VAPB ER-Aggregates, A Possible New Biomarker in ALS Pathology
Abstract
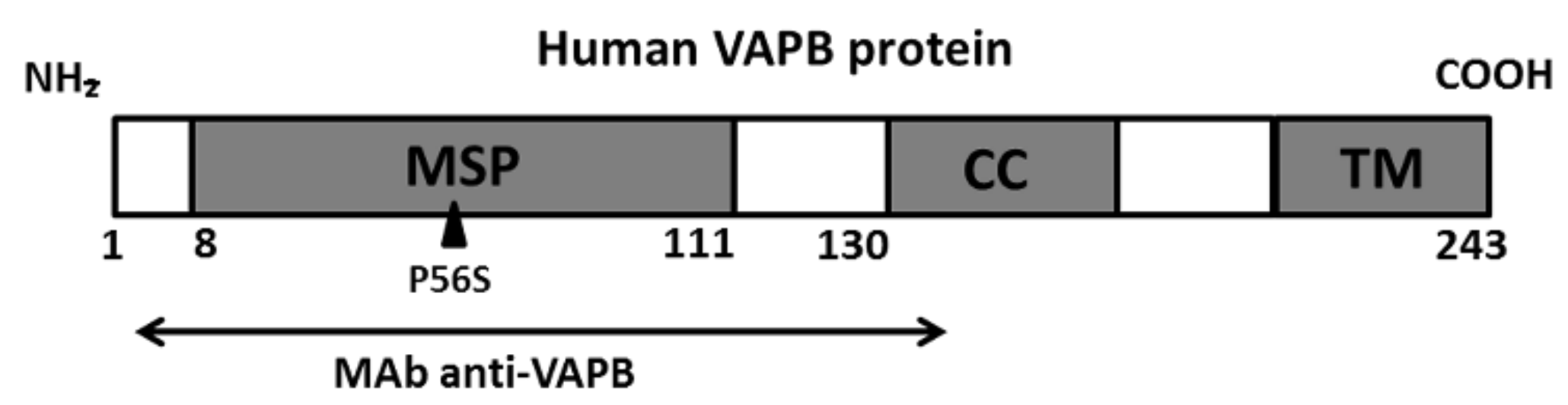
1. Introduction
2. Materials and Methods
2.1. HeLa Cells Transfection and Cultures
2.2. Ethical Committee Approval
2.3. Patients and Methods
2.4. PBMC Isolation
2.5. Western Blot Analysis
2.6. Primary Cutaneous Fibroblasts Cells Cultures
2.7. Immunofluorescence
2.8. Flow Cytometry Assays
2.9. VAPB mRNA Expression Gene
2.10. Genomic Analysis
2.11. Statistical Analysis
3. Results
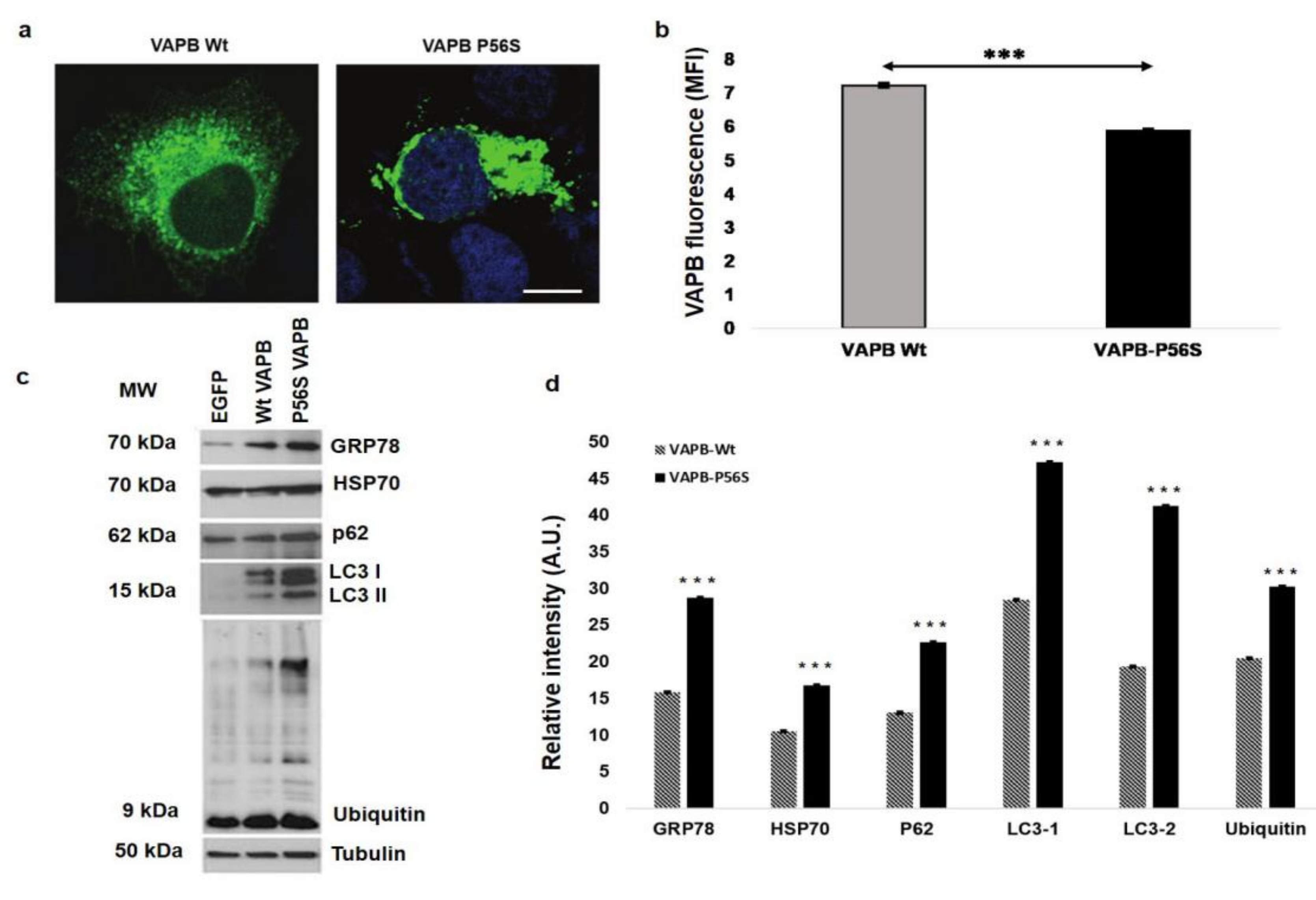
3.1. Cellular Model Analysis
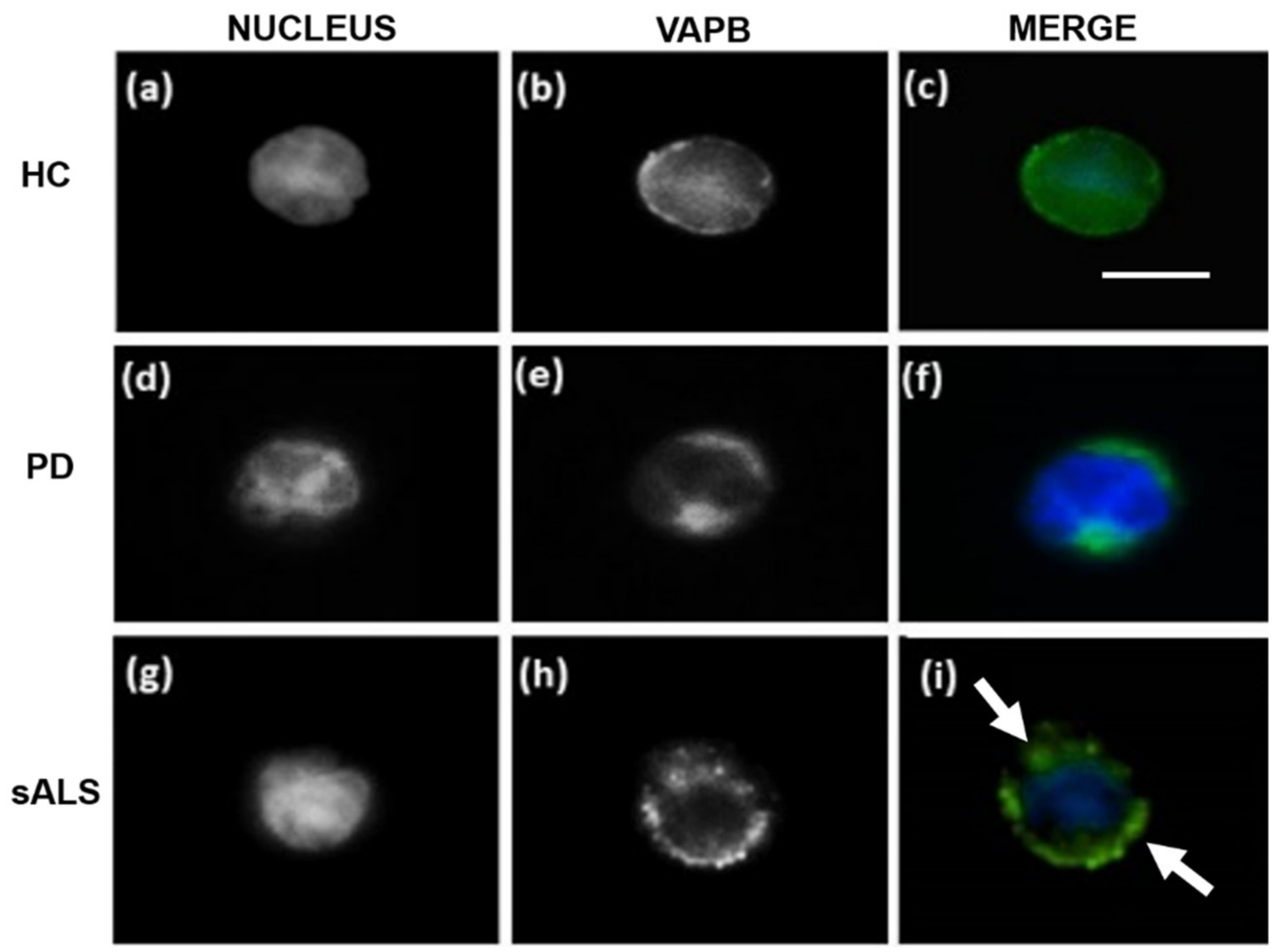
3.2. VAPB ER-Aggregates in sALS Patients PBMCs
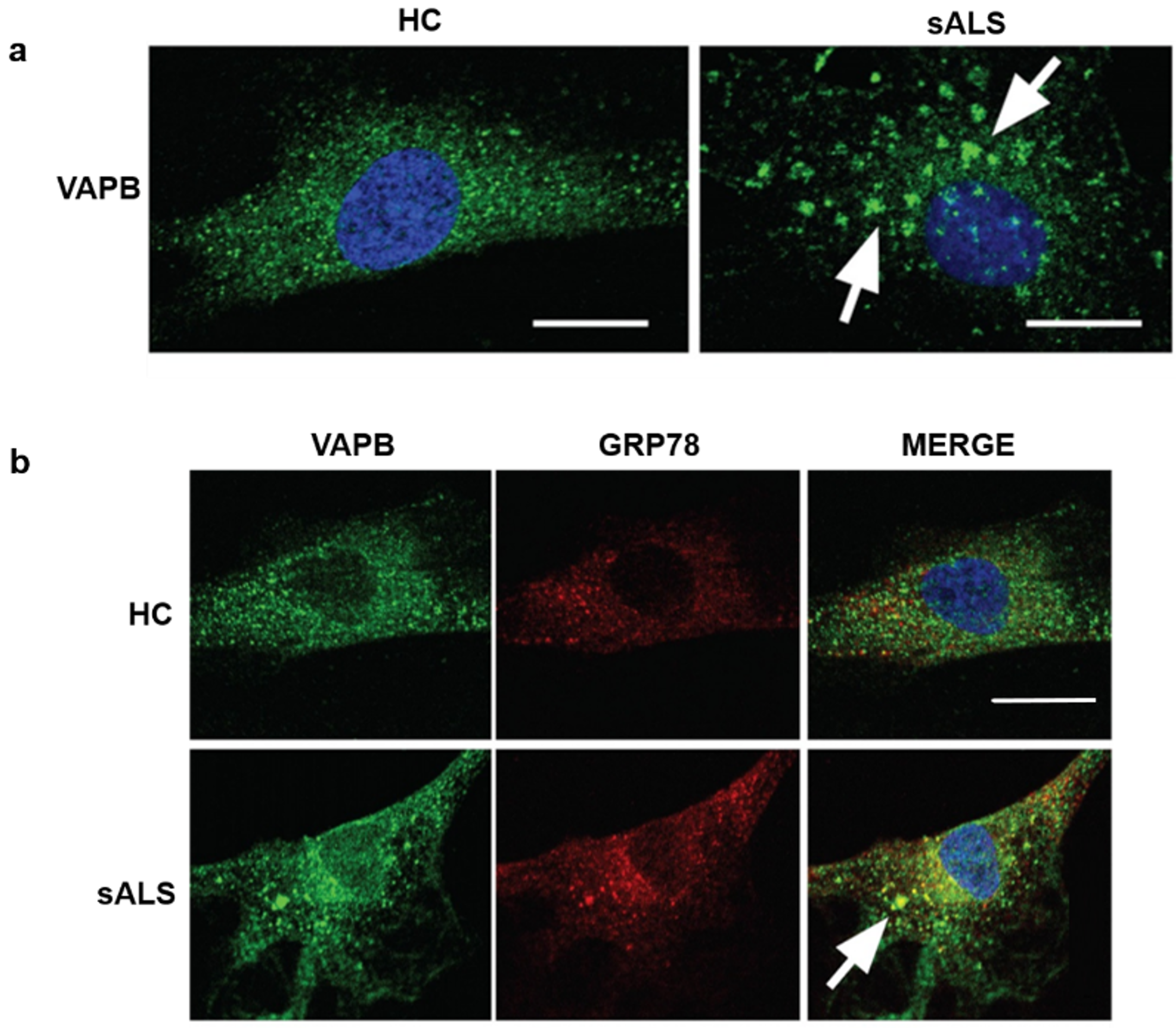
3.3. VAPB Accumulations in sALS Patients Fibroblasts
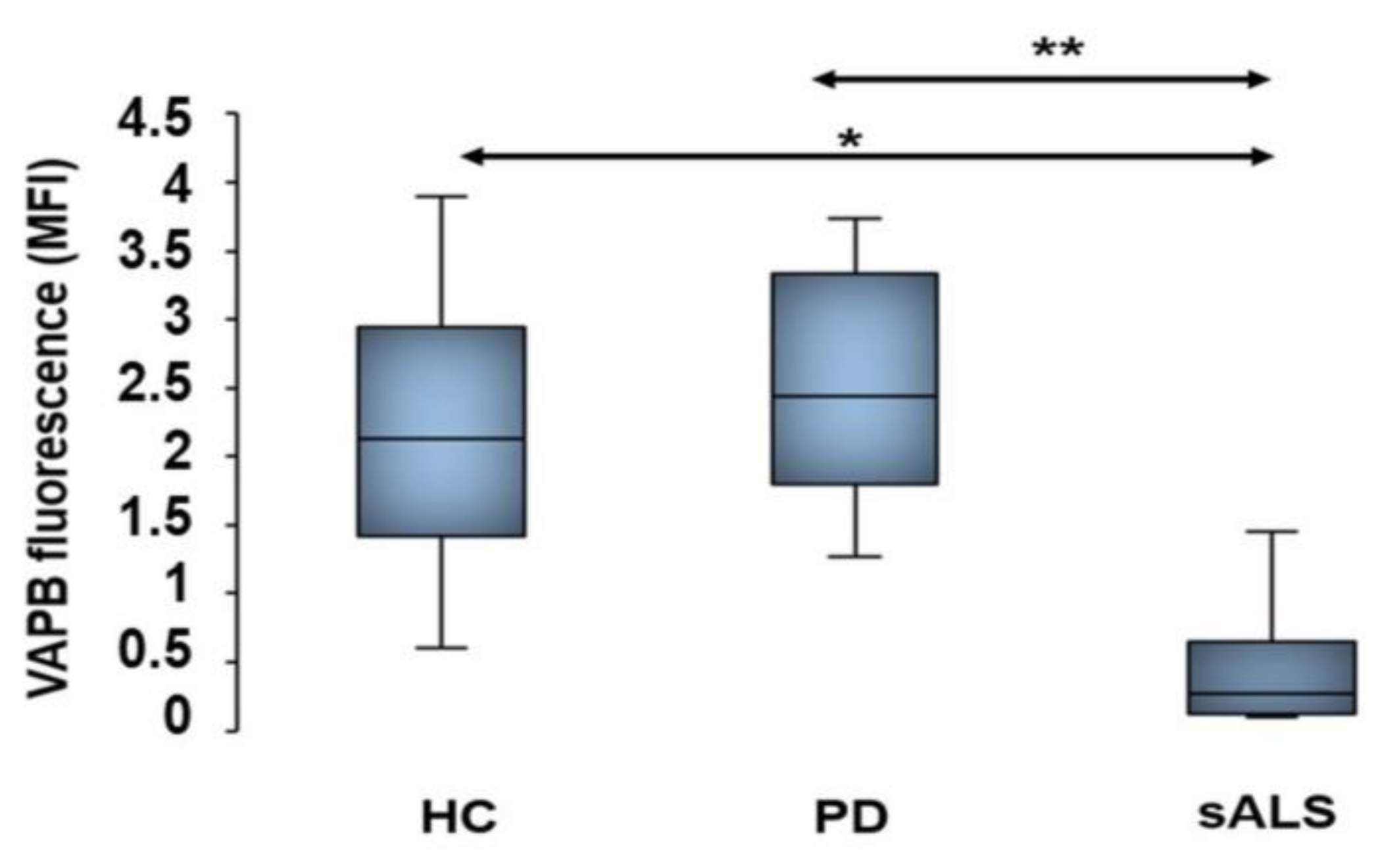
3.4. Decrease of VAPB Fluorescent Signals in sALS Patients
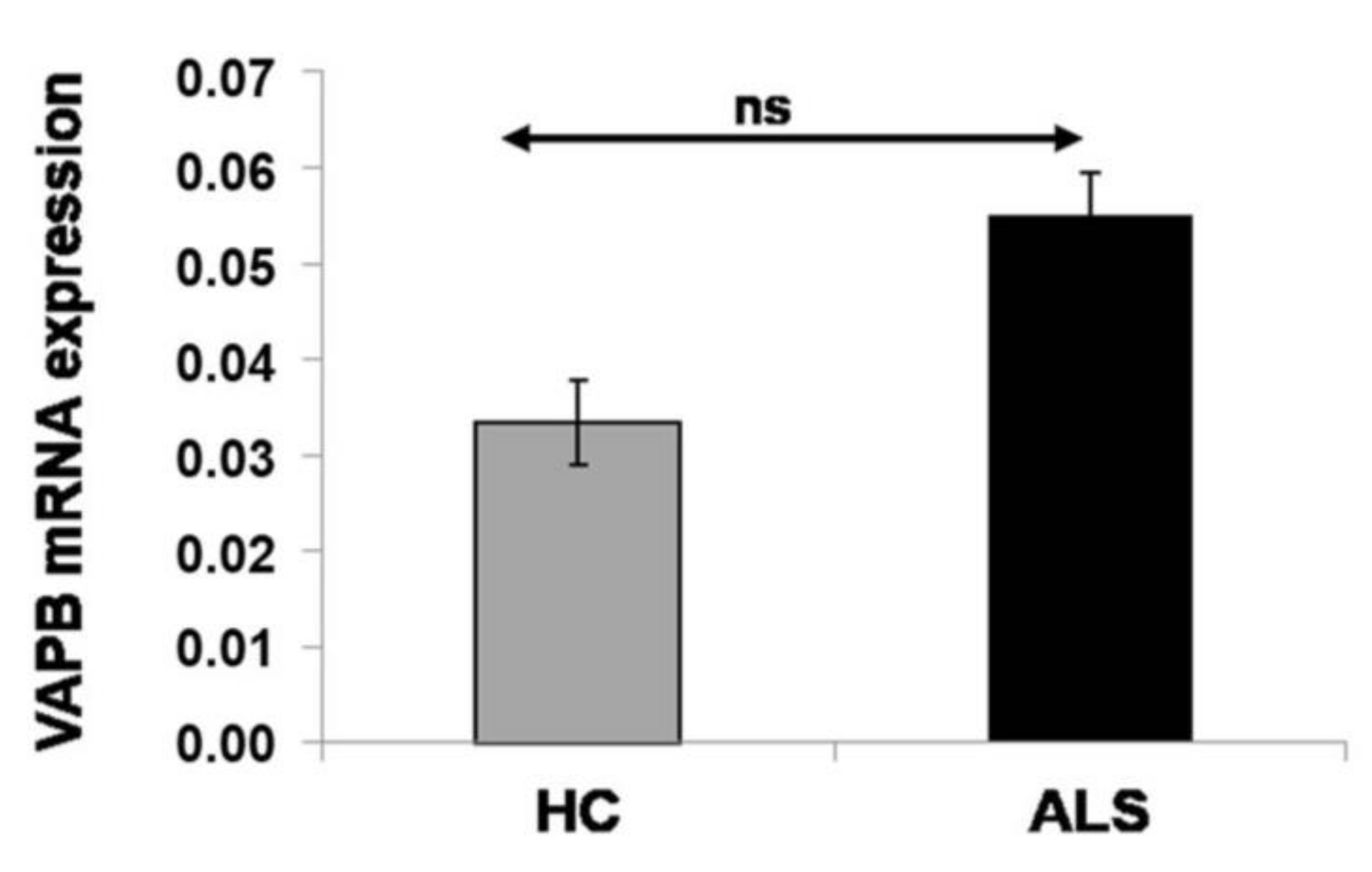
3.5. Nonalteration of VAPB mRNA Expression in sALS Patients
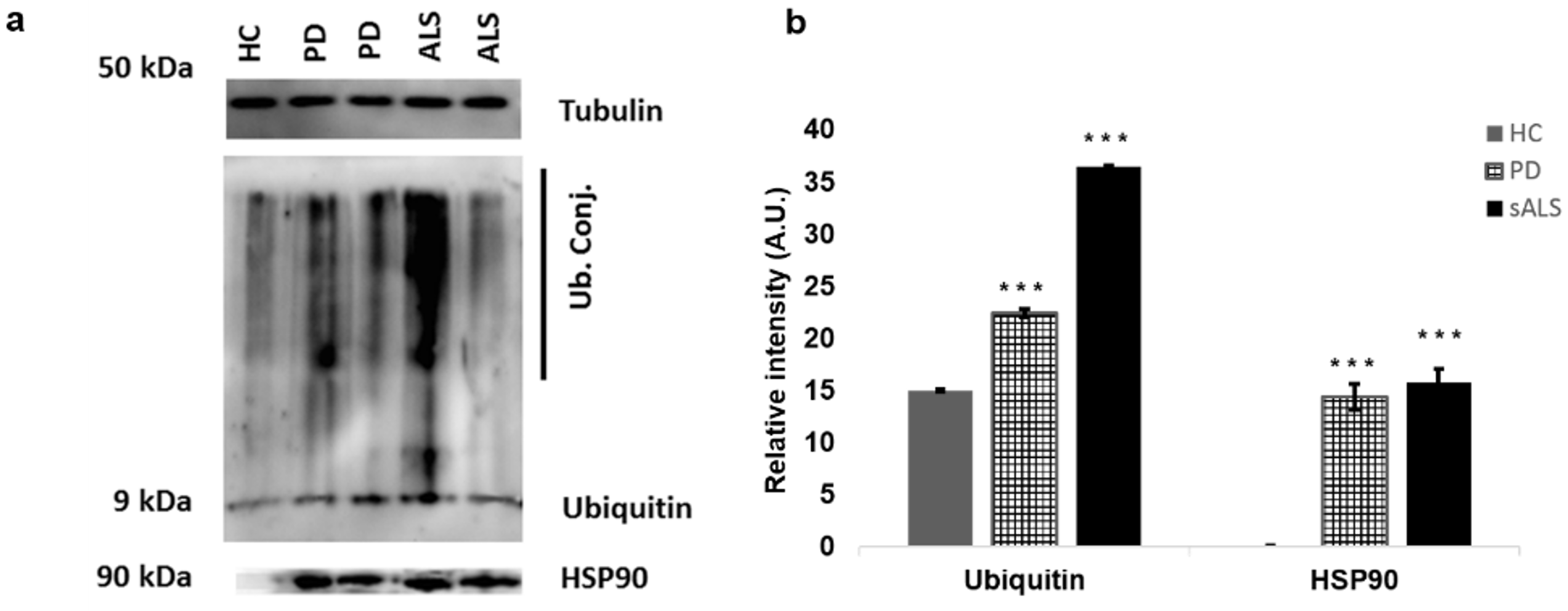
3.6. Western Blot Analysis of PBMCs
3.7. Genetic Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pansarasa, O.; Rossi, D.; Berardinelli, A.; Cereda, C. Amyotrophic lateral sclerosis and skeletal muscle: An update. Mol. Neurobiol. 2014, 49, 984–990. [Google Scholar] [CrossRef]
- Zarei, S.; Carr, K.; Reiley, L.; Diaz, K.; Guerra, O.; Altamirano, P.F.; Pagani, W.; Lodin, D.; Orozco, G.; Chinea, A. A comprehensive review of amyotrophic lateral sclerosis. Surg. Neurol. Int. 2015, 6, 171. [Google Scholar] [CrossRef]
- Saberi, S.; Stauffer, J.E.; Schulte, D.J.; Ravits, J. Neuropathology of amyotrophic lateral sclerosis and its variants. Neurol. Clin. 2015, 33, 855–876. [Google Scholar] [CrossRef]
- Kanekura, K.; Nishimoto, I.; Aiso, S.; Matsuoka, M. Characterization of amyotrophic lateral sclerosis-linked P56S mutation of vesicle-associated membrane protein-associated protein B (VAPB/ALS8). J. Biol. Chem. 2006, 281, 30223–30233. [Google Scholar] [CrossRef]
- Ince, P.G.; Highley, J.R.; Kirby, J.; Wharton, S.B.; Takahashi, H.; Strong, M.J.; Shaw, P.J. Molecular pathology and genetic advances in amyotrophic lateral sclerosis: An emerging molecular pathway and the significance of glial pathology. Acta Neuropathol. 2011, 122, 657–671. [Google Scholar] [CrossRef]
