pH-Dependent Aggregation in Intrinsically Disordered Proteins Is Determined by Charge and Lipophilicity
Abstract
1. Introduction
2. Materials and Methods
2.1. Generation of Lipophilicity Profiles
2.2. Solubility Modelling
2.3. Data Analysis and Fitting
3. Results
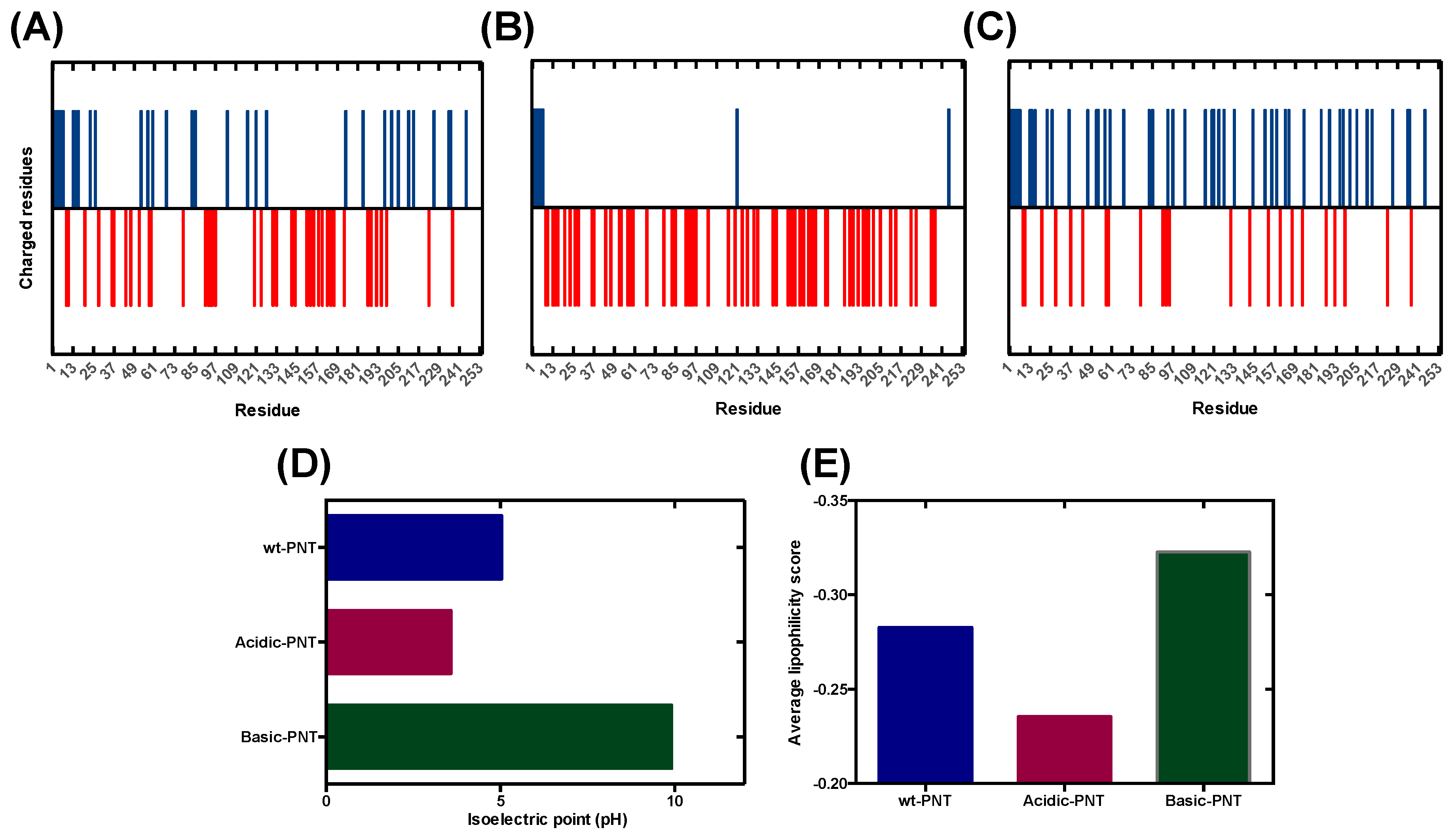
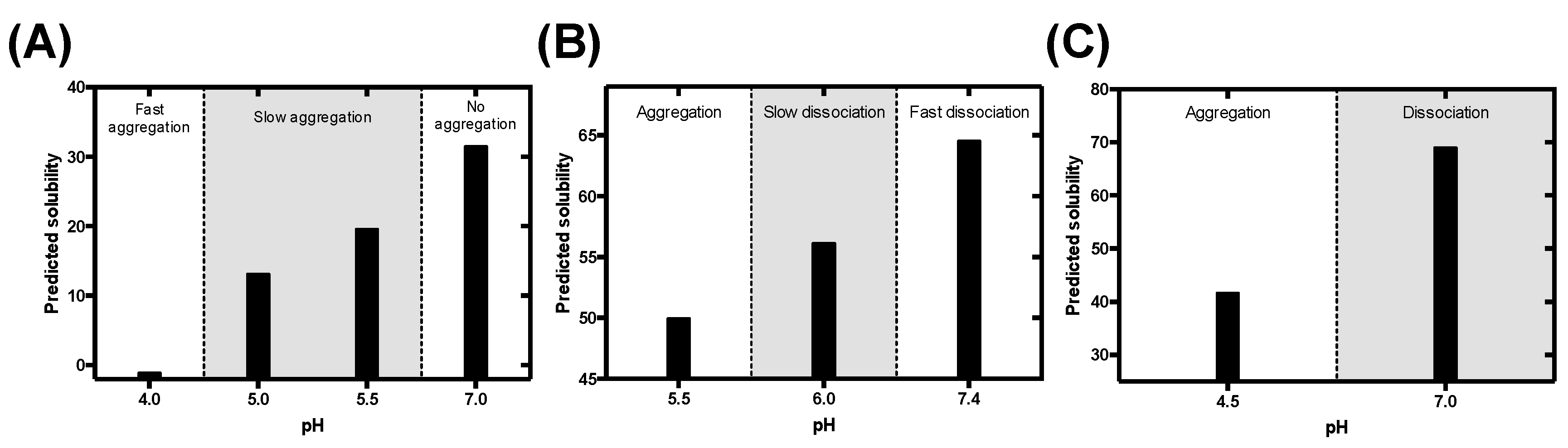
3.1. Rational Analysis of the Molecular Determinants behind pH-Associated Aggregation
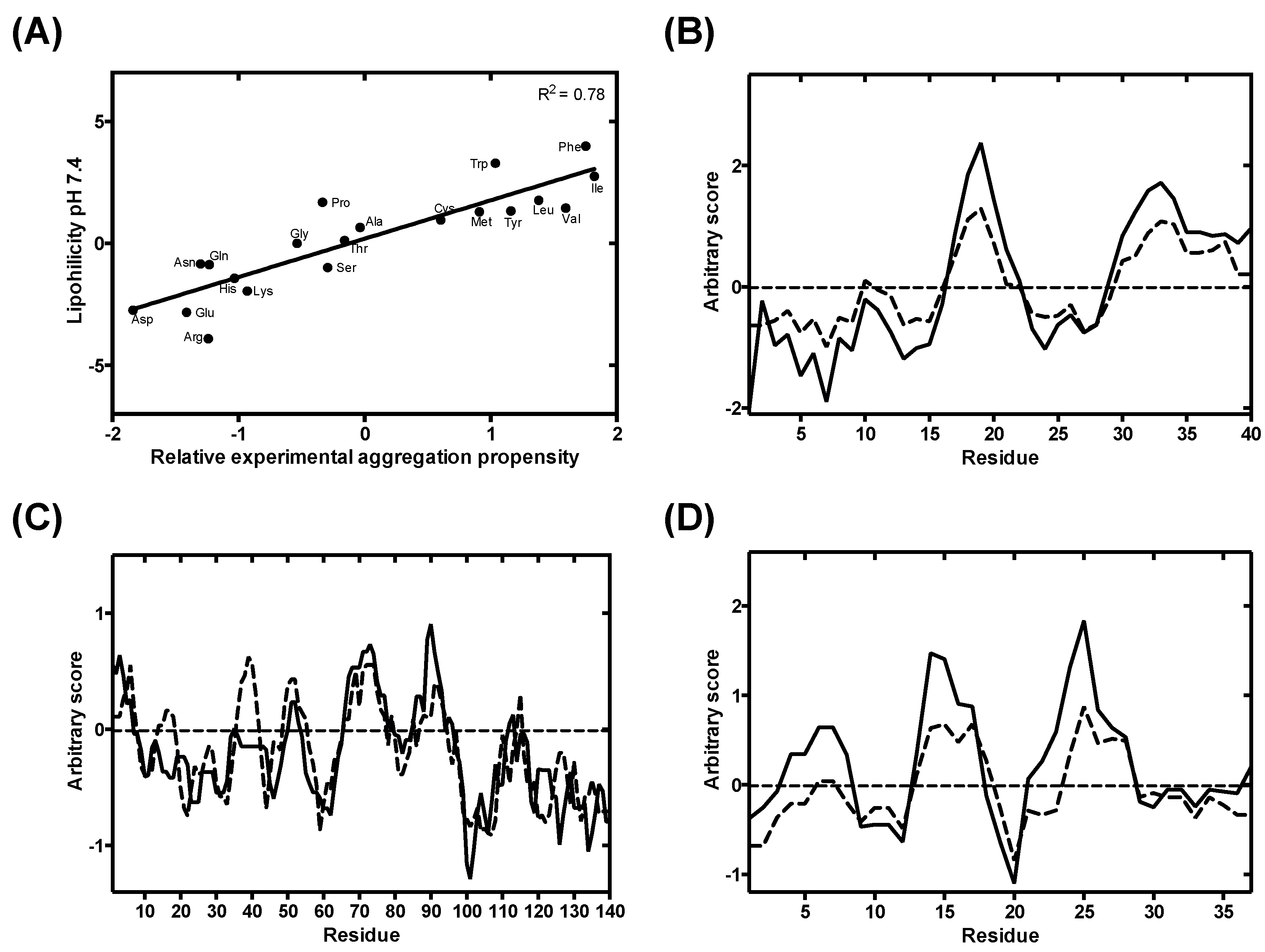
3.2. Analysis and Validation of the Lipophilicity Scale as a Proxy for Aggregation Prediction
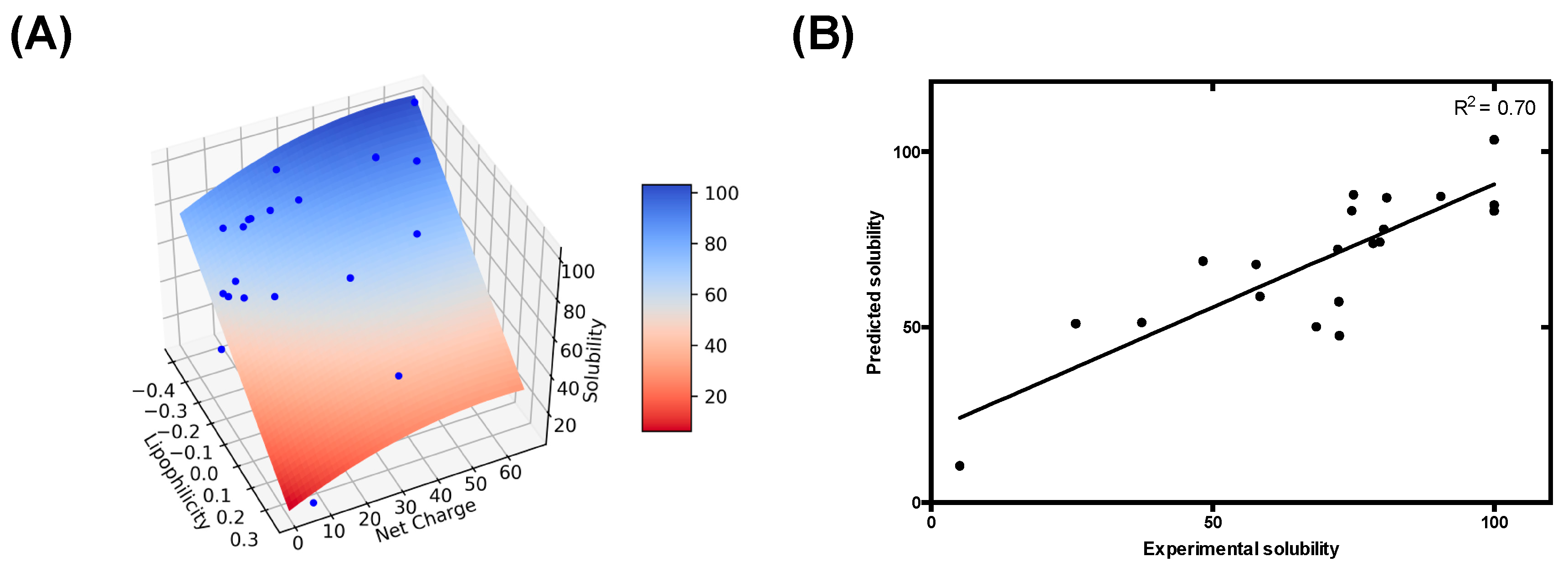
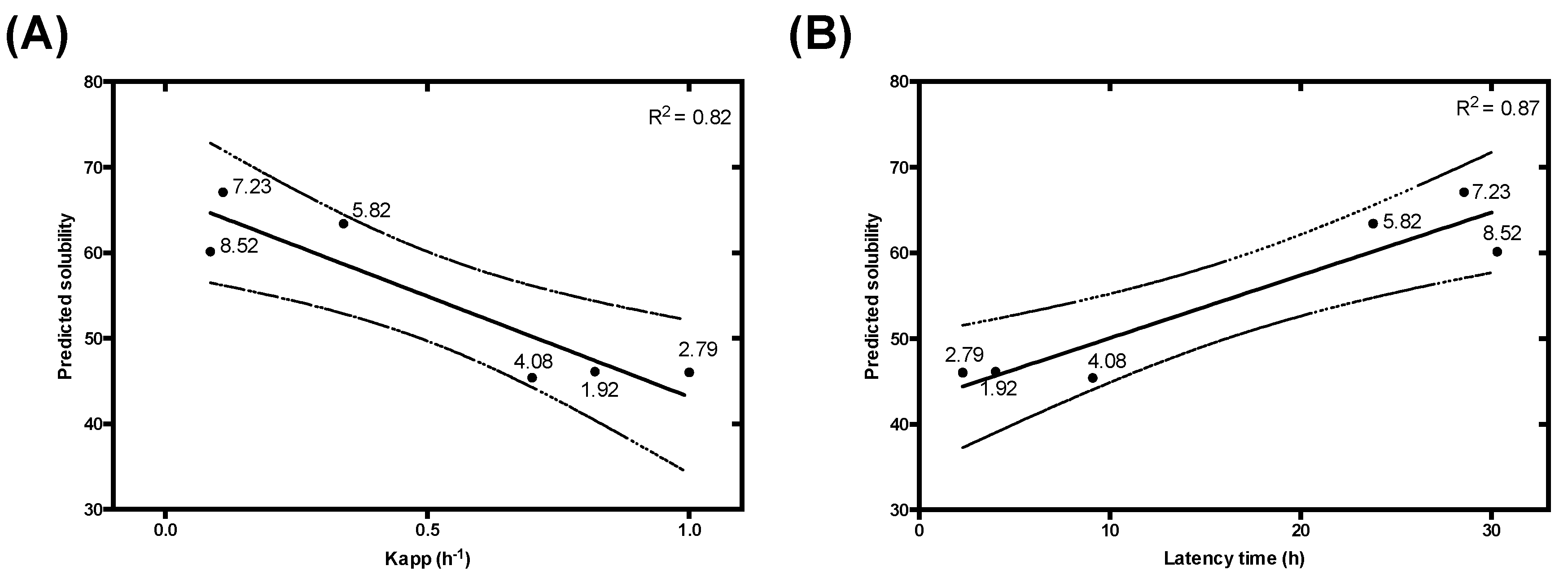
3.3. Modelling pH-Dependent Solubility usIng Lipophilicity and Net Charge
3.4. pH-Dependent Aggregation Prediction in Disease-Linked Proteins
3.4.1. α-Synuclein (α-S)
3.4.2. Islet Amyloid Polypeptide (IAPP)
3.4.3. Alzheimer’s Disease Related Proteins: Amyloid-Beta Peptides and Tau Protein
3.4.4. Use of a Lipophilicity Term Improves Accuracy in the Prediction of the pH-Dependent Aggregation of Disease-Linked Proteins
3.5. Predicting the Impact of pH on the Aggregation of Functional Amyloids: Context-Dependent Aggregation to Confine Functional Self-Assembly
3.5.1. Pigment Cell-Specific Melanosome Protein
3.5.2. Corticotropin-Releasing Hormone
3.5.3. B Domain of the Bap Protein
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chiti, F.; Dobson, C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.J.; Meyer, J.D.; Carpenter, J.F.; Manning, M.C. Stability of human serum albumin during bioprocessing: Denaturation and aggregation during processing of albumin paste. Pharm. Res. 2000, 17, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Cromwell, M.E.; Hilario, E.; Jacobson, F. Protein aggregation and bioprocessing. AAPS J. 2006, 8, 572–579. [Google Scholar] [CrossRef]
- Loquet, A.; Saupe, S.J.; Romero, D. Functional Amyloids in Health and Disease. J. Mol. Biol. 2018, 430, 3629–3630. [Google Scholar] [CrossRef]
