MicroRNAs: Biological Regulators in Pathogen–Host Interactions
Abstract
1. Introduction
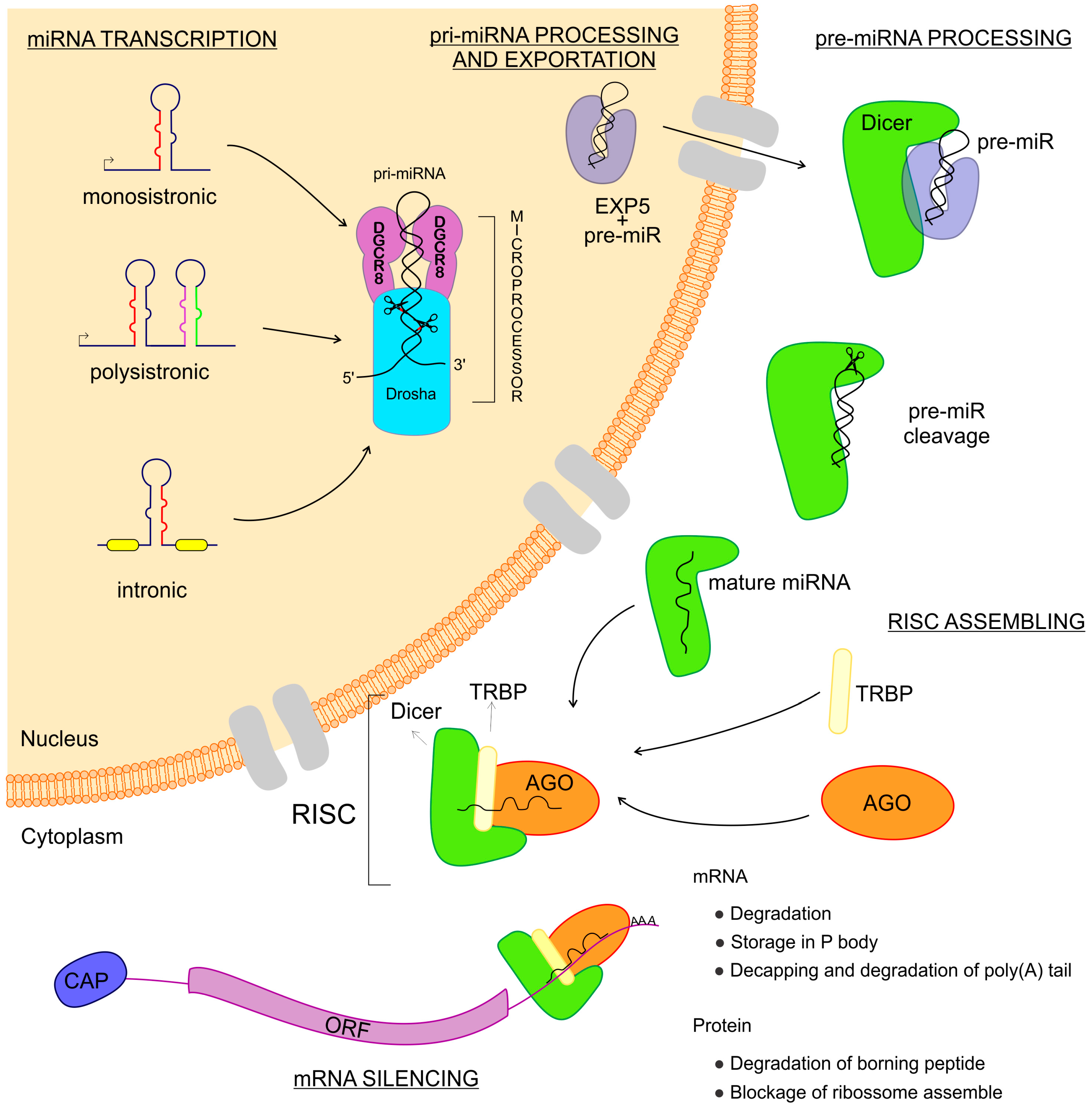
2. miRNAs—Biogenesis and Gene Expression Regulation
3. miRNA Expression in Infectious Diseases
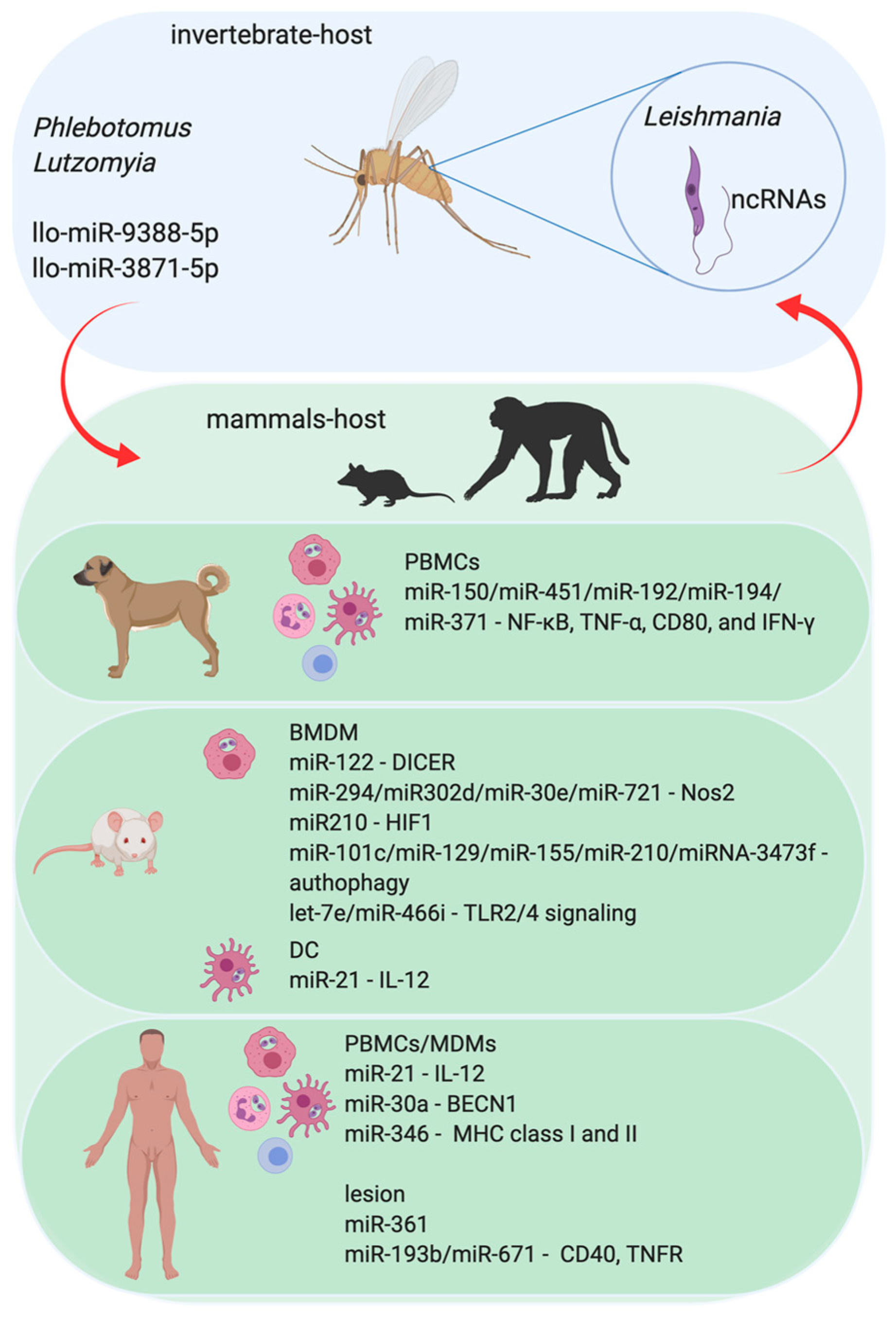
3.1. Leishmania–Host Interactions
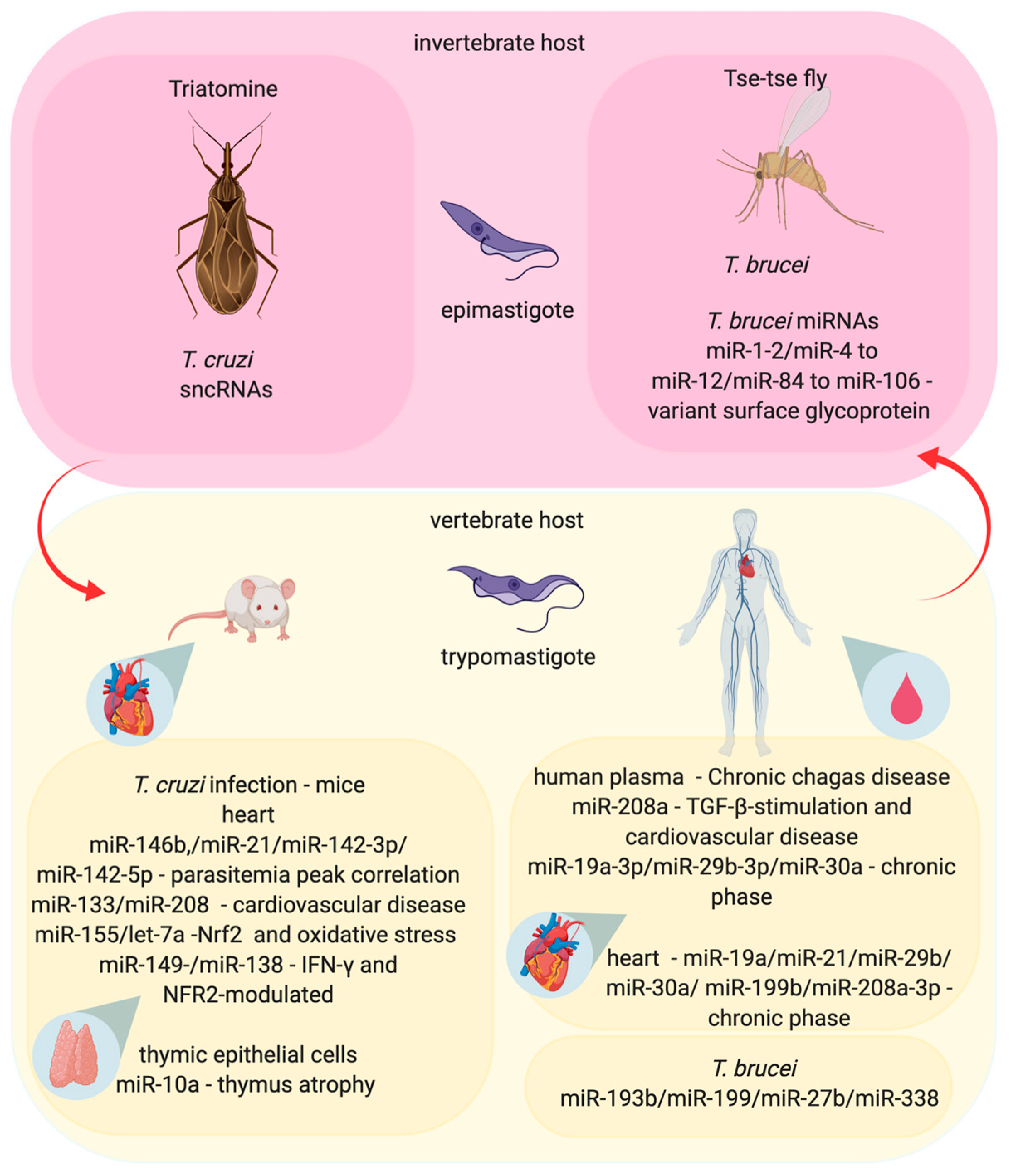
3.2. Trypanosoma–Host Interactions
3.3. Toxoplasma–Host Interaction
3.4. Plasmodium–Host Interactions
4. Novel Perspectives: MicroRNAs as Diagnostic Markers and Therapeutic Targets
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fernandes, J.C.R.; Acuna, S.M.; Aoki, J.I.; Floeter-Winter, L.M.; Muxel, S.M. Long non-coding RNAs in the regulation of gene expression: Physiology and disease. Non-Coding RNA 2019, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Wang, X.; Youmans, D.T.; Cech, T.R. How do lncRNAs regulate transcription? Sci. Adv. 2017, 3, eaao2110. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The gencode v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Ulitsky, I.; Bartel, D.P. LincRNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Bagga, S.; Bracht, J.; Hunter, S.; Massirer, K.; Holtz, J.; Eachus, R.; Pasquinelli, A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 2005, 122, 553–563. [Google Scholar] [CrossRef]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef]
- Ben-Hamo, R.; Efroni, S. MicroRNA regulation of molecular pathways as a generic mechanism and as a core disease phenotype. Oncotarget 2015, 6, 1594–1604. [Google Scholar] [CrossRef]
- Nussbacher, J.K.; Yeo, G.W. Systematic discovery of RNA binding proteins that regulate microRNA levels. Mol. Cell 2018, 69, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.; Duan, F.F.; Wang, Y. MicroRNAs and RNA binding protein regulators of microRNAs in the control of pluripotency and reprogramming. Curr. Opin. Genet. Dev. 2017, 46, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Nangia-Makker, P.; Farhana, L.; Majumdar, A.P.N. A novel mechanism of lncRNA and miRNA interaction: Ccat2 regulates mir-145 expression by suppressing its maturation process in colon cancer cells. Mol. Cancer 2017, 16, 155. [Google Scholar] [CrossRef]
- Baltimore, D.; Boldin, M.P.; O’Connell, R.M.; Rao, D.S.; Taganov, K.D. MicroRNAs: New regulators of immune cell development and function. Nat. Immunol. 2008, 9, 839–845. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, L.A.; Sheedy, F.J.; McCoy, C.E. MicroRNAs: The fine-tuners of toll-like receptor signalling. Nat. Rev. Immunol 2011, 11, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Labbaye, C.; Testa, U. The emerging role of mir-146a in the control of hematopoiesis, immune function and cancer. J. Hematol. Oncol. 2012, 5, 13. [Google Scholar] [CrossRef]
- Manzano-Roman, R.; Siles-Lucas, M. MicroRNAs in parasitic diseases: Potential for diagnosis and targeting. Mol. Biochem. Parasitol. 2012, 186, 81–86. [Google Scholar] [CrossRef]
- Petrini, E.; Caviglia, G.P.; Abate, M.L.; Fagoonee, S.; Smedile, A.; Pellicano, R. MicroRNAs in hbv-related hepatocellular carcinoma: Functions and potential clinical applications. Panminerva Med. 2015, 57, 201–209. [Google Scholar]
- Reynoso, R.; Laufer, N.; Hackl, M.; Skalicky, S.; Monteforte, R.; Turk, G.; Carobene, M.; Quarleri, J.; Cahn, P.; Werner, R.; et al. MicroRNAs differentially present in the plasma of hiv elite controllers reduce hiv infection in vitro. Sci. Rep. 2014, 4, 5915. [Google Scholar] [CrossRef]
- Fu, Y.; Yi, Z.; Wu, X.; Li, J.; Xu, F. Circulating microRNAs in patients with active pulmonary tuberculosis. J. Clin. Microbiol. 2011, 49, 4246–4251. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Lawrie, C.H.; Gal, S.; Dunlop, H.M.; Pushkaran, B.; Liggins, A.P.; Pulford, K.; Banham, A.H.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S.; et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large b-cell lymphoma. Br. J. Haematol. 2008, 141, 672–675. [Google Scholar] [CrossRef] [PubMed]
- Ozer, J.; Ratner, M.; Shaw, M.; Bailey, W.; Schomaker, S. The current state of serum biomarkers of hepatotoxicity. Toxicology 2008, 245, 194–205. [Google Scholar] [CrossRef] [PubMed]
- Fichtlscherer, S.; De Rosa, S.; Fox, H.; Schwietz, T.; Fischer, A.; Liebetrau, C.; Weber, M.; Hamm, C.W.; Roxe, T.; Muller-Ardogan, M.; et al. Circulating microRNAs in patients with coronary artery disease. Circ. Res. 2010, 107, 677–684. [Google Scholar] [CrossRef]
