Detection of Androgen Receptor Variant 7 (ARV7) mRNA Levels in EpCAM-Enriched CTC Fractions for Monitoring Response to Androgen Targeting Therapies in Prostate Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Cancer Cell Lines
2.2. Blood Collection and Processing
2.3. Spiking of Healthy Donor Blood
2.4. Immunocytochemical Stainings on Cell Culture Plates
2.5. Immunocytochemical Stainings on Cytospins
2.6. Western Blots
2.7. RNA Extraction and cDNA Synthesis
2.8. Polymerase-Chain Reaction (PCR) Analysis
2.9. Quantitative Polymerase-Chain Reaction (qPCR) Analysis
3. Results
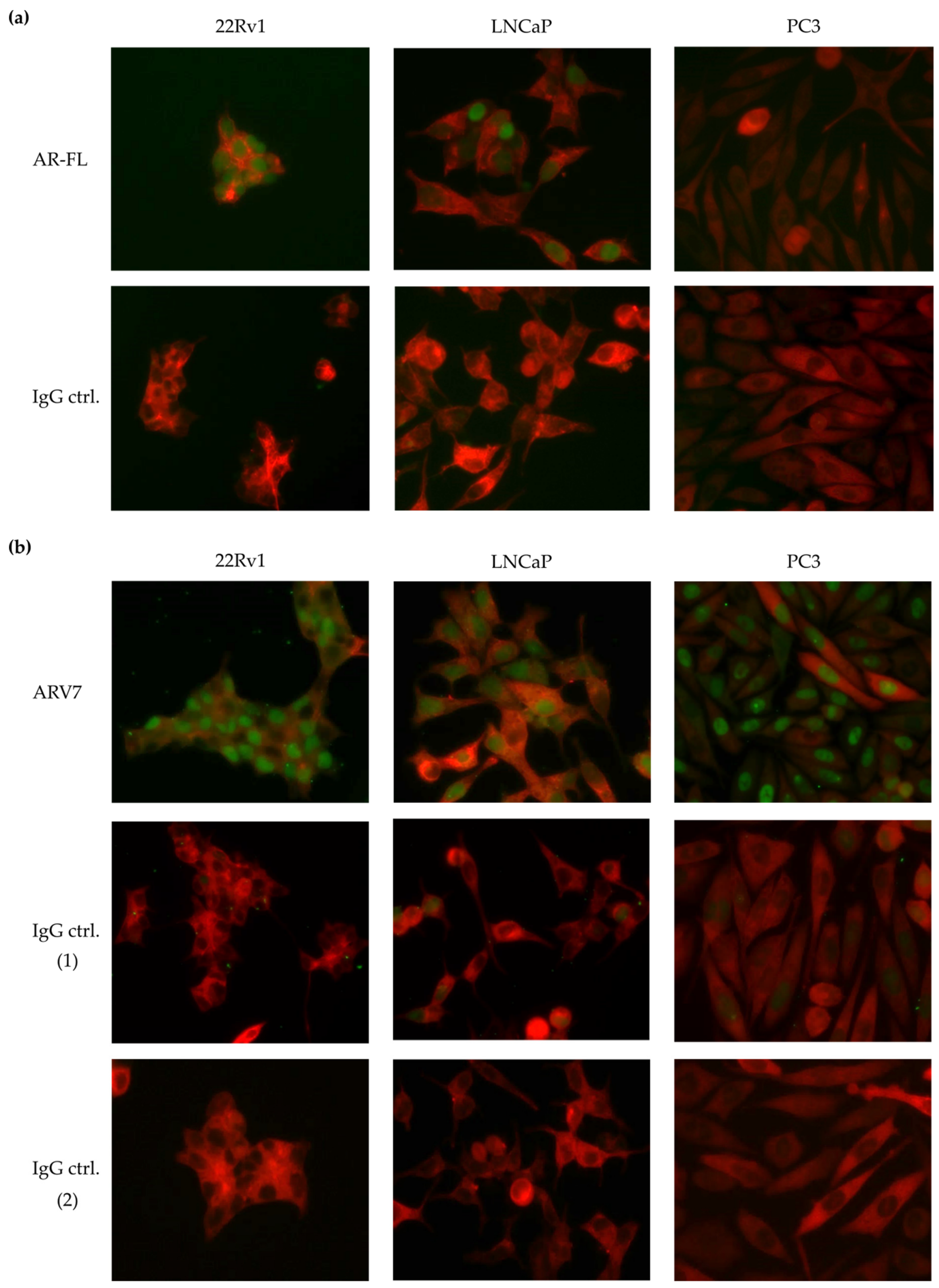
3.1. Test of Commercially Available ARV7 Antibodies for Fourth CellSearch® Channel
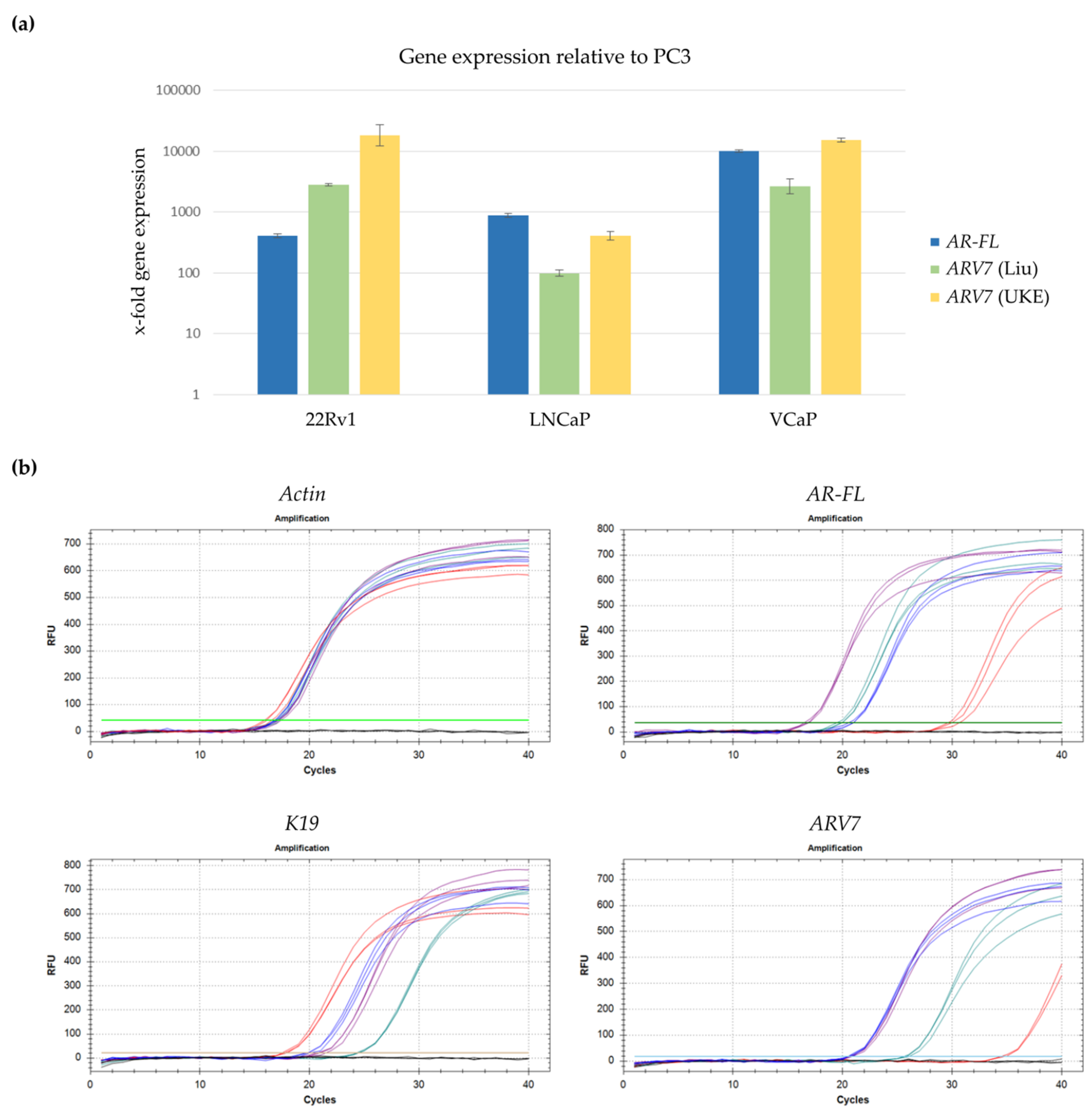
3.2. Development of a qPCR Based Assay to Detect ARV7 mRNA
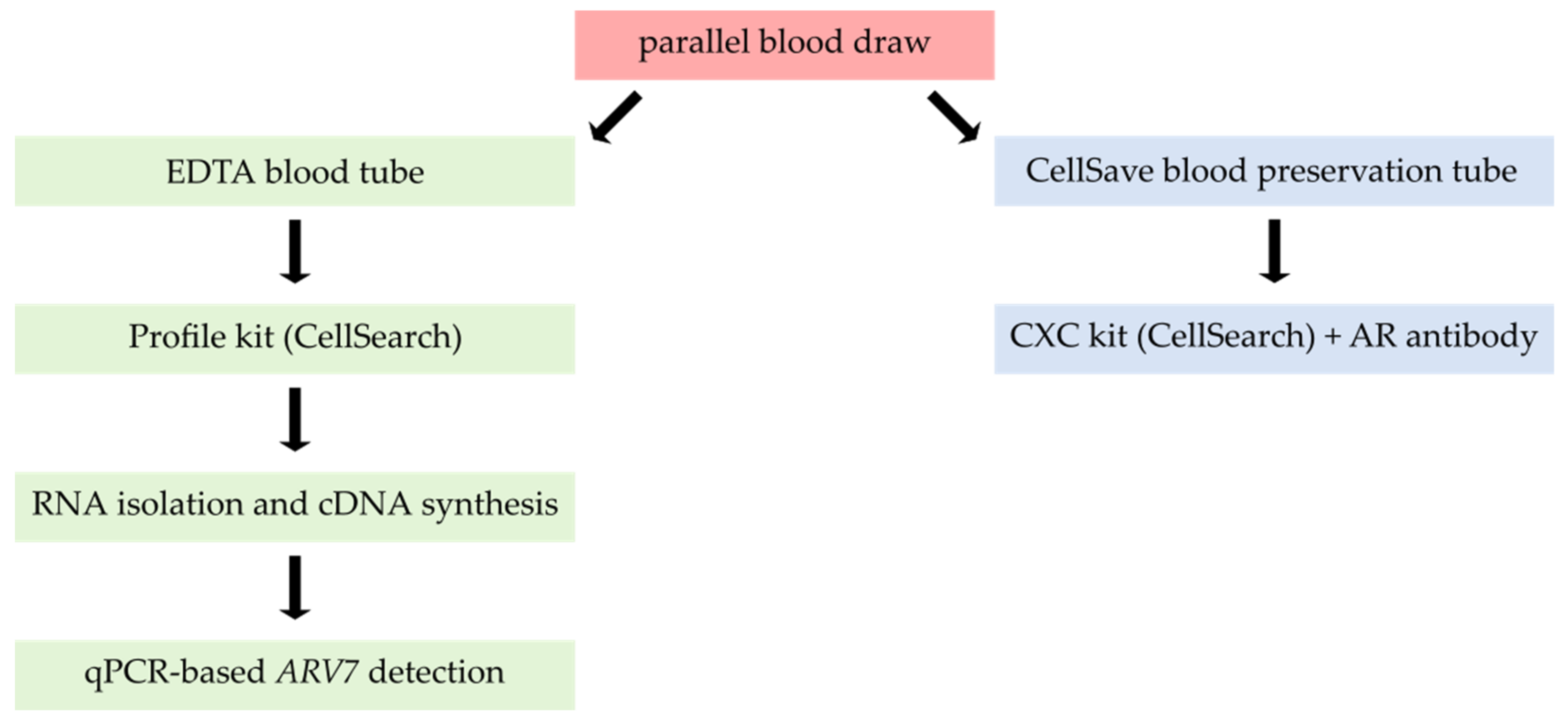
3.3. Combining Profile-Kit-Based CTC Enrichment with ARV7 mRNA Detection
3.4. Assessment of Sample Storage Parameters
3.5. Clinical Feasibility of the Complete ARV7 Detection Workflow
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Cancer Research Fund Inthernational, American Institute for Cancer Research. Prostate Cancer. 2018. Available online: https://www.wcrf.org/dietandcancer/prostate-cancer (accessed on 30 July 2019).