- Camara-Almiron, J.; Caro-Astorga, J.; de Vicente, A.; Romero, D. Beyond the expected: The structural and functional diversity of bacterial amyloids. Crit. Rev. Microbiol. 2018, 44, 653–666. [Google Scholar] [CrossRef]
- McGlinchey, R.P.; Lee, J.C. Why Study Functional Amyloids? Lessons from the Repeat Domain of Pmel17. J. Mol. Biol. 2018, 430, 3696–3706. [Google Scholar] [CrossRef]
- Diaz-Caballero, M.; Navarro, S.; Fuentes, I.; Teixidor, F.; Ventura, S. Minimalist Prion-Inspired Polar Self-Assembling Peptides. ACS Nano 2018, 12, 5394–5407. [Google Scholar] [CrossRef]
- Diaz-Caballero, M.; Fernandez, M.R.; Navarro, S.; Ventura, S. Prion-based nanomaterials and their emerging applications. Prion 2018, 12, 266–272. [Google Scholar] [CrossRef]
- Knowles, T.P.; Mezzenga, R. Amyloid Fibrils as Building Blocks for Natural and Artificial Functional Materials. Adv. Mater. 2016, 28, 6546–6561. [Google Scholar] [CrossRef]
- Wei, G.; Su, Z.; Reynolds, N.P.; Arosio, P.; Hamley, I.W.; Gazit, E.; Mezzenga, R. Self-assembling peptide and protein amyloids: From structure to tailored function in nanotechnology. Chem. Soc. Rev. 2017, 46, 4661–4708. [Google Scholar] [CrossRef] [PubMed]
- Pallares, I.; Ventura, S. Advances in the prediction of protein aggregation propensity. Curr. Med. Chem. 2017, 26, 3911–3920. [Google Scholar] [CrossRef] [PubMed]
- Graña-Montes, R.; Pujol, J.P.; Gómez-Picanyol, C.; Ventura, S. Prediction of Protein Aggregation and Amyloid Formation. In From Protein Structure to Function with Bioinformatics; Rigden, D.J., Ed.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 205–263. [Google Scholar] [CrossRef]
- Conchillo-Sole, O.; de Groot, N.S.; Aviles, F.X.; Vendrell, J.; Daura, X.; Ventura, S. AGGRESCAN: A server for the prediction and evaluation of “hot spots” of aggregation in polypeptides. BMC Bioinform. 2007, 8, 65. [Google Scholar] [CrossRef] [PubMed]
- Sanchez de Groot, N.; Pallares, I.; Aviles, F.X.; Vendrell, J.; Ventura, S. Prediction of “hot spots” of aggregation in disease-linked polypeptides. BMC Struct. Biol. 2005, 5, 18. [Google Scholar] [CrossRef]