- Abd-El-Fattah, A.A.; Sadik, N.A.; Shaker, O.G.; Aboulftouh, M.L. Differential microRNAs expression in serum of patients with lung cancer, pulmonary tuberculosis, and pneumonia. Cell Biochem. Biophys. 2013, 67, 875–884. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef]
- Lai, F.; Gardini, A.; Zhang, A.; Shiekhattar, R. Integrator mediates the biogenesis of enhancer RNAs. Nature 2015, 525, 399–403. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Yalcin, A.; Meyer, J.; Lendeckel, W.; Tuschl, T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002, 12, 735–739. [Google Scholar] [CrossRef]
- Lai, E.C.; Tomancak, P.; Williams, R.W.; Rubin, G.M. Computational identification of drosophila microRNA genes. Genome Biol. 2003, 4, R42. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S.; et al. The nuclear RNAse iii drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Han, J.; Pedersen, J.S.; Kwon, S.C.; Belair, C.D.; Kim, Y.K.; Yeom, K.H.; Yang, W.Y.; Haussler, D.; Blelloch, R.; Kim, V.N. Posttranscriptional crossregulation between drosha and dgcr8. Cell 2009, 136, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Yamazawa, R.; Jiko, C.; Choi, S.; Park, I.Y.; Nakagawa, A.; Yamashita, E.; Lee, S.J. Structural basis for selective binding of export cargoes by exportin-5. Structure 2018, 26, 1393–1398 e1392. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef] [PubMed]
- Hutvagner, G.; McLachlan, J.; Pasquinelli, A.E.; Balint, E.; Tuschl, T.; Zamore, P.D. A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838. [Google Scholar] [CrossRef]
- Schwarz, D.S.; Hutvagner, G.; Du, T.; Xu, Z.; Aronin, N.; Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115, 199–208. [Google Scholar] [CrossRef]
- Wang, B.; Li, S.; Qi, H.H.; Chowdhury, D.; Shi, Y.; Novina, C.D. Distinct passenger strand and mRNA cleavage activities of human argonaute proteins. Nat. Struct. Mol. Biol. 2009, 16, 1259–1266. [Google Scholar] [CrossRef]
- Vaucheret, H.; Vazquez, F.; Crete, P.; Bartel, D.P. The action of argonaute1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004, 18, 1187–1197. [Google Scholar] [CrossRef]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef]
- Pillai, R.S.; Bhattacharyya, S.N.; Artus, C.G.; Zoller, T.; Cougot, N.; Basyuk, E.; Bertrand, E.; Filipowicz, W. Inhibition of translational initiation by let-7 microRNA in human cells. Science 2005, 309, 1573–1576. [Google Scholar] [CrossRef]
- Humphreys, D.T.; Westman, B.J.; Martin, D.I.; Preiss, T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4e/cap and poly(a) tail function. Proc. Natl. Acad. Sci. USA 2005, 102, 16961–16966. [Google Scholar] [CrossRef]
- Richter, J.D.; Sonenberg, N. Regulation of cap-dependent translation by eif4e inhibitory proteins. Nature 2005, 433, 477–480. [Google Scholar] [CrossRef] [PubMed]
- Chendrimada, T.P.; Finn, K.J.; Ji, X.; Baillat, D.; Gregory, R.I.; Liebhaber, S.A.; Pasquinelli, A.E.; Shiekhattar, R. MicroRNA silencing through risc recruitment of eif6. Nature 2007, 447, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Maroney, P.A.; Yu, Y.; Fisher, J.; Nilsen, T.W. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat. Struct. Mol. Biol. 2006, 13, 1102–1107. [Google Scholar] [CrossRef] [PubMed]
- Parker, R.; Song, H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004, 11, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Eulalio, A.; Behm-Ansmant, I.; Izaurralde, E. P bodies: At the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007, 8, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Makarova, J.A.; Shkurnikov, M.U.; Wicklein, D.; Lange, T.; Samatov, T.R.; Turchinovich, A.A.; Tonevitsky, A.G. Intracellular and extracellular microRNA: An update on localization and biological role. Prog. Histochem. Cytochem. 2016, 51, 33–49. [Google Scholar] [CrossRef]
- Burza, S.; Croft, S.L.; Boelaert, M. Leishmaniasis. Lancet 2018, 392, 951–970. [Google Scholar] [CrossRef]
- Schewach-Millet, M.; Kahana, M.; Ronnen, M.; Yuzuk, S. Mucosal involvement of cutaneous leishmaniasis. Int. J. Dermatol. 1986, 25, 113–114. [Google Scholar] [CrossRef]
- WHO. Available online: http://www.who.int/en/news-room/fact-sheets/detail/leishmaniasis (accessed on 14 March 2019).
- Sacks, D.; Kamhawi, S. Molecular aspects of parasite-vector and vector-host interactions in leishmaniasis. Annu. Rev. Microbiol. 2001, 55, 453–483. [Google Scholar] [CrossRef]
- Teixeira, D.E.; Benchimol, M.; Rodrigues, J.C.; Crepaldi, P.H.; Pimenta, P.F.; de Souza, W. The cell biology of leishmania: How to teach using animations. PLoS Pathog. 2013, 9, e1003594. [Google Scholar] [CrossRef]