- Ladjevardi, S.; Auer, G.; Castro, J.; Ericsson, C.; Zetterberg, A.; Häggman, M.; Wiksell, H.; Jorulf, H. Prostate Biopsy Sampling Causes Hematogenous Dissemination of Epithelial Cellular Material. Dis. Markers 2014, 2014, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabières, C.; Pantel, K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov. 2016, 6, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Pantel, K.; Speicher, M.R. The biology of circulating tumor cells. Oncogene 2016, 35, 1216–1224. [Google Scholar] [CrossRef] [PubMed]
- Bardelli, A.; Pantel, K. Liquid Biopsies, What We Do Not Know (Yet). Cancer Cell 2017, 31, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabieres, C.; Pantel, K. Circulating tumor cells: Liquid biopsy of cancer. Clin. Chem. 2013, 59, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Hille, C.; Pantel, K. Circulating tumour cells in prostate cancer. Nat. Rev. Urol. 2018, 15, 265–266. [Google Scholar] [CrossRef] [PubMed]
- De Bono, J.S.; Scher, H.I.; Montgomery, R.B.; Parker, C.; Miller, M.C.; Tissing, H.; Doyle, G.V.; Terstappen, L.W.W.M.; Pienta, K.J.; Raghavan, D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 2008, 14, 6302–6309. [Google Scholar] [CrossRef] [PubMed]
- Scher, H.I.; Jia, X.Y.; de Bono, J.S.; Fleisher, M.; Pienta, K.J.; Raghavan, D.; Heller, G. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: A reanalysis of IMMC38 trial data. Lancet. Oncol. 2009, 10, 233–239. [Google Scholar] [CrossRef]
- Scher, H.I.; Heller, G.; Molina, A.; Attard, G.; Danila, D.C.; Jia, X.; Peng, W.; Sandhu, S.K.; Olmos, D.; Riisnaes, R.; et al. Circulating Tumor Cell Biomarker Panel As an Individual-Level Surrogate for Survival in Metastatic Castration-Resistant Prostate Cancer. J. Clin. Oncol. 2015, 33, 1348–1355. [Google Scholar] [CrossRef]
- Heller, G.; Fizazi, K.; McCormack, R.; Molina, A.; MacLean, D.; Webb, I.J.; Saad, F.; de Bono, J.S.; Scher, H.I. The Added Value of Circulating Tumor Cell Enumeration to Standard Markers in Assessing Prognosis in a Metastatic Castration-Resistant Prostate Cancer Population. Clin. Cancer Res. 2017, 23, 1967–1973. [Google Scholar] [CrossRef]
- Miller, M.C.; Doyle, G.V.; Terstappen, L.W. Significance of Circulating Tumor Cells Detected by the CellSearch System in Patients with Metastatic Breast Colorectal and Prostate Cancer. J. Oncol. 2010, 2010, 617421. [Google Scholar] [CrossRef] [PubMed]
- Goodman, O.B.; Symanowski, J.T.; Loudyi, A.; Fink, L.M.; Ward, D.C.; Vogelzang, N.J. Circulating Tumor Cells as a Predictive Biomarker in Patients With Hormone-sensitive Prostate Cancer. Clin. Genitourin. Cancer 2011, 9, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Danila, D.C.; Fleisher, M.; Scher, H.I. Circulating tumor cells as biomarkers in prostate cancer. Clin. Cancer Res. 2011, 17, 3903–3912. [Google Scholar] [CrossRef] [PubMed]
- Gorges, T.M.; Pantel, K. Circulating tumor cells as therapy-related biomarkers in cancer patients. Cancer Immunol. Immunother. 2013, 62, 931–939. [Google Scholar] [CrossRef] [PubMed]
- Singhal, U.; Wang, Y.; Henderson, J.; Niknafs, Y.S.; Qiao, Y.; Gursky, A.; Zaslavsky, A.; Chung, J.-S.; Smith, D.C.; Karnes, R.J.; et al. Multigene Profiling of CTCs in mCRPC Identifies a Clinically Relevant Prognostic Signature. Mol. Cancer Res. 2018, 16, 643–654. [Google Scholar] [CrossRef]