- Garbuzynskiy, S.O.; Lobanov, M.Y.; Galzitskaya, O.V. FoldAmyloid: A method of prediction of amyloidogenic regions from protein sequence. Bioinformatics 2010, 26, 326–332. [Google Scholar] [CrossRef]
- Tsolis, A.C.; Papandreou, N.C.; Iconomidou, V.A.; Hamodrakas, S.J. A consensus method for the prediction of ‘aggregation-prone’ peptides in globular proteins. PLoS ONE 2013, 8, e54175. [Google Scholar] [CrossRef]
- O’Donnell, C.W.; Waldispuhl, J.; Lis, M.; Halfmann, R.; Devadas, S.; Lindquist, S.; Berger, B. A method for probing the mutational landscape of amyloid structure. Bioinformatics 2011, 27, 34–42. [Google Scholar] [CrossRef]
- Walsh, I.; Seno, F.; Tosatto, S.C.; Trovato, A. PASTA 2.0: An improved server for protein aggregation prediction. Nucleic Acids Res. 2014, 42, 301–307. [Google Scholar] [CrossRef]
- Fernandez-Escamilla, A.M.; Rousseau, F.; Schymkowitz, J.; Serrano, L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 2004, 22, 1302–1306. [Google Scholar] [CrossRef]
- Rousseau, F.; Schymkowitz, J.; Serrano, L. Protein aggregation and amyloidosis: Confusion of the kinds? Curr. Opin. Struct. Biol. 2006, 16, 118–126. [Google Scholar] [CrossRef]
- Maurer-Stroh, S.; Debulpaep, M.; Kuemmerer, N.; Lopez de la Paz, M.; Martins, I.C.; Reumers, J.; Morris, K.L.; Copland, A.; Serpell, L.; Serrano, L.; et al. Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat. Methods 2010, 7, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Tartaglia, G.G.; Pawar, A.P.; Campioni, S.; Dobson, C.M.; Chiti, F.; Vendruscolo, M. Prediction of aggregation-prone regions in structured proteins. J. Mol. Biol. 2008, 380, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Singh, S.; Zeng, D.L.; King, K.; Nema, S. Antibody structure, instability, and formulation. J. Pharm. Sci. 2007, 96, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Roberts, C.J. Therapeutic protein aggregation: Mechanisms, design, and control. Trends Biotechnol. 2014, 32, 372–380. [Google Scholar] [CrossRef]
- Jha, R.K.; Leaver-Fay, A.; Yin, S.; Wu, Y.; Butterfoss, G.L.; Szyperski, T.; Dokholyan, N.V.; Kuhlman, B. Computational design of a PAK1 binding protein. J. Mol. Biol. 2010, 400, 257–270. [Google Scholar] [CrossRef]