- Gregory, D.J.; Olivier, M. Subversion of host cell signalling by the protozoan parasite leishmania. Parasitology 2005, 130, S27–S35. [Google Scholar] [CrossRef] [PubMed]
- Nathan, C.; Shiloh, M.U. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. USA 2000, 97, 8841–8848. [Google Scholar] [CrossRef] [PubMed]
- Mosser, D.M.; Edwards, J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008, 8, 958–969. [Google Scholar] [CrossRef] [PubMed]
- Scott, P.; Novais, F.O. Cutaneous leishmaniasis: Immune responses in protection and pathogenesis. Nat. Rev. Immunol. 2016, 16, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Frank, B.; Marcu, A.; de Oliveira Almeida Petersen, A.L.; Weber, H.; Stigloher, C.; Mottram, J.C.; Scholz, C.J.; Schurigt, U. Autophagic digestion of leishmania major by host macrophages is associated with differential expression of bnip3, ctse, and the miRNAs mir-101c, mir-129, and mir-210. Parasites Vectors 2015, 8, 404. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, B.; Paul, J.; Mukherjee, S.; Mukhopadhyay, R.; Das, S.; Naskar, K.; Sundar, S.; Dujardin, J.C.; Saha, B.; Roy, S. Antimony-resistant leishmania donovani exploits mir-466i to deactivate host myd88 for regulating il-10/il-12 levels during early hours of infection. J. Immunol. 2015, 195, 2731–2742. [Google Scholar] [CrossRef]
- Lemaire, J.; Mkannez, G.; Guerfali, F.Z.; Gustin, C.; Attia, H.; Sghaier, R.M.; Sysco, C.; Dellagi, K.; Laouini, D.; Renard, P. MicroRNA expression profile in human macrophages in response to leishmania major infection. PLoS Negl. Trop. Dis. 2013, 7, e2478. [Google Scholar] [CrossRef]
- Ghosh, J.; Bose, M.; Roy, S.; Bhattacharyya, S.N. Leishmania donovani targets dicer1 to downregulate mir-122, lower serum cholesterol, and facilitate murine liver infection. Cell Host Microbe 2013, 13, 277–288. [Google Scholar] [CrossRef]
- Geraci, N.S.; Tan, J.C.; McDowell, M.A. Characterization of microRNA expression profiles in leishmania-infected human phagocytes. Parasite Immunol. 2015, 37, 43–51. [Google Scholar] [CrossRef]
- Muxel, S.M.; Laranjeira-Silva, M.F.; Zampieri, R.A.; Floeter-Winter, L.M. Leishmania (leishmania) amazonensis induces macrophage mir-294 and mir-721 expression and modulates infection by targeting nos2 and l-arginine metabolism. Sci. Rep. 2017, 7, 44141. [Google Scholar] [CrossRef]
- Bragato, J.P.; Melo, L.M.; Venturin, G.L.; Rebech, G.T.; Garcia, L.E.; Lopes, F.L.; de Lima, V.M.F. Relationship of peripheral blood mononuclear cells miRNA expression and parasitic load in canine visceral leishmaniasis. PLoS ONE 2018, 13, e0206876. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, N.; Kumar, V.; Gedda, M.R.; Singh, A.K.; Singh, V.K.; Gannavaram, S.; Singh, S.P.; Singh, R.K. Identification and characterization of miRNAs in response to leishmania donovani infection: Delineation of their roles in macrophage dysfunction. Front. Microbiol. 2017, 8, 314. [Google Scholar] [CrossRef] [PubMed]
- Corraliza, I.M.; Soler, G.; Eichmann, K.; Modolell, M. Arginase induction by suppressors of nitric oxide synthesis (IL-4, IL-10 and PGE2) in murine bone-marrow-derived macrophages. Biochem. Biophys. Res. Commun. 1995, 206, 667–673. [Google Scholar] [CrossRef] [PubMed]
- Munder, M.; Eichmann, K.; Modolell, M. Alternative metabolic states in murine macrophages reflected by the nitric oxide synthase/arginase balance: Competitive regulation by cd4+ t cells correlates with th1/th2 phenotype. J. Immunol. 1998, 160, 5347–5354. [Google Scholar] [PubMed]
- Wanasen, N.; Soong, L. L-arginine metabolism and its impact on host immunity against leishmania infection. Immunol. Res. 2008, 41, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Liang, H.; Zen, K. Molecular mechanisms that influence the macrophage m1-m2 polarization balance. Front. Immunol. 2014, 5, 614. [Google Scholar] [CrossRef]
- Hrabak, A.; Bajor, T.; Temesi, A.; Meszaros, G. The inhibitory effect of nitrite, a stable product of nitric oxide (no) formation, on arginase. FEBS Lett. 1996, 390, 203–206. [Google Scholar] [CrossRef]
- Boucher, J.L.; Moali, C.; Tenu, J.P. Nitric oxide biosynthesis, nitric oxide synthase inhibitors and arginase competition for l-arginine utilization. Cell Mol. Life Sci. 1999, 55, 1015–1028. [Google Scholar] [CrossRef]
- Verreck, F.A.; de Boer, T.; Langenberg, D.M.; Hoeve, M.A.; Kramer, M.; Vaisberg, E.; Kastelein, R.; Kolk, A.; de Waal-Malefyt, R.; Ottenhoff, T.H. Human il-23-producing type 1 macrophages promote but il-10-producing type 2 macrophages subvert immunity to (myco)bacteria. Proc. Natl. Acad. Sci. USA 2004, 101, 4560–4565. [Google Scholar] [CrossRef]