- Yamashita, S.; Ando, Y. Genotype-phenotype relationship in hereditary amyotrophic lateral sclerosis. Transl. Neurodegener. 2015, 4, 13. [Google Scholar] [CrossRef] [PubMed]
- Bozzoni, V.; Pansarasa, O.; Diamanti, L.; Nosari, G.; Cereda, C.; Ceroni, M. Amyotrophic lateral sclerosis and environmental factors. Funct. Neurol. 2016, 31, 7–19. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Cao, C.; Qin, X.Y.; Yu, Y.; Yuan, J.; Zhao, Y.; Cheng, Y. Increased peripheral blood inflammatory cytokine levels in amyotrophic lateral sclerosis: A meta-analysis study. Sci. Rep. 2017, 7, 12–15. [Google Scholar] [CrossRef] [PubMed]
- Carrì, M.T.; D’Ambrosi, N.; Cozzolino, M. Pathways to mitochondrial dysfunction in ALS pathogenesis. Biochem. Biophys. Res. Commun. 2017, 483, 1187–1193. [Google Scholar] [CrossRef]
- Groen, E.J.N.; Pasterkamp, R.J.; van den Berg, L.H.; Koppers, M.; Blokhuis, A.M. Protein aggregation in amyotrophic lateral sclerosis. Acta Neuropathol. 2013, 125, 777–794. [Google Scholar]
- Fasana, E.; Fossati, M.; Ruggiano, A.; Brambillasca, S.; Hoogenraad, C.C.; Navone, F.; Francolini, M.; Borgese, N. A VAPB mutant linked to amyotrophic lateral sclerosis generates a novel form of organized smooth endoplasmic reticulum. FASEB J. 2010, 24, 1419–1430. [Google Scholar] [CrossRef]
- Mitne-Neto, M.; Machado-Costa, M.; Marchetto, M.C.N.; Bengtson, M.H.; Joazeiro, C.A.; Tsuda, H.; Bellen, H.J.; Silva, H.C.A.; Oliveira, A.S.B.; Lazar, M.; et al. Downregulation of VAPB expression in motor neurons derived from induced pluripotent stem cells of ALS8 patients. Hum. Mol. Genet. 2011, 20, 3642–3652. [Google Scholar] [CrossRef] [PubMed]
- Teuling, E.; Ahmed, S.; Haasdijk, E.; Demmers, J.; Steinmetz, M.O.; Akhmanova, A.; Jaarsma, D.; Hoogenraad, C.C. Motor neuron disease-associated mutant vesicle-associated membrane protein-associated protein (VAP) B recruits wild-type VAPs into endoplasmic reticulum-derived tubular aggregates. J. Neurosci. 2007, 27, 9801–9815. [Google Scholar] [CrossRef] [PubMed]
