santambrogio, G.; Catena, M.; Viale, E.; Veglia, F.; Donadini, A.; Biffo, S.; Marchisio, P.C. Expression of a highly conserved protein, p27BBP, during the progression of human colorectal cancer. Cancer Res. 2000, 60, 510–516. [Google Scholar] [PubMed]
- Brina, D.; Grosso, S.; Miluzio, A.; Biffo, S. Translational control by 80S formation and 60S availability: The central role of eIF6, a rate limiting factor in cell cycle progression and tumorigenesis. Cell Cycle 2011, 10, 3441–3446. [Google Scholar] [CrossRef] [PubMed]
- Miluzio, A.; Beugnet, A.; Volta, V.; Biffo, S. Eukaryotic initiation factor 6 mediates a continuum between 60S ribosome biogenesis and translation. Embo Rep. 2009, 10, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Ceci, M.; Gaviraghi, C.; Gorrini, C.; Sala, L.A.; Offenhauser, N.; Marchisio, P.C.; Biffo, S. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature 2003, 426, 579–584. [Google Scholar] [CrossRef]
- Menne, T.F.; Goyenechea, B.; Sanchez-Puig, N.; Wong, C.C.; Tonkin, L.M.; Ancliff, P.J.; Brost, R.L.; Costanzo, M.; Boone, C.; Warren, A.J. The Shwachman-Bodian-Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nat. Genet. 2007, 39, 486–495. [Google Scholar] [CrossRef]
- Boocock, G.R.; Morrison, J.A.; Popovic, M.; Richards, N.; Ellis, L.; Durie, P.R.; Rommens, J.M. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat. Genet. 2003, 33, 97–101. [Google Scholar] [CrossRef]
- Senger, B.; Lafontaine, D.L.; Graindorge, J.S.; Gadal, O.; Camasses, A.; Sanni, A.; Garnier, J.M.; Breitenbach, M.; Hurt, E.; Fasiolo, F. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol. Cell 2001, 8, 1363–1373. [Google Scholar] [CrossRef]
- Wong, C.C.; Traynor, D.; Basse, N.; Kay, R.R.; Warren, A.J. Defective ribosome assembly in Shwachman-Diamond syndrome. Blood 2011, 118, 4305–4312. [Google Scholar] [CrossRef]
- Weis, F.; Giudice, E.; Churcher, M.; Jin, L.; Hilcenko, C.; Wong, C.C.; Traynor, D.; Kay, R.R.; Warren, A.J. Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nat. Struct. Mol. Biol. 2015, 22, 914–919. [Google Scholar] [CrossRef] [PubMed]
- Kargas, V.; Castro-Hartmann, P.; Escudero-Urquijo, N.; Dent, K.; Hilcenko, C.; Sailer, C.; Zisser, G.; Marques-Carvalho, M.J.; Pellegrino, S.; Wawiorka, L.; et al. Mechanism of completion of peptidyltransferase centre assembly in eukaryotes. eLife 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Finch, A.J.; Hilcenko, C.; Basse, N.; Drynan, L.F.; Goyenechea, B.; Menne, T.F.; Gonzalez Fernandez, A.; Simpson, P.; D’Santos, C.S.; Arends, M.J.; et al. Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman-Diamond syndrome. Genes Dev. 2011, 25, 917–929. [Google Scholar] [CrossRef] [PubMed]
- Pesce, E.; Minici, C.; Babetaler, J.; Hurt, E.; Degano, M.; Calamita, P.; Biffo, S. Direct and high throughput (HT) interactions on the ribosomal surface by iRIA. Sci. Rep. 2015, 5, 15401. [Google Scholar] [CrossRef]
- Pignatti, E.; Mascheroni, L.; Sabelli, M.; Barelli, S.; Biffo, S.; Pietrangelo, A. Ferroportin is a monomer in vivo in mice. Blood Cells Mol. Dis. 2006, 36, 26–32. [Google Scholar] [CrossRef]