- Martinez, F.O.; Helming, L.; Gordon, S. Alternative activation of macrophages: An immunologic functional perspective. Annu. Rev. Immunol. 2009, 27, 451–483. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Jiang, T.; Li, M.Q.; Zheng, X.L.; Zhao, G.J. Transcriptional regulation of macrophages polarization by microRNAs. Front. Immunol. 2018, 9, 1175. [Google Scholar] [CrossRef] [PubMed]
- Lima-Junior, D.S.; Costa, D.L.; Carregaro, V.; Cunha, L.D.; Silva, A.L.; Mineo, T.W.; Gutierrez, F.R.; Bellio, M.; Bortoluci, K.R.; Flavell, R.A.; et al. Inflammasome-derived il-1beta production induces nitric oxide-mediated resistance to leishmania. Nat. Med. 2013, 19, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Aoki, J.I.; Muxel, S.M.; Zampieri, R.A.; Müller, K.E.; Nerland, A.H.; Floeter-Winter, L.M. Differential immune response modulation in early leishmania amazonensis infection of balb/c and c57bl/6 macrophages based on transcriptome profiles. Sci. Rep. 2019, 9, 19841. [Google Scholar] [CrossRef] [PubMed]
- Zea, A.H.; Rodriguez, P.C.; Culotta, K.S.; Hernandez, C.P.; DeSalvo, J.; Ochoa, J.B.; Park, H.J.; Zabaleta, J.; Ochoa, A.C. L-arginine modulates cd3zeta expression and t cell function in activated human t lymphocytes. Cell Immunol. 2004, 232, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, P.C.; Zea, A.H.; DeSalvo, J.; Culotta, K.S.; Zabaleta, J.; Quiceno, D.G.; Ochoa, J.B.; Ochoa, A.C. L-arginine consumption by macrophages modulates the expression of cd3 zeta chain in t lymphocytes. J. Immunol. 2003, 171, 1232–1239. [Google Scholar] [CrossRef] [PubMed]
- Kishikawa, T.; Otsuka, M.; Tan, P.S.; Ohno, M.; Sun, X.; Yoshikawa, T.; Shibata, C.; Takata, A.; Kojima, K.; Takehana, K.; et al. Decreased mir122 in hepatocellular carcinoma leads to chemoresistance with increased arginine. Oncotarget 2015, 6, 8339–8352. [Google Scholar] [CrossRef]
- Dunand-Sauthier, I.; Irla, M.; Carnesecchi, S.; Seguin-Estevez, Q.; Vejnar, C.E.; Zdobnov, E.M.; Santiago-Raber, M.L.; Reith, W. Repression of arginase-2 expression in dendritic cells by microRNA-155 is critical for promoting t cell proliferation. J. Immunol. 2014, 193, 1690–1700. [Google Scholar] [CrossRef]
- Fernandes, J.C.R.; Aoki, J.I.; Acuña, S.M.; Zampieri, R.A.; Markus, R.P.; Floeter-Winter, L.M.; Muxel, S.M. Melatonin and leishmania amazonensis infection altered mir-294, mir-30e, and mir-302d impacting on tnf, mcp-1, and nos2 expression. Front. Cell. Infect. Microbiol. 2019, 9, 60. [Google Scholar] [CrossRef]
- Guerfali, F.Z.; Laouini, D.; Guizani-Tabbane, L.; Ottones, F.; Ben-Aissa, K.; Benkahla, A.; Manchon, L.; Piquemal, D.; Smandi, S.; Mghirbi, O.; et al. Simultaneous gene expression profiling in human macrophages infected with leishmania major parasites using sage. BMC Genom. 2008, 9, 238. [Google Scholar] [CrossRef]
- Degrossoli, A.; Bosetto, M.C.; Lima, C.B.; Giorgio, S. Expression of hypoxia-inducible factor 1alpha in mononuclear phagocytes infected with leishmania amazonensis. Immunol. Lett. 2007, 114, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Kumar, A.; Das, S.; Kumar, A.; Abhishek, K.; Verma, S.; Mandal, A.; Singh, R.K.; Das, P. Leishmania donovani activates hypoxia inducible factor-1alpha and mir-210 for survival in macrophages by downregulation of nf-kappab mediated pro-inflammatory immune response. Front. Microbiol. 2018, 9, 385. [Google Scholar] [CrossRef] [PubMed]
- Ismail, N.; Kaul, A.; Bhattacharya, P.; Gannavaram, S.; Nakhasi, H.L. Immunization with live attenuated leishmania donovani centrin−/− parasites is efficacious in asymptomatic infection. Front. Immunol. 2017, 8, 1788. [Google Scholar] [CrossRef]
- Lago, T.S.; Silva, J.A.; Lago, E.L.; Carvalho, E.M.; Zanette, D.L.; Castellucci, L.C. The miRNA 361-3p, a regulator of gzmb and tnf is associated with therapeutic failure and longer time healing of cutaneous leishmaniasis caused by l. (viannia) braziliensis. Front. Immunol. 2018, 9, 2621. [Google Scholar] [CrossRef] [PubMed]