- Pantel, K.; Hille, C.; Scher, H.I. Circulating Tumor Cells in Prostate Cancer: From Discovery to Clinical Utility. Clin. Chem. 2019, 65, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Attard, G.; Balk, S.P.; Bevan, C.; Burnstein, K.; Cato, L.; Cherkasov, A.; De Bono, J.S.; Dong, Y.; Gao, A.C.; et al. Role of Androgen Receptor Variants in Prostate Cancer: Report from the 2017 Mission Androgen Receptor Variants Meeting. Eur. Urol. 2018, 73, 715–723. [Google Scholar] [CrossRef]
- Antonarakis, E.S.; Armstrong, A.J.; Dehm, S.M.; Luo, J. Androgen receptor variant-driven prostate cancer: Clinical implications and therapeutic targeting. Prostate Cancer Prostatic Dis. 2016, 19, 231–241. [Google Scholar] [CrossRef]
- Antonarakis, E.S.; Lu, C.; Wang, H.; Luber, B.; Nakazawa, M.; Roeser, J.C.; Chen, Y.; Mohammad, T.A.; Chen, Y.; Fedor, H.L.; et al. AR-V7 and Resistance to Enzalutamide and Abiraterone in Prostate Cancer. New Engl. J. Med. 2014, 371, 1028–1038. [Google Scholar] [CrossRef]
- Antonarakis, E.S.; Luo, J. Blood Based Detection of Androgen Receptor Splice Variants in Patients with Advanced Prostate Cancer. J. Urol. 2016, 196, 1606–1607. [Google Scholar] [CrossRef]
- Lokhandwala, P.M.; Riel, S.L.; Haley, L.; Lu, C.; Chen, Y.; Silberstein, J.; Zhu, Y.; Zheng, G.; Lin, M.-T.; Gocke, C.D.; et al. Analytical Validation of Androgen Receptor Splice Variant 7 Detection in a Clinical Laboratory Improvement Amendments (CLIA) Laboratory Setting. J. Mol. Diagn. 2017, 19, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Antonarakis, E.S.; Lu, C.; Luber, B.; Wang, H.; Chen, Y.; Zhu, Y.; Silberstein, J.L.; Taylor, M.N.; Maughan, B.L.; Denmeade, S.R.; et al. Clinical Significance of Androgen Receptor Splice Variant-7 mRNA Detection in Circulating Tumor Cells of Men With Metastatic Castration-Resistant Prostate Cancer Treated With First- and Second-Line Abiraterone and Enzalutamide. J. Clin. Oncol. 2017, 35, 2149–2156. [Google Scholar] [CrossRef]
- Scher, H.I.; Graf, R.P.; Schreiber, N.A.; Jayaram, A.; Winquist, E.; McLaughlin, B.; Lu, D.; Fleisher, M.; Orr, S.; Lowes, L.; et al. Assessment of the Validity of Nuclear-Localized Androgen Receptor Splice Variant 7 in Circulating Tumor Cells as a Predictive Biomarker for Castration-Resistant Prostate Cancer. JAMA Oncol. 2018, 4, 1179–1186. [Google Scholar] [CrossRef]
- Scher, H.I.; Lu, D.; Schreiber, N.A.; Louw, J.; Graf, R.P.; Vargas, H.A.; Johnson, A.; Jendrisak, A.; Bambury, R.; Danila, D.; et al. Association of AR-V7 on Circulating Tumor Cells as a Treatment-Specific Biomarker With Outcomes and Survival in Castration-Resistant Prostate Cancer. JAMA Oncol. 2016, 2, 1441–1449. [Google Scholar] [CrossRef] [PubMed]
- Onstenk, W.; Sieuwerts, A.M.; Kraan, J.; Van, M.; Nieuweboer, A.J.; Mathijssen, R.H.; Hamberg, P.; Meulenbeld, H.J.; De Laere, B.; Dirix, L.Y.; et al. Efficacy of Cabazitaxel in Castration-resistant Prostate Cancer Is Independent of the Presence of AR-V7 in Circulating Tumor Cells. Eur. Urol. 2015, 68, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Antonarakis, E.S.; Lu, C.; Luber, B.; Wang, H.; Chen, Y.; Nakazawa, M.; Nadal, R.; Paller, C.J.; Denmeade, S.R.; Carducci, M.A.; et al. Androgen Receptor Splice Variant 7 and Efficacy of Taxane Chemotherapy in Patients With Metastatic Castration-Resistant Prostate Cancer. JAMA Oncol. 2015, 1, 582–591. [Google Scholar] [CrossRef] [PubMed]