- Lev, S.; Halevy, D.B.; Peretti, D.; Dahan, N. The VAP protein family: From cellular functions to motor neuron disease. Trends Cell Biol. 2008, 18, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Leal, S.S.; Ben Halevy, D.; Gomes, C.M.; Lev, S. Structural requirements for VAP-B oligomerization and their implication in amyotrophic lateral sclerosis-associated VAP-B(P56S) neurotoxicity. J. Biol. Chem. 2010, 285, 13839–13849. [Google Scholar] [CrossRef] [PubMed]
- Gkogkas, C.; Middleton, S.; Kremer, A.M.; Wardrope, C.; Hannah, M.; Gillingwater, T.H.; Skehel, P. VAPB interacts with and modulates the activity of ATF6. Hum. Mol. Genet. 2008, 17, 1517–1526. [Google Scholar] [CrossRef]
- Mórotz, G.M.; De Vos, K.J.; Vagnoni, A.; Ackerley, S.; Shaw, C.E.; Miller, C.C.J. Amyotrophic lateral sclerosis-associated mutant VAPBP56s perturbs calcium homeostasis to disrupt axonal transport of mitochondria. Hum. Mol. Genet. 2012, 21, 1979–1988. [Google Scholar] [CrossRef]
- Suzuki, H.; Kanekura, K.; Levine, T.P.; Kohno, K.; Olkkonen, V.M.; Aiso, S.; Matsuoka, M. ALS-linked P56S-VAPB, an aggregated loss-of-function mutant of VAPB, predisposes motor neurons to ER stress-related death by inducing aggregation of co-expressed wild-type VAPB. J. Neurochem. 2009, 108, 973–985. [Google Scholar] [CrossRef]
- Nishimura, A.L.; Mitne-Neto, M.; Silva, H.C.A.; Richieri-Costa, A.; Middleton, S.; Cascio, D.; Kok, F.; Oliveira, J.R.M.; Gillingwater, T.; Webb, J.; et al. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am. J. Hum. Genet. 2004, 75, 822–831. [Google Scholar] [CrossRef]
- Marques, V.D.; Barreira, A.A.; Davis, M.B.; Abou-Sleiman, P.M.; Silva, W.A.; Zago, M.A.; Sobreira, C.; Fazan, V.; Marques, W. Expanding the phenotypes of the Pro56Ser VAPB mutation: Proximal SMA with dysautonomia. Muscle Nerve 2006, 34, 731–739. [Google Scholar] [CrossRef]
- Chen, H.-J.; Anagnostou, G.; Chai, A.; Withers, J.; Morris, A.; Adhikaree, J.; Pennetta, G.; de Belleroche, J.S. Characterization of the properties of a novel mutation in VAPB in familial amyotrophic lateral sclerosis. J. Biol. Chem. 2010, 285, 40266–40281. [Google Scholar] [CrossRef]