- Crauwels, P.; Bohn, R.; Thomas, M.; Gottwalt, S.; Jackel, F.; Kramer, S.; Bank, E.; Tenzer, S.; Walther, P.; Bastian, M.; et al. Apoptotic-like leishmania exploit the host’s autophagy machinery to reduce t-cell-mediated parasite elimination. Autophagy 2015, 11, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Dias, B.R.S.; de Souza, C.S.; Almeida, N.J.; Lima, J.G.B.; Fukutani, K.F.; Dos Santos, T.B.S.; Franca-Cost, J.; Brodskyn, C.I.; de Menezes, J.P.B.; Colombo, M.I.; et al. Autophagic induction greatly enhances leishmania major intracellular survival compared to leishmania amazonensis in cba/j-infected macrophages. Front. Microbiol. 2018, 9, 1890. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Pandey, R.K.; Shaha, C.; Madhubala, R. MicroRNA expression profiling of leishmania donovani-infected host cells uncovers the regulatory role of mir30a-3p in host autophagy. Autophagy 2016, 12, 1817–1831. [Google Scholar] [CrossRef]
- Tolouei, S.; Hejazi, S.H.; Ghaedi, K.; Khamesipour, A.; Hasheminia, S.J. Tlr2 and tlr4 in cutaneous leishmaniasis caused by leishmania major. Scand. J. Immunol. 2013, 78, 478–484. [Google Scholar] [CrossRef]
- Muxel, S.M.; Acuna, S.M.; Aoki, J.I.; Zampieri, R.A.; Floeter-Winter, L.M. Toll-like receptor and miRNA-let-7e expression alter the inflammatory response in leishmania amazonensis-infected macrophages. Front. Immunol. 2018, 9, 2792. [Google Scholar] [CrossRef]
- Elizabeth, M.C.; Hernandez de la Cruz, O.N.; Mauricio, C.A. Infection of j774a.1 with different mycobacterium species induces differential immune and miRNA-related responses. Microbiol. Immunol. 2016, 60, 356–363. [Google Scholar] [CrossRef]
- Kalantari, P.; Harandi, O.F.; Agarwal, S.; Rus, F.; Kurt-Jones, E.A.; Fitzgerald, K.A.; Caffrey, D.R.; Golenbock, D.T. Mir-718 represses proinflammatory cytokine production through targeting phosphatase and tensin homolog (pten). J. Biol. Chem. 2017, 292, 5634–5644. [Google Scholar] [CrossRef] [PubMed]
- Colineau, L.; Lambertz, U.; Fornes, O.; Wasserman, W.W.; Reiner, N.E. C-myc is a novel leishmania virulence factor by proxy that targets the host miRNA system and is essential for survival in human macrophages. J. Biol.Chem. 2018, 293, 12805–12819. [Google Scholar] [CrossRef] [PubMed]
- Abrudan, J.; Ramalho-Ortigao, M.; O’Neil, S.; Stayback, G.; Wadsworth, M.; Bernard, M.; Shoue, D.; Emrich, S.; Lawyer, P.; Kamhawi, S.; et al. The characterization of the phlebotomus papatasi transcriptome. Insect Mol. Biol. 2013, 22, 211–232. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, F.V.; Aguiar, E.; Olmo, R.P.; de Oliveira, K.P.V.; Silva, E.G.; Sant’Anna, M.R.V.; Gontijo, N.F.; Kroon, E.G.; Imler, J.L.; Marques, J.T. The small non-coding RNA response to virus infection in the leishmania vector lutzomyia longipalpis. PLoS Negl. Trop. Dis. 2018, 12, e0006569. [Google Scholar] [CrossRef] [PubMed]
- Rossi, E.; Bongiorno, G.; Ciolli, E.; Di Muccio, T.; Scalone, A.; Gramiccia, M.; Gradoni, L.; Maroli, M. Seasonal phenology, host-blood feeding preferences and natural leishmania infection of phlebotomus perniciosus (diptera, psychodidae) in a high-endemic focus of canine leishmaniasis in Rome province, Italy. Acta Trop. 2008, 105, 158–165. [Google Scholar] [CrossRef]
- Yang, Z.; Wu, Y. Improved annotation of lutzomyia longipalpis genome using bioinformatics analysis. PeerJ 2019, 7, e7862. [Google Scholar] [CrossRef]
- Petrella, V.; Aceto, S.; Musacchia, F.; Colonna, V.; Robinson, M.; Benes, V.; Cicotti, G.; Bongiorno, G.; Gradoni, L.; Volf, P.; et al. De novo assembly and sex-specific transcriptome profiling in the sand fly phlebotomus perniciosus (diptera, phlebotominae), a major old world vector of leishmania infantum. BMC Genom. 2015, 16, 847. [Google Scholar] [CrossRef][Green Version]
- Robinson, K.A.; Beverley, S.M. Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite leishmania. Mol. Biochem. Parasitol. 2003, 128, 217–228. [Google Scholar] [CrossRef]
- Rastrojo, A.; Carrasco-Ramiro, F.; Martin, D.; Crespillo, A.; Reguera, R.M.; Aguado, B.; Requena, J.M. The transcriptome of leishmania major in the axenic promastigote stage: Transcript annotation and relative expression levels by RNA-seq. BMC Genom. 2013, 14, 223. [Google Scholar] [CrossRef]
- Fernandes, M.C.; Dillon, L.A.; Belew, A.T.; Bravo, H.C.; Mosser, D.M.; El-Sayed, N.M. Dual transcriptome profiling of leishmania-infected human macrophages reveals distinct reprogramming signatures. MBio 2016, 7. [Google Scholar] [CrossRef]
- Aoki, J.I.; Muxel, S.M.; Zampieri, R.A.; Laranjeira-Silva, M.F.; Muller, K.E.; Nerland, A.H.; Floeter-Winter, L.M. RNA-seq transcriptional profiling of leishmania amazonensis reveals an arginase-dependent gene expression regulation. PLoS Negl. Trop. Dis. 2017, 11, e0006026. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Available online: http://www.who.int/wer/2015/wer9006.pdf?ua=1 (accessed on 6 November 2015).