- Biffo, S.; Sanvito, F.; Costa, S.; Preve, L.; Pignatelli, R.; Spinardi, L.; Marchisio, P.C. Isolation of a novel beta4 integrin-binding protein (p27(BBP)) highly expressed in epithelial cells. J. Biol. Chem. 1997, 272, 30314–30321. [Google Scholar] [CrossRef]
- Biffo, S.; Verdun di Cantogno, L.; Fasolo, A. Double labeling with non-isotopic in situ hybridization and BrdU immunohistochemistry: Calmodulin (CaM) mRNA expression in post-mitotic neurons of the olfactory system. J. Histochem. Cytochem. 1992, 40, 535–540. [Google Scholar] [CrossRef]
- Calamita, P.; Miluzio, A.; Russo, A.; Pesce, E.; Ricciardi, S.; Khanim, F.; Cheroni, C.; Alfieri, R.; Mancino, M.; Gorrini, C.; et al. SBDS-Deficient Cells Have an Altered Homeostatic Equilibrium due to Translational Inefficiency Which Explains their Reduced Fitness and Provides a Logical Framework for Intervention. PLoS Genet. 2017, 13, e1006552. [Google Scholar] [CrossRef]
- Bielefeld-Sevigny, M. AlphaLISA immunoassay platform- the “no-wash” high-throughput alternative to ELISA. Assay Drug Dev. Technol. 2009, 7, 90–92. [Google Scholar] [CrossRef]
- Zhang, J.H.; Chung, T.D.; Oldenburg, K.R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol Screen 1999, 4, 67–73. [Google Scholar] [CrossRef]
- Ye, F.; Zhang, W.; Lu, W.; Xie, Y.; Jiang, H.; Jin, J.; Luo, C. Identification of Novel Inhibitors against Coactivator Associated Arginine Methyltransferase 1 Based on Virtual Screening and Biological Assays. Biomed. Res. Int. 2016, 2016, 7086390. [Google Scholar] [CrossRef] [PubMed]
- Pradet-Balade, B.; Boulme, F.; Beug, H.; Mullner, E.W.; Garcia-Sanz, J.A. Translation control: Bridging the gap between genomics and proteomics? Trends Biochem. Sci. 2001, 26, 225–229. [Google Scholar] [CrossRef]
- Roux, P.P.; Topisirovic, I. Signaling Pathways Involved in the Regulation of mRNA Translation. Mol. Cell Biol. 2018, 38. [Google Scholar] [CrossRef] [PubMed]
- Ricciardi, S.; Manfrini, N.; Alfieri, R.; Calamita, P.; Crosti, M.C.; Gallo, S.; Muller, R.; Pagani, M.; Abrignani, S.; Biffo, S. The Translational Machinery of Human CD4(+) T Cells Is Poised for Activation and Controls the Switch from Quiescence to Metabolic Remodeling. Cell Metab. 2018, 28, 895–906. [Google Scholar] [CrossRef]
- Xu, Y.; Poggio, M.; Jin, H.Y.; Shi, Z.; Forester, C.M.; Wang, Y.; Stumpf, C.R.; Xue, L.; Devericks, E.; So, L.; et al. Translation control of the immune checkpoint in cancer and its therapeutic targeting. Nat. Med. 2019, 25, 301–311. [Google Scholar] [CrossRef]
- Santini, E.; Huynh, T.N.; MacAskill, A.F.; Carter, A.G.; Pierre, P.; Ruggero, D.; Kaphzan, H.; Klann, E. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature 2013, 493, 411–415. [Google Scholar] [CrossRef]
- De la Parra, C.; Walters, B.A.; Geter, P.; Schneider, R.J. Translation initiation factors and their relevance in cancer. Curr. Opin. Genet. Dev. 2018, 48, 82–88. [Google Scholar] [CrossRef]
- Sidrauski, C.; Acosta-Alvear, D.; Khoutorsky, A.; Vedantham, P.; Hearn, B.R.; Li, H.; Gamache, K.; Gallagher, C.M.; Ang, K.K.; Wilson, C.; et al. Pharmacological brake-release of mRNA translation enhances cognitive memory. eLife 2013, 2, e00498. [Google Scholar] [CrossRef]