- Anagnostou, G.; Paul, P.; Akbar, M.T.; Steiner, T.J.; Angelinetta, C.; de Belleroche, J. Vesicle associated membrane protein B (VAPB) is decreased in ALS spinal cord. Neurobiol. Aging 2010, 31, 969–985. [Google Scholar] [CrossRef]
- Qiu, L.; Qiao, T.; Beers, M.; Tan, W.; Wang, H.; Yang, B.; Xu, Z. Widespread aggregation of mutant VAPB associated with ALS does not cause motor neuron degeneration or modulate mutant SOD1 aggregation and toxicity in mice. Mol. Neurodegener. 2013, 8, 1. [Google Scholar] [CrossRef]
- Tudor, E.L.; Galtrey, C.M.; Perkinton, M.S.; Lau, K.-F.; De Vos, K.J.; Mitchell, J.C.; Ackerley, S.; Hortobágyi, T.; Vámos, E.; Leigh, P.N.; et al. Amyotrophic lateral sclerosis mutant vesicle-associated membrane protein-associated protein-B transgenic mice develop TAR-DNA-binding protein-43 pathology. Neuroscience 2010, 167, 774–785. [Google Scholar] [CrossRef] [PubMed]
- van Blitterswijk, M.; van Es, M.A.; Koppers, M.; van Rheenen, W.; Medic, J.; Schelhaas, H.J.; van der Kooi, A.J.; de Visser, M.; Veldink, J.H.; van den Berg, L.H. VAPB and C9orf72 mutations in 1 familial amyotrophic lateral sclerosis patient. Neurobiol. Aging 2012, 33, 2950-e1. [Google Scholar] [CrossRef] [PubMed]
- Liew, C.C.; Ma, J.; Tang, H.C.; Zheng, R.; Dempsey, A.A. The peripheral blood transcriptome dynamically reflects system wide biology: A potential diagnostic tool. J. Lab. Clin. Med. 2006, 147, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Cereda, C.; Leoni, E.; Milani, P.; Pansarasa, O.; Mazzini, G.; Guareschi, S.; Alvisi, E.; Ghiroldi, A.; Diamanti, L.; Bernuzzi, S.; et al. Altered Intracellular Localization of SOD1 in Leukocytes from Patients with Sporadic Amyotrophic Lateral Sclerosis. PLoS ONE 2013, 8, e75916. [Google Scholar] [CrossRef]
- Deidda, I.; Galizzi, G.; Passantino, R.; Cascio, C.; Russo, D.; Colletti, T.; La Bella, V.; Guarneri, P. Expression of vesicle-associated membrane-protein-associated protein B cleavage products in peripheral blood leukocytes and cerebrospinal fluid of patients with sporadic amyotrophic lateral sclerosis. Eur. J. Neurol. 2014, 21, 478–485. [Google Scholar] [CrossRef]