- Bonney, K.M. Chagas disease in the 21st century: A public health success or an emerging threat? Parasite 2014, 21, 11. [Google Scholar] [CrossRef] [PubMed]
- Lidani, K.C.F.; Andrade, F.A.; Bavia, L.; Damasceno, F.S.; Beltrame, M.H.; Messias-Reason, I.J.; Sandri, T.L. Chagas disease: From discovery to a worldwide health problem. Front. Public. Health 2019, 7, 166. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.Y.; Bacon, K.M.; Bottazzi, M.E.; Hotez, P.J. Global economic burden of chagas disease: A computational simulation model. Lancet Infect. Dis. 2013, 13, 342–348. [Google Scholar] [CrossRef]
- Nagajyothi, F.; Machado, F.S.; Burleigh, B.A.; Jelicks, L.A.; Scherer, P.E.; Mukherjee, S.; Lisanti, M.P.; Weiss, L.M.; Garg, N.J.; Tanowitz, H.B. Mechanisms of trypanosoma cruzi persistence in chagas disease. Cell. Microbiol. 2012, 14, 634–643. [Google Scholar] [CrossRef]
- Rassi, A., Jr.; Rassi, A.; Marin-Neto, J.A. Chagas disease. Lancet 2010, 375, 1388–1402. [Google Scholar] [CrossRef]
- Basso, B. Modulation of immune response in experimental chagas disease. World J. Exp. Med. 2013, 3, 1–10. [Google Scholar] [CrossRef]
- Zingales, B.; Andrade, S.G.; Briones, M.R.; Campbell, D.A.; Chiari, E.; Fernandes, O.; Guhl, F.; Lages-Silva, E.; Macedo, A.M.; Machado, C.R.; et al. A new consensus for trypanosoma cruzi intraspecific nomenclature: Second revision meeting recommends tci to tcvi. Mem. Inst. Oswaldo Cruz 2009, 104, 1051–1054. [Google Scholar] [CrossRef]
- Kratz, J.M.; Garcia Bournissen, F.; Forsyth, C.J.; Sosa-Estani, S. Clinical and pharmacological profile of benznidazole for treatment of chagas disease. Expert Rev. Clin. Pharmacol. 2018, 11, 943–957. [Google Scholar] [CrossRef]
- Ferreira, L.R.; Frade, A.F.; Baron, M.A.; Navarro, I.C.; Kalil, J.; Chevillard, C.; Cunha-Neto, E. Interferon-gamma and other inflammatory mediators in cardiomyocyte signaling during chagas disease cardiomyopathy. World J. Cardiol. 2014, 6, 782–790. [Google Scholar] [CrossRef]
- Andrade, D.V.; Gollob, K.J.; Dutra, W.O. Acute chagas disease: New global challenges for an old neglected disease. PLoS Negl. Trop. Dis. 2014, 8, e3010. [Google Scholar] [CrossRef] [PubMed]
- Abel, L.C.; Rizzo, L.V.; Ianni, B.; Albuquerque, F.; Bacal, F.; Carrara, D.; Bocchi, E.A.; Teixeira, H.C.; Mady, C.; Kalil, J.; et al. Chronic chagas’ disease cardiomyopathy patients display an increased ifn-gamma response to trypanosoma cruzi infection. J. Autoimmun. 2001, 17, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, L.G.; Santos, R.H.; Fiorelli, A.I.; Mairena, E.C.; Benvenuti, L.A.; Bocchi, E.A.; Stolf, N.A.; Kalil, J.; Cunha-Neto, E. Myocardial gene expression of T-bet, GATA-3, Ror-γt, FoxP3, and hallmark cytokines in chronic chagas disease cardiomyopathy: An essentially unopposed TH1-type response. Mediat. Inflamm. 2014, 2014, 914326. [Google Scholar] [CrossRef] [PubMed]
- Navarro, I.C.; Ferreira, F.M.; Nakaya, H.I.; Baron, M.A.; Vilar-Pereira, G.; Pereira, I.R.; Silva, A.M.; Real, J.M.; De Brito, T.; Chevillard, C.; et al. MicroRNA transcriptome profiling in heart of trypanosoma cruzi-infected mice: Parasitological and cardiological outcomes. PLoS Negl. Trop. Dis. 2015, 9, e0003828. [Google Scholar] [CrossRef]
- Ferreira, L.R.P.; Ferreira, F.M.; Laugier, L.; Cabantous, S.; Navarro, I.C.; da Silva Candido, D.; Rigaud, V.C.; Real, J.M.; Pereira, G.V.; Pereira, I.R.; et al. Integration of miRNA and gene expression profiles suggest a role for miRNAs in the pathobiological processes of acute trypanosoma cruzi infection. Sci. Rep. 2017, 7, 17990. [Google Scholar] [CrossRef]
- Cunha-Neto, E.; Dzau, V.J.; Allen, P.D.; Stamatiou, D.; Benvenutti, L.; Higuchi, M.L.; Koyama, N.S.; Silva, J.S.; Kalil, J.; Liew, C.C. Cardiac gene expression profiling provides evidence for cytokinopathy as a molecular mechanism in chagas’ disease cardiomyopathy. Am. J. Pathol. 2005, 167, 305–313. [Google Scholar] [CrossRef]
- Meng, L.D.; Meng, A.C.; Zhu, Q.; Jia, R.Y.; Kong, Q.Z. Effect of microRNA-208a on mitochondrial apoptosis of cardiomyocytes of neonatal rats. Asian Pac. J. Trop. Med. 2015, 8, 747–751. [Google Scholar] [CrossRef]
- Shyu, K.G.; Wang, B.W.; Cheng, W.P.; Lo, H.M. MicroRNA-208a increases myocardial endoglin expression and myocardial fibrosis in acute myocardial infarction. Can. J. Cardiol. 2015, 31, 679–690. [Google Scholar] [CrossRef]
- Nonaka, C.K.V.; Macedo, C.T.; Cavalcante, B.R.R.; Alcantara, A.C.; Silva, D.N.; Bezerra, M.D.R.; Caria, A.C.I.; Tavora, F.R.F.; Neto, J.D.S.; Noya-Rabelo, M.M.; et al. Circulating miRNAs as potential biomarkers associated with cardiac remodeling and fibrosis in chagas disease cardiomyopathy. Int. J. Mol. Sci. 2019, 20, 4064. [Google Scholar] [CrossRef]
- Zhou, X.; Li, Q.; Xu, J.; Zhang, X.; Zhang, H.; Xiang, Y.; Fang, C.; Wang, T.; Xia, S.; Zhang, Q.; et al. The aberrantly expressed mir-193b-3p contributes to preeclampsia through regulating transforming growth factor-beta signaling. Sci. Rep. 2016, 6, 19910. [Google Scholar] [CrossRef]
- Gu, N.; You, L.; Shi, C.; Yang, L.; Pang, L.; Cui, X.; Ji, C.; Zheng, W.; Guo, X. Expression of mir-199a-3p in human adipocytes is regulated by free fatty acids and adipokines. Mol. Med. Rep. 2016, 14, 1180–1186. [Google Scholar] [CrossRef] [PubMed]
- Savino, W. Intrathymic t cell migration is a multivectorial process under a complex neuroendocrine control. Neuroimmunomodulation 2010, 17, 142–145. [Google Scholar] [CrossRef] [PubMed]
- Linhares-Lacerda, L.; Palu, C.C.; Ribeiro-Alves, M.; Paredes, B.D.; Morrot, A.; Garcia-Silva, M.R.; Cayota, A.; Savino, W. Differential expression of microRNAs in thymic epithelial cells from trypanosoma cruzi acutely infected mice: Putative role in thymic atrophy. Front. Immunol. 2015, 6, 428. [Google Scholar] [CrossRef] [PubMed]
- Bayer-Santos, E.; Lima, F.M.; Ruiz, J.C.; Almeida, I.C.; da Silveira, J.F. Characterization of the small RNA content of trypanosoma cruzi extracellular vesicles. Mol. Biochem. Parasitol. 2014, 193, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Silva, M.R.; Cabrera-Cabrera, F.; das Neves, R.F.; Souto-Padron, T.; de Souza, W.; Cayota, A. Gene expression changes induced by trypanosoma cruzi shed microvesicles in mammalian host cells: Relevance of tRNA-derived halves. Biomed. Res. Int. 2014, 2014, 305239. [Google Scholar] [CrossRef] [PubMed]
- Franzen, O.; Arner, E.; Ferella, M.; Nilsson, D.; Respuela, P.; Carninci, P.; Hayashizaki, Y.; Aslund, L.; Andersson, B.; Daub, C.O. The short non-coding transcriptome of the protozoan parasite trypanosoma cruzi. PLoS Negl. Trop. Dis. 2011, 5, e1283. [Google Scholar] [CrossRef]
- Fernandez-Calero, T.; Garcia-Silva, R.; Pena, A.; Robello, C.; Persson, H.; Rovira, C.; Naya, H.; Cayota, A. Profiling of small RNA cargo of extracellular vesicles shed by trypanosoma cruzi reveals a specific extracellular signature. Mol. Biochem. Parasitol. 2015, 199, 19–28. [Google Scholar] [CrossRef]
- World Health Organization. Available online: http://www.who.int/ith/diseases/trypanosomiasis/en/ (accessed on 19 December 2019).