- Ventura, S.; Zurdo, J.; Narayanan, S.; Parreno, M.; Mangues, R.; Reif, B.; Chiti, F.; Giannoni, E.; Dobson, C.M.; Aviles, F.X.; et al. Short amino acid stretches can mediate amyloid formation in globular proteins: The Src homology 3 (SH3) case. Proc. Natl. Acad. Sci. USA 2004, 101, 7258–7263. [Google Scholar] [CrossRef]
- Riek, R.; Eisenberg, D.S. The activities of amyloids from a structural perspective. Nature 2016, 539, 227–235. [Google Scholar] [CrossRef]
- Jahn, T.R.; Radford, S.E. Folding versus aggregation: Polypeptide conformations on competing pathways. Arch. Biochem. Biophys. 2008, 469, 100–117. [Google Scholar] [CrossRef]
- Castillo, V.; Grana-Montes, R.; Sabate, R.; Ventura, S. Prediction of the aggregation propensity of proteins from the primary sequence: Aggregation properties of proteomes. Biotechnol. J. 2011, 6, 674–685. [Google Scholar] [CrossRef]
- Simm, S.; Einloft, J.; Mirus, O.; Schleiff, E. 50 years of amino acid hydrophobicity scales: Revisiting the capacity for peptide classification. Biol. Res. 2016, 49, 31. [Google Scholar] [CrossRef]
- MacCallum, J.L.; Tieleman, D.P. Hydrophobicity scales: A thermodynamic looking glass into lipid-protein interactions. Trends Biochem. Sci. 2011, 36, 653–662. [Google Scholar] [CrossRef] [PubMed]
- Tedeschi, G.; Mangiagalli, M.; Chmielewska, S.; Lotti, M.; Natalello, A.; Brocca, S. Aggregation properties of a disordered protein are tunable by pH and depend on its net charge per residue. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 2543–2550. [Google Scholar] [CrossRef] [PubMed]
- Shaw, K.L.; Grimsley, G.R.; Yakovlev, G.I.; Makarov, A.A.; Pace, C.N. The effect of net charge on the solubility, activity, and stability of ribonuclease Sa. Protein Sci. 2001, 10, 1206–1215. [Google Scholar] [CrossRef] [PubMed]
- Zamora, W.J.; Campanera, J.M.; Luque, F.J. Development of a Structure-Based, pH-Dependent Lipophilicity Scale of Amino Acids from Continuum Solvation Calculations. J. Phys. Chem. Lett. 2019, 10, 883–889. [Google Scholar] [CrossRef] [PubMed]