- Bernemann, C.; Schnoeller, T.J.; Luedeke, M.; Steinestel, K.; Boegemann, M.; Schrader, A.J.; Steinestel, J. Expression of AR-V7 in Circulating Tumour Cells Does Not Preclude Response to Next Generation Androgen Deprivation Therapy in Patients with Castration Resistant Prostate Cancer. Eur. Urol. 2017, 71, 1–3. [Google Scholar] [CrossRef]
- Nakazawa, M.; Lu, C.; Chen, Y.; Paller, C.J.; Carducci, M.A.; Eisenberger, M.A.; Luo, J.; Antonarakis, E.S. Serial blood-based analysis of AR-V7 in men with advanced prostate cancer. Ann. Oncol. 2015, 26, 1859–1865. [Google Scholar] [CrossRef]
- De Laere, B.; Van Dam, P.-J.; Whitington, T.; Mayrhofer, M.; Diaz, E.H.; Eynden, G.V.D.; Vandebroek, J.; Del-Favero, J.; Van Laere, S.; Dirix, L.; et al. Comprehensive Profiling of the Androgen Receptor in Liquid Biopsies from Castration-resistant Prostate Cancer Reveals Novel Intra-AR Structural Variation and Splice Variant Expression Patterns. Eur. Urol. 2017, 72, 192–200. [Google Scholar] [CrossRef]
- El-Heliebi, A.; Attard, G.; Balk, S.P.; Bevan, C.; Burnstein, K.; Cato, L.; Cherkasov, A.; De Bono, J.S.; Dong, Y.; Gao, A.C.; et al. In Situ Detection and Quantification of AR-V7, AR-FL, PSA, and KRAS Point Mutations in Circulating Tumor Cells. Clin. Chem. 2018, 64. [Google Scholar] [CrossRef]
- Sharp, A.; Welti, J.C.; Lambros, M.B.; Dolling, D.; Rodrigues, D.N.; Pope, L.; Aversa, C.; Figueiredo, I.; Fraser, J.; Ahmad, Z.; et al. Clinical Utility of Circulating Tumour Cell Androgen Receptor Splice Variant-7 Status in Metastatic Castration-resistant Prostate Cancer. Eur. Urol. 2019. [Google Scholar] [CrossRef]
- Riethdorf, S.; O’Flaherty, L.; Hille, C.; Pantel, K. Clinical applications of the CellSearch platform in cancer patients. Adv. Drug Deliv. Rev. 2018, 125, 102–121. [Google Scholar] [CrossRef] [PubMed]
- Riethdorf, S.; Fritsche, H.; Müller, V.; Rau, T.; Schindlbeck, C.; Rack, B.; Janni, W.; Coith, C.; Beck, K.; Jänicke, F.; et al. Detection of Circulating Tumor Cells in Peripheral Blood of Patients with Metastatic Breast Cancer: A Validation Study of the CellSearch System. Clin. Cancer Res. 2007, 13, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Yang, X.; Sun, F.; Jiang, R.; Linn, D.E.; Chen, H.; Chen, H.; Kong, X.; Melamed, J.; Tepper, C.G.; et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009, 69, 2305–2313. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.L.; Xie, N.; Sun, S.; Plymate, S.; Mostaghel, E.; Dong, X. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene 2014, 33, 3140–3150. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Stathopoulou, A.; Angelopoulou, K.; Perraki, M.; Georgoulias, V.; Malamos, N.; Lianidou, E. Quantitative RT-PCR luminometric hybridization assay with an RNA internal standard for cytokeratin-19 mRNA in peripheral blood of patients with breast cancer. Clin. Biochem. 2001, 34, 651–659. [Google Scholar] [CrossRef]