- Ralston, K.S.; Kabututu, Z.P.; Melehani, J.H.; Oberholzer, M.; Hill, K.L. The trypanosoma brucei flagellum: Moving parasites in new directions. Annu. Rev. Microbiol. 2009, 63, 335–362. [Google Scholar] [CrossRef]
- Lythgoe, K.A.; Morrison, L.J.; Read, A.F.; Barry, J.D. Parasite-intrinsic factors can explain ordered progression of trypanosome antigenic variation. Proc. Natl. Acad. Sci. USA 2007, 104, 8095–8100. [Google Scholar] [CrossRef]
- Rico, E.; Rojas, F.; Mony, B.M.; Szoor, B.; Macgregor, P.; Matthews, K.R. Bloodstream form pre-adaptation to the tsetse fly in trypanosoma brucei. Front. Cell. Infect. Microbiol. 2013, 3, 78. [Google Scholar] [CrossRef]
- Lueong, S.; Leong, S.; Simo, G.; Camara, M.; Jamonneau, V.; Kabore, J.; Ilboudo, H.; Bucheton, B.; Hoheisel, J.D.; Clayton, C. The miRNA and mRNA signatures of peripheral blood cells in humans infected with trypanosoma brucei gambiense. PLoS ONE 2013, 8, e67312. [Google Scholar] [CrossRef]
- Kolev, N.G.; Franklin, J.B.; Carmi, S.; Shi, H.; Michaeli, S.; Tschudi, C. The transcriptome of the human pathogen trypanosoma brucei at single-nucleotide resolution. PLoS Pathog. 2010, 6, e1001090. [Google Scholar] [CrossRef] [PubMed]
- Liniger, M.; Bodenmuller, K.; Pays, E.; Gallati, S.; Roditi, I. Overlapping sense and antisense transcription units in trypanosoma brucei. Mol. Microbiol. 2001, 40, 869–878. [Google Scholar] [CrossRef] [PubMed]
- Mallick, B.; Ghosh, Z.; Chakrabarti, J. MicroRNA switches in trypanosoma brucei. Biochem. Biophys. Res. Commun. 2008, 372, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Boothroyd, J.C.; Grigg, M.E. Population biology of toxoplasma gondii and its relevance to human infection: Do different strains cause different disease? Curr. Opin. Microbiol. 2002, 5, 438–442. [Google Scholar] [CrossRef]
- Montoya, J.G.; Liesenfeld, O. Toxoplasmosis. Lancet 2004, 363, 1965–1976. [Google Scholar] [CrossRef]
- Parlog, A.; Schluter, D.; Dunay, I.R. Toxoplasma gondii-induced neuronal alterations. Parasite Immunol. 2015, 37, 159–170. [Google Scholar] [CrossRef]
- Ehmen, H.G.; Luder, C.G.K. Long-term impact of toxoplasma gondii infection on human monocytes. Front. Cell. Infect. Microbiol. 2019, 9, 235. [Google Scholar] [CrossRef]
- Hargrave, K.E.; Woods, S.; Millington, O.; Chalmers, S.; Westrop, G.D.; Roberts, C.W. Multi-omics studies demonstrate toxoplasma gondii-induced metabolic reprogramming of murine dendritic cells. Front. Cell. Infect. Microbiol. 2019, 9, 309. [Google Scholar] [CrossRef]
- Saadatnia, G.; Golkar, M. A review on human toxoplasmosis. Scand. J. Infect. Dis. 2012, 44, 805–814. [Google Scholar] [CrossRef]
- Da Silva, R.C.; Langoni, H. Toxoplasma gondii: Host-parasite interaction and behavior manipulation. Parasitol. Res. 2009, 105, 893–898. [Google Scholar] [CrossRef] [PubMed]
- Zeiner, G.M.; Norman, K.L.; Thomson, J.M.; Hammond, S.M.; Boothroyd, J.C. Toxoplasma gondii infection specifically increases the levels of key host microRNAs. PLoS ONE 2010, 5, e8742. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, L.; Strasser, A.; O’Reilly, L.A.; Hausmann, G.; Adams, J.M.; Cory, S.; Huang, D.C. Bim: A novel member of the bcl-2 family that promotes apoptosis. EMBO J. 1998, 17, 384–395. [Google Scholar] [CrossRef] [PubMed]
- Goebel, S.; Gross, U.; Luder, C.G. Inhibition of host cell apoptosis by toxoplasma gondii is accompanied by reduced activation of the caspase cascade and alterations of poly(adp-ribose) polymerase expression. J. Cell Sci. 2001, 114, 3495–3505. [Google Scholar] [PubMed]
- Ota, A.; Tagawa, H.; Karnan, S.; Tsuzuki, S.; Karpas, A.; Kira, S.; Yoshida, Y.; Seto, M. Identification and characterization of a novel gene, c13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004, 64, 3087–3095. [Google Scholar] [CrossRef] [PubMed]
- Kuo, G.; Wu, C.Y.; Yang, H.Y. Mir-17-92 cluster and immunity. J. Formos. Med. Assoc. 2019, 118, 2–6. [Google Scholar] [CrossRef]
- O’Donnell, K.A.; Wentzel, E.A.; Zeller, K.I.; Dang, C.V.; Mendell, J.T. C-myc-regulated microRNAs modulate e2f1 expression. Nature 2005, 435, 839–843. [Google Scholar] [CrossRef]
- Baumjohann, D. Diverse functions of mir-17-92 cluster microRNAs in t helper cells. Cancer Lett. 2018, 423, 147–152. [Google Scholar] [CrossRef]
- Xiao, C.; Srinivasan, L.; Calado, D.P.; Patterson, H.C.; Zhang, B.; Wang, J.; Henderson, J.M.; Kutok, J.L.; Rajewsky, K. Lymphoproliferative disease and autoimmunity in mice with increased mir-17-92 expression in lymphocytes. Nat. Immunol. 2008, 9, 405–414. [Google Scholar] [CrossRef]
- Koralov, S.B.; Muljo, S.A.; Galler, G.R.; Krek, A.; Chakraborty, T.; Kanellopoulou, C.; Jensen, K.; Cobb, B.S.; Merkenschlager, M.; Rajewsky, N.; et al. Dicer ablation affects antibody diversity and cell survival in the b lymphocyte lineage. Cell 2008, 132, 860–874. [Google Scholar] [CrossRef]
- Hou, Z.; Liu, D.; Su, S.; Wang, L.; Zhao, Z.; Ma, Y.; Li, Q.; Jia, C.; Xu, J.; Zhou, Y.; et al. Comparison of splenocyte microRNA expression profiles of pigs during acute and chronic toxoplasmosis. BMC Genom. 2019, 20, 97. [Google Scholar] [CrossRef] [PubMed]
- Cannella, D.; Brenier-Pinchart, M.P.; Braun, L.; van Rooyen, J.M.; Bougdour, A.; Bastien, O.; Behnke, M.S.; Curt, R.L.; Curt, A.; Saeij, J.P.; et al. Mir-146a and mir-155 delineate a microRNA fingerprint associated with toxoplasma persistence in the host brain. Cell Rep. 2014, 6, 928–937. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Chang, Z.; Wei, X.; Lu, H.; Yin, J.; Jiang, N.; Chen, Q. Plasma microRNAs are promising novel biomarkers for the early detection of toxoplasma gondii infection. Parasites Vectors 2014, 7, 433. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.S.; He, J.J.; Elsheikha, H.M.; Zhang, F.K.; Zou, Y.; Zhao, G.H.; Cong, W.; Zhu, X.Q. Differential brain microRNA expression profiles after acute and chronic infection of mice with toxoplasma gondii oocysts. Front. Microbiol. 2018, 9, 2316. [Google Scholar] [CrossRef]
- He, J.J.; Ma, J.; Wang, J.L.; Xu, M.J.; Zhu, X.Q. Analysis of miRNA expression profiling in mouse spleen affected by acute toxoplasma gondii infection. Infect. Genet. Evol. 2016, 37, 137–142. [Google Scholar] [CrossRef]
- Xiao, J.; Li, Y.; Prandovszky, E.; Karuppagounder, S.S.; Talbot, C.C., Jr.; Dawson, V.L.; Dawson, T.M.; Yolken, R.H. MicroRNA-132 dysregulation in toxoplasma gondii infection has implications for dopamine signaling pathway. Neuroscience 2014, 268, 128–138. [Google Scholar] [CrossRef]
- Ngo, H.M.; Zhou, Y.; Lorenzi, H.; Wang, K.; Kim, T.K.; Zhou, Y.; El Bissati, K.; Mui, E.; Fraczek, L.; Rajagopala, S.V.; et al. Toxoplasma modulates signature pathways of human epilepsy, neurodegeneration & cancer. Sci. Rep. 2017, 7, 11496. [Google Scholar]
- Cong, W.; Zhang, X.X.; He, J.J.; Li, F.C.; Elsheikha, H.M.; Zhu, X.Q. Global miRNA expression profiling of domestic cat livers following acute toxoplasma gondii infection. Oncotarget 2017, 8, 25599–25611. [Google Scholar] [CrossRef]
- World Health Organization. Available online: http://www.who.int/news-room/fact-sheets/detail/malaria (accessed on 27 March 2019).