- Nardo, G.; Pozzi, S.; Pignataro, M.; Lauranzano, E.; Spano, G.; Garbelli, S.; Mantovani, S.; Marinou, K.; Papetti, L.; Monteforte, M.; et al. Amyotrophic lateral sclerosis multiprotein biomarkers in peripheral blood mononuclear cells. PLoS ONE 2011, 6, e25545. [Google Scholar] [CrossRef]
- Silani, V.; Ludolph, A.; Fornai, F. The emerging picture of ALS: A multisystem, not only a “motor neuron disease”. Arch. Ital. Biol. 2017, 155, 153–158. [Google Scholar]
- Vijayakumar, U.G.; Milla, V.; Stafford, M.Y.C.; Bjourson, A.J.; Duddy, W.; Duguez, S.M.R. A systematic review of suggested molecular strata, biomarkers and their tissue sources in ALS. Front. Neurol. 2019, 10, 400. [Google Scholar] [CrossRef] [PubMed]
- Musarò, A. Understanding ALS: New therapeutic approaches. FEBS J. 2013, 280, 4315–4322. [Google Scholar] [CrossRef] [PubMed]
- Nachreiner, T.; Esser, M.; Tenten, V.; Troost, D.; Weis, J.; Krüttgen, A. Novel splice variants of the amyotrophic lateral sclerosis-associated gene VAPB expressed in human tissues. Biochem. Biophys. Res. Commun. 2010, 394, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Jesse, C.M.; Bushuven, E.; Tripathi, P.; Chandrasekar, A.; Simon, C.M.; Drepper, C.; Yamoah, A.; Dreser, A.; Katona, I.; Johann, S.; et al. ALS-Associated Endoplasmic Reticulum Proteins in Denervated Skeletal Muscle: Implications for Motor Neuron Disease Pathology. Brain Pathol. 2017, 27, 781–794. [Google Scholar] [CrossRef] [PubMed]
- Funke, A.D.; Esser, M.; Krüttgen, A.; Weis, J.; Mitne-Neto, M.; Lazar, M.; Nishimura, A.L.; Sperfeld, A.D.; Trillenberg, P.; Senderek, J.; et al. The p.P56S mutation in the VAPB gene is not due to a single founder: The first European case. Clin. Genet. 2010, 77, 302–303. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinf. 2012, 13, 134. [Google Scholar] [CrossRef]
- DeJesus-Hernandez, M.; Mackenzie, I.R.; Boeve, B.F.; Boxer, B.F.; Baker, M.; Rutherford, N.J.; Nicholson, A.M.; Finch, N.A.; Flynn, H.; Adamson, J.; et al. Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron 2011, 72, 245–256. [Google Scholar] [CrossRef]