- Putnam, C. Protein Calculator. Available online: http://protcalc.sourceforge.net/ (accessed on 19 June 2019).
- Morris, A.M.; Watzky, M.A.; Finke, R.G. Protein aggregation kinetics, mechanism, and curve-fitting: A review of the literature. Biochim. Biophys. Acta 2009, 1794, 375–397. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Li, J.; Fink, A.L. Evidence for a partially folded intermediate in alpha-synuclein fibril formation. J. Biol. Chem. 2001, 276, 10737–10744. [Google Scholar] [CrossRef] [PubMed]
- Jha, S.; Snell, J.M.; Sheftic, S.R.; Patil, S.M.; Daniels, S.B.; Kolling, F.W.; Alexandrescu, A.T. pH dependence of amylin fibrillization. Biochemistry 2014, 53, 300–310. [Google Scholar] [CrossRef]
- Jeganathan, S.; von Bergen, M.; Mandelkow, E.M.; Mandelkow, E. The natively unfolded character of tau and its aggregation to Alzheimer-like paired helical filaments. Biochemistry 2008, 47, 10526–10539. [Google Scholar] [CrossRef]
- Hortschansky, P.; Schroeckh, V.; Christopeit, T.; Zandomeneghi, G.; Fandrich, M. The aggregation kinetics of Alzheimer’s beta-amyloid peptide is controlled by stochastic nucleation. Protein Sci. 2005, 14, 1753–1759. [Google Scholar] [CrossRef]
- Pfefferkorn, C.M.; McGlinchey, R.P.; Lee, J.C. Effects of pH on aggregation kinetics of the repeat domain of a functional amyloid, Pmel17. Proc. Natl. Acad. Sci. USA 2010, 107, 21447–21452. [Google Scholar] [CrossRef]
- Maji, S.K.; Perrin, M.H.; Sawaya, M.R.; Jessberger, S.; Vadodaria, K.; Rissman, R.A.; Singru, P.S.; Nilsson, K.P.; Simon, R.; Schubert, D.; et al. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 2009, 325, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Taglialegna, A.; Navarro, S.; Ventura, S.; Garnett, J.A.; Matthews, S.; Penades, J.R.; Lasa, I.; Valle, J. Staphylococcal Bap Proteins Build Amyloid Scaffold Biofilm Matrices in Response to Environmental Signals. PLoS Pathog. 2016, 12, e1005711. [Google Scholar] [CrossRef] [PubMed]
- Soper, D.S. p-Value Calculator for Correlation Coefficients. Available online: http://www.danielsoper.com/statcalc (accessed on 20 June 2018).