- Zyryanova, A.F.; Weis, F.; Faille, A.; Alard, A.A.; Crespillo-Casado, A.; Sekine, Y.; Harding, H.P.; Allen, F.; Parts, L.; Fromont, C.; et al. Binding of ISRIB reveals a regulatory site in the nucleotide exchange factor eIF2B. Science 2018, 359, 1533–1536. [Google Scholar] [CrossRef]
- Chu, J.; Zhang, W.; Cencic, R.; Devine, W.G.; Beglov, D.; Henkel, T.; Brown, L.E.; Vajda, S.; Porco, J.A., Jr.; Pelletier, J. Amidino-Rocaglates: A Potent Class of eIF4A Inhibitors. Cell Chem. Biol. 2019. [Google Scholar] [CrossRef]
- Lo, K.Y.; Li, Z.; Bussiere, C.; Bresson, S.; Marcotte, E.M.; Johnson, A.W. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol. Cell 2010, 39, 196–208. [Google Scholar] [CrossRef]
- Brina, D.; Miluzio, A.; Ricciardi, S.; Biffo, S. eIF6 anti-association activity is required for ribosome biogenesis, translational control and tumor progression. Biochim. Biophys. Acta 2015, 1849, 830–835. [Google Scholar] [CrossRef]
- Ricciardi, S.; Miluzio, A.; Brina, D.; Clarke, K.; Bonomo, M.; Aiolfi, R.; Guidotti, L.G.; Falciani, F.; Biffo, S. Eukaryotic translation initiation factor 6 is a novel regulator of reactive oxygen species-dependent megakaryocyte maturation. J. Thromb Haemost 2015, 13, 2108–2118. [Google Scholar] [CrossRef]
- Manfrini, N.; Ricciardi, S.; Miluzio, A.; Fedeli, M.; Scagliola, A.; Gallo, S.; Brina, D.; Adler, T.; Busch, D.H.; Gailus-Durner, V.; et al. High levels of eukaryotic Initiation Factor 6 (eIF6) are required for immune system homeostasis and for steering the glycolytic flux of TCR-stimulated CD4(+) T cells in both mice and humans. Dev. Comp. Immunol. 2017, 77, 69–76. [Google Scholar] [CrossRef]
- Klinge, S.; Voigts-Hoffmann, F.; Leibundgut, M.; Arpagaus, S.; Ban, N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 2011, 334, 941–948. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pesce, E.; Miluzio, A.; Turcano, L.; Minici, C.; Cirino, D.; Calamita, P.; Manfrini, N.; Oliveto, S.; Ricciardi, S.; Grifantini, R.; et al. Discovery and Preliminary Characterization of Translational Modulators that Impair the Binding of eIF6 to 60S Ribosomal Subunits. Cells 2020, 9, 172. https://doi.org/10.3390/cells9010172
Pesce E, Miluzio A, Turcano L, Minici C, Cirino D, Calamita P, Manfrini N, Oliveto S, Ricciardi S, Grifantini R, et al. Discovery and Preliminary Characterization of Translational Modulators that Impair the Binding of eIF6 to 60S Ribosomal Subunits. Cells. 2020; 9(1):172. https://doi.org/10.3390/cells9010172
Chicago/Turabian StylePesce, Elisa, Annarita Miluzio, Lorenzo Turcano, Claudia Minici, Delia Cirino, Piera Calamita, Nicola Manfrini, Stefania Oliveto, Sara Ricciardi, Renata Grifantini, and et al. 2020. "Discovery and Preliminary Characterization of Translational Modulators that Impair the Binding of eIF6 to 60S Ribosomal Subunits" Cells 9, no. 1: 172. https://doi.org/10.3390/cells9010172
APA StylePesce, E., Miluzio, A., Turcano, L., Minici, C., Cirino, D., Calamita, P., Manfrini, N., Oliveto, S., Ricciardi, S., Grifantini, R., Degano, M., Bresciani, A., & Biffo, S. (2020). Discovery and Preliminary Characterization of Translational Modulators that Impair the Binding of eIF6 to 60S Ribosomal Subunits. Cells, 9(1), 172. https://doi.org/10.3390/cells9010172