- Papiani, G.; Ruggiano, A.; Fossati, M.; Raimondi, A.; Bertoni, G.; Francolini, M.; Benfante, R.; Navone, F.; Borgese, N. Restructured endoplasmic reticulum generated by mutant amyotrophic lateral sclerosis-linked VAPB is cleared by the proteasome. J. Cell Sci. 2012, 125, 3601–3611. [Google Scholar] [CrossRef]
- Kiriyama, Y.; Nochi, H. The Function of Autophagy in Neurodegenerative Diseases. Int. J. Mol. Sci. 2015, 16, 26797–26812. [Google Scholar] [CrossRef]
- Ciechanover, A.; Brundin, P. The ubiquitin proteasome system in neurodegenerative diseases: Sometimes the chicken, sometimes the egg. Neuron 2003, 40, 427–446. [Google Scholar] [CrossRef]
- Jaronen, M.; Goidsteins, G.; Koistinaho, J. ER stress and unfolded protein response in amyotrophic lateral sclerosis—A controversial role of protein disulphide isomerase. Front. Cell. Neurosci. 2014, 8, 402. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wey, S.; Zhang, Y.; Ye, R.; Lee, A.S. Role of the unfolded protein response regulator GRP78/BiP in development, cancer, and neurological disorders. Antioxid. Redox Signal. 2009, 11, 2307–2316. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, A.; Chen, A.W.; Varner, J.D. A review of the mammalian unfolded protein response. Biotechnol. Bioeng. 2011, 108, 2777–2793. [Google Scholar] [CrossRef] [PubMed]
- Tanida, I.; Ueno, T.; Kominami, E. LC3 and autophagy. Methods Mol. Biol. 2008, 445, 77–88. [Google Scholar]
- Nedelsky, N.B.; Todd, P.K.; Taylor, J.P. Autophagy and the ubiquitin-proteasome system: Collaborators in neuroprotection. Biochim. Biophys. Acta Mol. Basis Dis. 2008, 1782, 691–699. [Google Scholar] [CrossRef]
- Régal, L.; Vanopdenbosch, L.; Tilkin, P.; Van Den Bosch, L.; Thijs, V.; Sciot, R.; Robberecht, W. The G93C mutation in superoxide dismutase 1: Clinicopathologic phenotype and prognosis. Arch. Neurol. 2006, 63, 262–267. [Google Scholar] [CrossRef]
- Doyle, K.M.; Kennedy, D.; Gorman, A.M.; Gupta, S.; Healy, S.J.M.; Samali, A. Unfolded proteins and endoplasmic reticulum stress in neurodegenerative disorders. J. Cell. Mol. Med. 2011, 15, 2025–2039. [Google Scholar] [CrossRef]