- De Groot, N.S.; Aviles, F.X.; Vendrell, J.; Ventura, S. Mutagenesis of the central hydrophobic cluster in Abeta42 Alzheimer’s peptide. Side-chain properties correlate with aggregation propensities. FEBS J. 2006, 273, 658–668. [Google Scholar] [CrossRef] [PubMed]
- Fink, A.L. Protein aggregation: Folding aggregates, inclusion bodies and amyloid. Fold. Des. 1998, 3, 9–23. [Google Scholar] [CrossRef]
- Belli, M.; Ramazzotti, M.; Chiti, F. Prediction of amyloid aggregation in vivo. EMBO Rep. 2011, 12, 657–663. [Google Scholar] [CrossRef]
- Emamzadeh, F.N. Alpha-synuclein structure, functions, and interactions. J. Res. Med. Sci. 2016, 21, 29. [Google Scholar] [CrossRef]
- Goedert, M.; Spillantini, M.G.; Del Tredici, K.; Braak, H. 100 years of Lewy pathology. Nat. Rev. Neurol. 2013, 9, 13–24. [Google Scholar] [CrossRef]
- Spillantini, M.G.; Schmidt, M.L.; Lee, V.M.; Trojanowski, J.Q.; Jakes, R.; Goedert, M. Alpha-synuclein in Lewy bodies. Nature 1997, 388, 839–840. [Google Scholar] [CrossRef]
- Villar-Pique, A.; Lopes da Fonseca, T.; Outeiro, T.F. Structure, function and toxicity of alpha-synuclein: The Bermuda triangle in synucleinopathies. J. Neurochem. 2016, 139, 240–255. [Google Scholar] [CrossRef]
- Lashuel, H.A.; Overk, C.R.; Oueslati, A.; Masliah, E. The many faces of alpha-synuclein: From structure and toxicity to therapeutic target. Nat. Rev. Neurosci. 2013, 14, 38–48. [Google Scholar] [CrossRef]
- Lassen, L.B.; Reimer, L.; Ferreira, N.; Betzer, C.; Jensen, P.H. Protein Partners of alpha-Synuclein in Health and Disease. Brain Pathol. 2016, 26, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Westermark, G.T.; Westermark, P.; Berne, C.; Korsgren, O.; Nordic Network for Clinical Islet Transplantation. Widespread amyloid deposition in transplanted human pancreatic islets. N Engl. J. Med. 2008, 359, 977–979. [Google Scholar] [CrossRef] [PubMed]
- Denroche, H.C.; Verchere, C.B. IAPP and type 1 diabetes: Implications for immunity, metabolism and islet transplants. J. Mol. Endocrinol. 2018, 60, 57–75. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Morales-Scheihing, D.; Salvadores, N.; Moreno-Gonzalez, I.; Gonzalez, C.; Taylor-Presse, K.; Mendez, N.; Shahnawaz, M.; Gaber, A.O.; Sabek, O.M.; et al. Induction of IAPP amyloid deposition and associated diabetic abnormalities by a prion-like mechanism. J. Exp. Med. 2017, 214, 2591–2610. [Google Scholar] [CrossRef]