- Stathopoulou, A.; Gizi, A.; Perraki, M.; Apostolaki, S.; Malamos, N.; Mavroudis, D.; Georgoulias, V.; Lianidou, E.S. Real-time quantification of CK-19 mRNA-positive cells in peripheral blood of breast cancer patients using the lightcycler system. Clin. Cancer Res. 2003, 9, 5145–5151. [Google Scholar]
- Gorges, T.M.; Kuske, A.; Röck, K.; Mauermann, O.; Müller, V.; Peine, S.; Verpoort, K.; Novosadova, V.; Kubista, M.; Riethdorf, S.; et al. Accession of Tumor Heterogeneity by Multiplex Transcriptome Profiling of Single Circulating Tumor Cells. Clin. Chem. 2016, 62, 1504–1515. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. New Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.J.; Punt, C.J.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C.; et al. Relationship of Circulating Tumor Cells to Tumor Response, Progression-Free Survival, and Overall Survival in Patients With Metastatic Colorectal Cancer. J. Clin. Oncol. 2008, 26, 3213–3221. [Google Scholar] [CrossRef] [PubMed]
- Ligthart, S.T.; Coumans, F.A.W.; Attard, G.; Cassidy, A.M.; De Bono, J.S.; Terstappen, L.W.M.M. Unbiased and Automated Identification of a Circulating Tumour Cell Definition That Associates with Overall Survival. PLoS ONE 2011, 6, e27419. [Google Scholar] [CrossRef] [PubMed]
- Scholtens, T.M.; Schreuder, F.; Ligthart, S.T.; Swennenhuis, J.F.; Greve, J.; Terstappen, L.W. Automated identification of circulating tumor cells by image cytometry. Cytometry A 2012, 81A, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Markowski, M.C.; Silberstein, J.L.; Eshleman, J.R.; Eisenberger, M.A.; Luo, J.; Antonarakis, E.S. Clinical Utility of CLIA-Grade AR-V7 Testing in Patients With Metastatic Castration-Resistant Prostate Cancer. JCO Precis. Oncol. 2017, 1–9. [Google Scholar]
- Todenhöfer, T.; Azad, A.; Stewart, C.; Gao, J.; Eigl, B.J.; Gleave, M.E.; Joshua, A.M.; Black, P.C.; Chi, K.N. AR-V7 Transcripts in Whole Blood RNA of Patients with Metastatic Castration Resistant Prostate Cancer Correlate with Response to Abiraterone Acetate. J. Urol. 2017, 197, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Tommasi, S.; Pilato, B.; Carella, C.; Lasorella, A.; Danza, K.; Vallini, I.; De Summa, S.; Naglieri, E. Standardization of CTC AR-V7 PCR assay and evaluation of its role in castration resistant prostate cancer progression. Prostate 2018, 79, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Sieuwerts, A.M.; Mostert, B.; Van Der Vlugt-Daane, M.; Kraan, J.; Beaufort, C.M.; Van, M.; Prager, W.J.; De Laere, B.; Beije, N.; Hamberg, P.; et al. An In-Depth Evaluation of the Validity and Logistics Surrounding the Testing of AR-V7 mRNA Expression in Circulating Tumor Cells. J. Mol. Diagn. 2018, 20, 316–325. [Google Scholar] [CrossRef]
- Gorges, T.M.; Riethdorf, S.; Von Ahsen, O.; Nastały, P.; Röck, K.; Boede, M.; Peine, S.; Kuske, A.; Schmid, E.; Kneip, C.; et al. Heterogeneous PSMA expression on circulating tumor cells—A potential basis for stratification and monitoring of PSMA-directed therapies in prostate cancer. Oncotarget 2016, 7, 34930–34941. [Google Scholar] [CrossRef] [PubMed]