- Burkhardt, M.F.; Martinez, F.J.; Wright, S.; Ramos, C.; Volfson, D.; Mason, M.; Garnes, J.; Dang, V.; Lievers, J.; Shoukat-Mumtaz, U.; et al. A cellular model for sporadic ALS using patient-derived induced pluripotent stem cells. Mol. Cell. Neurosci. 2013, 56, 355–364. [Google Scholar] [CrossRef]
- Gkogkas, C.; Wardrope, C.; Hannah, M.; Skehel, P. The ALS8-associated mutant VAPBP56S is resistant to proteolysis in neurons. J. Neurochem. 2011, 117, 286–294. [Google Scholar] [CrossRef]
- Orrù, S.; Manolakos, E.; Orrù, N.; Kokotas, H.; Mascia, V.; Carcassi, C.; Petersen, M. High frequency of the TARDBP p.Ala382Thr mutation in Sardinian patients with amyotrophic lateral sclerosis. Clin. Genet. 2012, 81, 172–178. [Google Scholar] [CrossRef]
- Sabatelli, M.; Conforti, F.L.; Zollino, M.; Mora, G.; Monsurrò, M.R.; Volanti, P.; Marinou, K.; Salvi, F.; Corbo, M.; Giannini, F.; et al. C9ORF72 hexanucleotide repeat expansions in the Italian sporadic ALS population. Neurobiol. Aging 2012, 33, 1848–e15. [Google Scholar] [CrossRef] [PubMed]
- Aliaga, L.; Lai, C.; Yu, J.; Chub, N.; Shim, H.; Sun, L.; Xie, C.; Yang, W.J.; Lin, X.; O’Donovan, M.J.; et al. Amyotrophic lateral sclerosis-related VAPB P56S mutation differentially affects the function and survival of corticospinal and spinal motor neurons. Hum. Mol. Genet. 2013, 22, 4293–4305. [Google Scholar] [CrossRef] [PubMed]
- Prause, J.; Goswami, A.; Katona, I.; Roos, A.; Schnizler, M.; Bushuven, E.; Dreier, A.; Buchkremer, S.; Johann, S.; Beyer, C.; et al. Altered localization, abnormal modification and loss of function of sigma receptor-1 in amyotrophic lateral sclerosis. Hum. Mol. Genet. 2013, 22, 1581–1600. [Google Scholar] [CrossRef] [PubMed]
- Masala, A.; Sanna, S.; Esposito, S.; Rassu, M.; Galioto, M.; Zinellu, A.; Carru, C.; Carrì, M.T.; Iaccarino, C.; Crosio, C. Epigenetic Changes Associated with the Expression of Amyotrophic Lateral Sclerosis (ALS) Causing Genes. Neuroscience 2018, 390, 1–11. [Google Scholar] [CrossRef]
| sALS | PD | HC | |
|---|---|---|---|
| Number | 24 | 14 | 24 |
| Mean age ± SD | 66.1 ± 6 | 71.3 ± 9.6 | 63.2 ± 5.8 |
| Mean age at onset ± SD | 59.7 ± 10.6 | 63 ± 11.9 | |
| Males/Females ratio | 16/8 | 8/6 | 16/8 |