- Khemtemourian, L.; Domenech, E.; Doux, J.P.; Koorengevel, M.C.; Killian, J.A. Low pH acts as inhibitor of membrane damage induced by human islet amyloid polypeptide. J. Am. Chem. Soc. 2011, 133, 15598–15604. [Google Scholar] [CrossRef]
- Akter, R.; Cao, P.; Noor, H.; Ridgway, Z.; Tu, L.H.; Wang, H.; Wong, A.G.; Zhang, X.; Abedini, A.; Schmidt, A.M.; et al. Islet Amyloid Polypeptide: Structure, Function, and Pathophysiology. J. Diabetes Res. 2016, 2016, 2798269. [Google Scholar] [CrossRef]
- Lane, C.A.; Hardy, J.; Schott, J.M. Alzheimer’s disease. Eur. J. Neurol. 2018, 25, 59–70. [Google Scholar] [CrossRef]
- Meng, F.; Bellaiche, M.M.J.; Kim, J.Y.; Zerze, G.H.; Best, R.B.; Chung, H.S. Highly Disordered Amyloid-beta Monomer Probed by Single-Molecule FRET and MD Simulation. Biophys. J. 2018, 114, 870–884. [Google Scholar] [CrossRef]
- Eliezer, D.; Barre, P.; Kobaslija, M.; Chan, D.; Li, X.; Heend, L. Residual structure in the repeat domain of tau: Echoes of microtubule binding and paired helical filament formation. Biochemistry 2005, 44, 1026–1036. [Google Scholar] [CrossRef]
- Schweers, O.; Schonbrunn-Hanebeck, E.; Marx, A.; Mandelkow, E. Structural studies of tau protein and Alzheimer paired helical filaments show no evidence for beta-structure. J. Biol. Chem. 1994, 269, 24290–24297. [Google Scholar]
- Siddiqua, A.; Margittai, M. Three-and four-repeat Tau coassemble into heterogeneous filaments: An implication for Alzheimer disease. J. Biol. Chem. 2010, 285, 37920–37926. [Google Scholar] [CrossRef] [PubMed]
- Dinkel, P.D.; Siddiqua, A.; Huynh, H.; Shah, M.; Margittai, M. Variations in filament conformation dictate seeding barrier between three-and four-repeat tau. Biochemistry 2011, 50, 4330–4336. [Google Scholar] [CrossRef] [PubMed]
- Pham, C.L.; Kwan, A.H.; Sunde, M. Functional amyloid: Widespread in Nature, diverse in purpose. Essays Biochem. 2014, 56, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Otzen, D. Functional amyloid: Turning swords into plowshares. Prion 2010, 4, 256–264. [Google Scholar] [CrossRef]
- Jackson, M.P.; Hewitt, E.W. Why are Functional Amyloids Non-Toxic in Humans? Biomolecules 2017, 7, 71. [Google Scholar] [CrossRef]
- Franzmann, T.M.; Jahnel, M.; Pozniakovsky, A.; Mahamid, J.; Holehouse, A.S.; Nuske, E.; Richter, D.; Baumeister, W.; Grill, S.W.; Pappu, R.V.; et al. Phase separation of a yeast prion protein promotes cellular fitness. Science 2018, 359, 5654. [Google Scholar] [CrossRef]