- Engwerda, C.R.; Beattie, L.; Amante, F.H. The importance of the spleen in malaria. Trends Parasitol. 2005, 21, 75–80. [Google Scholar] [CrossRef]
- Wunderlich, F.; Al-Quraishy, S.; Dkhil, M.A. Liver-inherent immune system: Its role in blood-stage malaria. Front. Microbiol. 2014, 5, 559. [Google Scholar] [CrossRef]
- Hentzschel, F.; Hammerschmidt-Kamper, C.; Borner, K.; Heiss, K.; Knapp, B.; Sattler, J.M.; Kaderali, L.; Castoldi, M.; Bindman, J.G.; Malato, Y.; et al. Aav8-mediated in vivo overexpression of mir-155 enhances the protective capacity of genetically attenuated malarial parasites. Mol. Ther. 2014, 22, 2130–2141. [Google Scholar] [CrossRef] [PubMed]
- Dkhil, M.A.; Al-Quraishy, S.A.; Abdel-Baki, A.S.; Delic, D.; Wunderlich, F. Differential miRNA expression in the liver of balb/c mice protected by vaccination during crisis of plasmodium chabaudi blood-stage malaria. Front. Microbiol. 2016, 7, 2155. [Google Scholar] [CrossRef] [PubMed]
- Martin-Alonso, A.; Cohen, A.; Quispe-Ricalde, M.A.; Foronda, P.; Benito, A.; Berzosa, P.; Valladares, B.; Grau, G.E. Differentially expressed microRNAs in experimental cerebral malaria and their involvement in endocytosis, adherens junctions, foxo and tgf-beta signalling pathways. Sci. Rep. 2018, 8, 11277. [Google Scholar] [CrossRef] [PubMed]
- El-Assaad, F.; Hempel, C.; Combes, V.; Mitchell, A.J.; Ball, H.J.; Kurtzhals, J.A.; Hunt, N.H.; Mathys, J.M.; Grau, G.E. Differential microRNA expression in experimental cerebral and noncerebral malaria. Infect. Immun. 2011, 79, 2379–2384. [Google Scholar] [CrossRef] [PubMed]
- Chamnanchanunt, S.; Kuroki, C.; Desakorn, V.; Enomoto, M.; Thanachartwet, V.; Sahassananda, D.; Sattabongkot, J.; Jenwithisuk, R.; Fucharoen, S.; Svasti, S.; et al. Downregulation of plasma mir-451 and mir-16 in plasmodium vivax infection. Exp. Parasitol. 2015, 155, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Glinsky, G.V. Snp-guided microRNA maps (mirmaps) of 16 common human disorders identify a clinically accessible therapy reversing transcriptional aberrations of nuclear import and inflammasome pathways. Cell Cycle 2008, 7, 3564–3576. [Google Scholar] [CrossRef]
- van Loon, W.; Gai, P.P.; Hamann, L.; Bedu-Addo, G.; Mockenhaupt, F.P. MiRNA-146a polymorphism increases the odds of malaria in pregnancy. Malar. J. 2019, 18, 7. [Google Scholar] [CrossRef]
- Taganov, K.D.; Boldin, M.P.; Chang, K.J.; Baltimore, D. Nf-kappab-dependent induction of microRNA mir-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef]
- Longley, R.; Smith, C.; Fortin, A.; Berghout, J.; McMorran, B.; Burgio, G.; Foote, S.; Gros, P. Host resistance to malaria: Using mouse models to explore the host response. Mamm. Genome 2011, 22, 32–42. [Google Scholar] [CrossRef]
- Freitas do Rosario, A.P.; Lamb, T.; Spence, P.; Stephens, R.; Lang, A.; Roers, A.; Muller, W.; O’Garra, A.; Langhorne, J. Il-27 promotes il-10 production by effector th1 cd4+ t cells: A critical mechanism for protection from severe immunopathology during malaria infection. J. Immunol. 2012, 188, 1178–1190. [Google Scholar] [CrossRef]
- Stephens, R.; Culleton, R.L.; Lamb, T.J. The contribution of plasmodium chabaudi to our understanding of malaria. Trends Parasitol. 2012, 28, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Muxel, S.M.; Freitas do Rosario, A.P.; Zago, C.A.; Castillo-Mendez, S.I.; Sardinha, L.R.; Rodriguez-Malaga, S.M.; Camara, N.O.; Alvarez, J.M.; Lima, M.R. The spleen cd4+ t cell response to blood-stage plasmodium chabaudi malaria develops in two phases characterized by different properties. PLoS ONE 2011, 6, e22434. [Google Scholar] [CrossRef] [PubMed]
- Delic, D.; Dkhil, M.; Al-Quraishy, S.; Wunderlich, F. Hepatic miRNA expression reprogrammed by plasmodium chabaudi malaria. Parasitol. Res. 2011, 108, 1111–1121. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, G.; Isberg, R.R. Innate immunity to intracellular pathogens: Balancing microbial elimination and inflammation. Cell Host Microbe 2017, 22, 166–175. [Google Scholar] [CrossRef]
- Agop-Nersesian, C.; Niklaus, L.; Wacker, R.; Theo Heussler, V. Host cell cytosolic immune response during plasmodium liver stage development. FEMS Microbiol. Rev. 2018, 42, 324–334. [Google Scholar] [CrossRef]
- Vembar, S.S.; Scherf, A.; Siegel, T.N. Noncoding RNAs as emerging regulators of plasmodium falciparum virulence gene expression. Curr. Opin. Microbiol. 2014, 20, 153–161. [Google Scholar] [CrossRef]
- Guizetti, J.; Barcons-Simon, A.; Scherf, A. Trans-acting gc-rich non-coding RNA at var expression site modulates gene counting in malaria parasite. Nucleic Acids Res. 2016, 44, 9710–9718. [Google Scholar]
- Biryukova, I.; Ye, T.; Levashina, E. Transcriptome-wide analysis of microRNA expression in the malaria mosquito anopheles gambiae. BMC Genom. 2014, 15, 557. [Google Scholar] [CrossRef]
- Winter, F.; Edaye, S.; Huttenhofer, A.; Brunel, C. Anopheles gambiae miRNAs as actors of defence reaction against plasmodium invasion. Nucleic Acids Res. 2007, 35, 6953–6962. [Google Scholar] [CrossRef]
- Jain, S.; Rana, V.; Shrinet, J.; Sharma, A.; Tridibes, A.; Sunil, S.; Bhatnagar, R.K. Blood feeding and plasmodium infection alters the mirnome of anopheles stephensi. PLoS ONE 2014, 9, e98402. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Doss, C.G.P.; Lee, S.S. Therapeutic miRNA and siRNA: Moving from bench to clinic as next generation medicine. Mol. Ther. Nucleic Acids 2017, 8, 132–143. [Google Scholar] [CrossRef] [PubMed]
- Hewson, C.; Capraro, D.; Burdach, J.; Whitaker, N.; Morris, K.V. Extracellular vesicle associated long non-coding RNAs functionally enhance cell viability. Non-Coding RNA Res. 2016, 1, 3–11. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Acuña, S.M.; Floeter-Winter, L.M.; Muxel, S.M. MicroRNAs: Biological Regulators in Pathogen–Host Interactions. Cells 2020, 9, 113. https://doi.org/10.3390/cells9010113
Acuña SM, Floeter-Winter LM, Muxel SM. MicroRNAs: Biological Regulators in Pathogen–Host Interactions. Cells. 2020; 9(1):113. https://doi.org/10.3390/cells9010113
Chicago/Turabian StyleAcuña, Stephanie Maia, Lucile Maria Floeter-Winter, and Sandra Marcia Muxel. 2020. "MicroRNAs: Biological Regulators in Pathogen–Host Interactions" Cells 9, no. 1: 113. https://doi.org/10.3390/cells9010113
APA StyleAcuña, S. M., Floeter-Winter, L. M., & Muxel, S. M. (2020). MicroRNAs: Biological Regulators in Pathogen–Host Interactions. Cells, 9(1), 113. https://doi.org/10.3390/cells9010113