| Disease duration ± SD 1 | 4.6 ± 4.1 | 8.3 ± 4.8 | - |
| Signs at onset: spinal | 19 cases | - | - |
| Signs at onset: bulbar | 5 cases | - | - |
| H&Y score, median ± SD | - | 3 ± 1.2 | - |
| ALSFRS-R, median ± SD | 36 ± 5.1 | - | - |
| Gene | Exon | Forward Primer Sequence | Reverse Primer Sequence |
|---|---|---|---|
| TARDBP | 1–3 | 5′-3′ GGGGAGGTCAGCTCCTATAC | 5′-3′ TCATCTGTGTAACAGAAAGGACAGT |
| 4–6 | 5′-3′ CCACTGCATCCAGTTGAAACC | 5′-3′ TGCCCACCCAGTGTAATGTC | |
| FUS | 1–6 | 5′-3′ TAGTCCTGCCGAGGAGAGAG | 5′-3′ CTACACTGCGAGAAGAGCGG |
| 7–15 | 5′-3′ GTCAGACAAGGGGTGGTCAG | 5′-3′ CTGGCAGGGGAAGTAAACCT | |
| SOD | 1 | 5′-3′ TTTCGGGGTTCTGGACGTTT | 5′-3′ TAGGTTGTGCCAGCGTCTAC |
| VAPB | 1 | 5′-3′ TGGCTGTCGTAGCTGGACTA | 5′-3′ GTCCTTCCAGCACTGGGG |
| 2 | 5′-3′ GTGAGGTTTCAATGAGATGCCA | 5′-3′ TGGGACCCAGCTTTCTGAT | |
| 3 | 5′-3′ CAGCTCTGTCATGCGTGATTT | 5′-3′ ACAGCCTTCTAGAAAGTCACAAAA | |
| 4–6 | 5′-3′ TTTTGTTCAGTGGTGGGCCT | 5′-3′ AAACATGGCATGGAGGGAGG |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cadoni, M.P.L.; Biggio, M.L.; Arru, G.; Secchi, G.; Orrù, N.; Clemente, M.G.; Sechi, G.; Yamoah, A.; Tripathi, P.; Orrù, S.; et al. VAPB ER-Aggregates, A Possible New Biomarker in ALS Pathology. Cells 2020, 9, 164. https://doi.org/10.3390/cells9010164
Cadoni MPL, Biggio ML, Arru G, Secchi G, Orrù N, Clemente MG, Sechi G, Yamoah A, Tripathi P, Orrù S, et al. VAPB ER-Aggregates, A Possible New Biomarker in ALS Pathology. Cells. 2020; 9(1):164. https://doi.org/10.3390/cells9010164
Chicago/Turabian StyleCadoni, Maria Piera L, Maria Luigia Biggio, Giannina Arru, Giannina Secchi, Nicola Orrù, Maria Grazia Clemente, GianPietro Sechi, Alfred Yamoah, Priyanka Tripathi, Sandro Orrù, and et al. 2020. "VAPB ER-Aggregates, A Possible New Biomarker in ALS Pathology" Cells 9, no. 1: 164. https://doi.org/10.3390/cells9010164
APA StyleCadoni, M. P. L., Biggio, M. L., Arru, G., Secchi, G., Orrù, N., Clemente, M. G., Sechi, G., Yamoah, A., Tripathi, P., Orrù, S., Manetti, R., & Galleri, G. (2020). VAPB ER-Aggregates, A Possible New Biomarker in ALS Pathology. Cells, 9(1), 164. https://doi.org/10.3390/cells9010164