- Zambrano, R.; Jamroz, M.; Szczasiuk, A.; Pujols, J.; Kmiecik, S.; Ventura, S. AGGRESCAN3D (A3D): Server for prediction of aggregation properties of protein structures. Nucleic Acids Res. 2015, 43, 306–313. [Google Scholar] [CrossRef]
- Kuriata, A.; Iglesias, V.; Pujols, J.; Kurcinski, M.; Kmiecik, S.; Ventura, S. Aggrescan3D (A3D) 2.0: Prediction and engineering of protein solubility. Nucleic Acids Res. 2019, 47, 300–307. [Google Scholar] [CrossRef]
- Kuriata, A.; Iglesias, V.; Kurcinski, M.; Ventura, S.; Kmiecik, S. Aggrescan3D standalone package for structure-based prediction of protein aggregation properties. Bioinformatics 2019, 35, 3834–3835. [Google Scholar] [CrossRef]
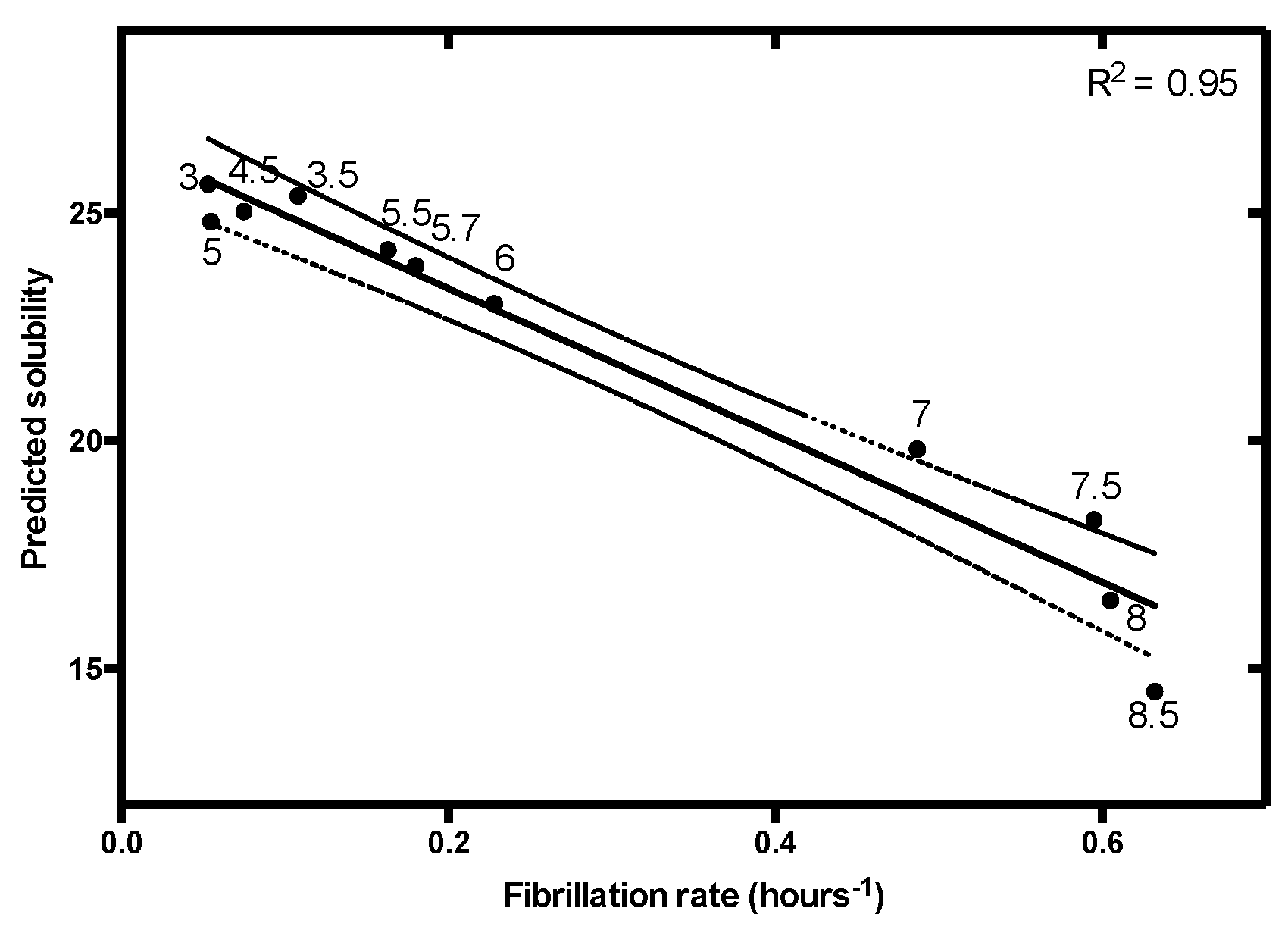
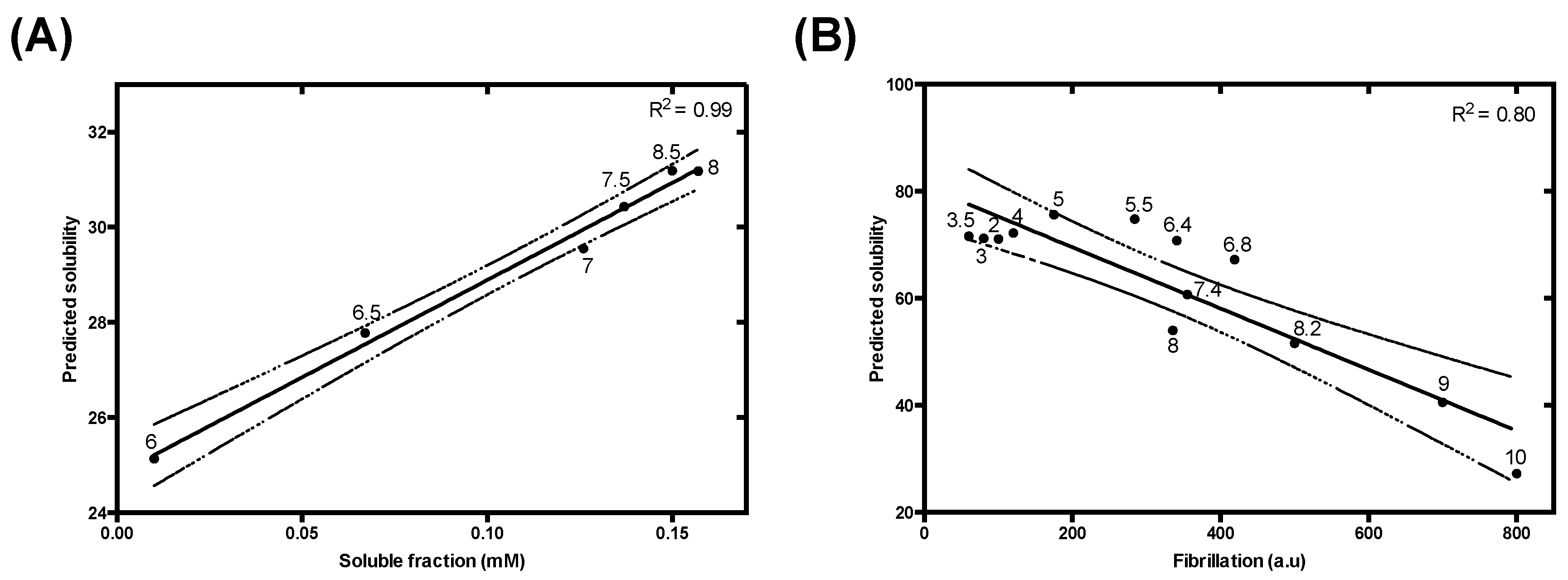
| Parameter | α | β | γ | δ |
|---|---|---|---|---|
| Values | −97.82 | −0.00747 | 0.8770 | 38.24 |
| Protein | PNTs | α-S (4.67) * | IAPP | Aβ40 | Tau K19 | ||
|---|---|---|---|---|---|---|---|
| Kapp | Tlag | (8.90) * | (5.31) * | (9.68) * | |||
| Charge | R2 | 0.20 | 0.47 | 0.50 | 0.86 | 0.93 | 0.80 |
| p-value | 0.048 | 0.13 | 0.12 | 0.000041 | 0.0019 | 0.000037 | |
| Charge and Lipophilicity | R2 | 0.70 | 0.82 | 0.87 | 0.95 | 0.99 | 0.80 |
| p-value | <0.00001 | 0.013 | 0.0066 | <0.00001 | 0.000039 | 0.000037 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, J.; Iglesias, V.; Santos-Suárez, J.; Mangiagalli, M.; Brocca, S.; Pallarès, I.; Ventura, S. pH-Dependent Aggregation in Intrinsically Disordered Proteins Is Determined by Charge and Lipophilicity. Cells 2020, 9, 145. https://doi.org/10.3390/cells9010145
Santos J, Iglesias V, Santos-Suárez J, Mangiagalli M, Brocca S, Pallarès I, Ventura S. pH-Dependent Aggregation in Intrinsically Disordered Proteins Is Determined by Charge and Lipophilicity. Cells. 2020; 9(1):145. https://doi.org/10.3390/cells9010145
Chicago/Turabian StyleSantos, Jaime, Valentín Iglesias, Juan Santos-Suárez, Marco Mangiagalli, Stefania Brocca, Irantzu Pallarès, and Salvador Ventura. 2020. "pH-Dependent Aggregation in Intrinsically Disordered Proteins Is Determined by Charge and Lipophilicity" Cells 9, no. 1: 145. https://doi.org/10.3390/cells9010145
APA StyleSantos, J., Iglesias, V., Santos-Suárez, J., Mangiagalli, M., Brocca, S., Pallarès, I., & Ventura, S. (2020). pH-Dependent Aggregation in Intrinsically Disordered Proteins Is Determined by Charge and Lipophilicity. Cells, 9(1), 145. https://doi.org/10.3390/cells9010145