- Scher, H.I.; Graf, R.P.; Schreiber, N.A.; McLaughlin, B.; Jendrisak, A.; Wang, Y.; Lee, J.; Greene, S.; Krupa, R.; Lu, D.; et al. Phenotypic Heterogeneity of Circulating Tumor Cells Informs Clinical Decisions between AR Signaling Inhibitors and Taxanes in Metastatic Prostate Cancer. Cancer Res. 2017, 77, 5687–5698. [Google Scholar] [CrossRef]
- Martín, M.; García-Sáenz, J.A.; Casas, M.L.M.D.L.; Vidaurreta, M.; Puente, J.; Veganzones, S.; Rodríguez-Lajusticia, L.; De La Orden, V.; Oliva, B.; De La Torre, J.-C.; et al. Circulating tumor cells in metastatic breast cancer: timing of blood extraction for analysis. Anticancer. Res. 2009, 29, 4185–4187. [Google Scholar] [PubMed]
- Tibbe, A.G.J.; Miller, M.C.; Terstappen, L.W.M.M. Statistical considerations for enumeration of circulating tumor cells. Cytom. Part. A 2007, 71, 154–162. [Google Scholar] [CrossRef]
- Miyamoto, D.T.; Lee, R.J.; Stott, S.L.; Ting, D.T.; Wittner, B.S.; Ulman, M.; Smas, M.E.; Lord, J.B.; Brannigan, B.W.; Trautwein, J.; et al. Androgen receptor signaling in circulating tumor cells as a marker of hormonally responsive prostate cancer. Cancer Discov. 2012, 2, 995–1003. [Google Scholar] [CrossRef] [PubMed]
- Crespo, M.; Van Dalum, G.; Ferraldeschi, R.; Zafeiriou, Z.; Sideris, S.; Lorente, D.; Bianchini, D.; Rodrigues, D.N.; Riisnaes, R.; Miranda, S.; et al. Androgen receptor expression in circulating tumour cells from castration-resistant prostate cancer patients treated with novel endocrine agents. Br. J. Cancer 2015, 112, 1166–1174. [Google Scholar] [CrossRef] [PubMed]
- Hvichia, G.; Parveen, Z.; Wagner, C.; Janning, M.; Quidde, J.; Stein, A.; Müller, V.; Loges, S.; Neves, R.; Stoecklein, N.; et al. A novel microfluidic platform for size and deformability based separation and the subsequent molecular characterization of viable circulating tumor cells. Int. J. Cancer 2016, 138, 2894–2904. [Google Scholar] [CrossRef]
- Services, C.f.M.M. 2018. Available online: https://med.noridianmedicare.com/documents/10546/6990981/MolDX+Circulating+Tumor+Cell+Marker+Assays+LCD/8eaf89f0-9970-455c-b048-ebaeaf42bd7d (accessed on 30 July 2019).
- Services C.f.M.M. 2018. Available online: https://www.cms.gov/medicare-coverage-database/details/lcd-details.aspx?LCDId=37914&ver=2&Cntrctr=All&UpdatePeriod=796&bc=AQAAEAAAAAAA& (accessed on 30 July 2019).


| Cell Line | ARV7 Status | Cell Amount | Target | Detection | Detection (n = 2) | Detection (n = 3) |
|---|---|---|---|---|---|---|
| HD | − | 0 | ARV7 | N/D | N/D | N/D |
| K19 | N/D | N/D | N/D | |||
| Actin | yes | yes | yes | |||
| PC3 | − | 50 | ARV7 | N/D | - | - |
| K19 | yes | - | - | |||
| Actin | yes | - | - | |||
| 100 | ARV7 | N/D | - | - | ||
| K19 | yes | - | - | |||
| Actin | yes | - | - | |||
| 22RV1 | + | 50 | ARV7 | yes | - | - |
| K19 | yes | - | - | |||
| Actin | yes | - | - | |||
| 20 | ARV7 | yes | - | - | ||
| K19 | yes | - | - | |||
| Actin | yes | - | - | |||
| 10 | ARV7 | yes | yes | yes | ||
| K19 | yes | yes | yes | |||
| Actin | yes | yes | yes | |||
| 5 | ARV7 | yes | yes | yes | ||
| K19 | yes | yes | yes | |||
| Actin | yes | yes | yes |
| Cell Line | ARV7 Status | Cell Amount | Target | Detection | N | Ratio [detection/N] |
|---|---|---|---|---|---|---|
| HD | − | 0 | ARV7 | N/D | 3 | 3/3 |
| K19 | N/D | |||||
| Actin | yes | |||||
| 22RV1 | + | 10 | ARV7 | yes | 1 | 1/1 |
| K19 | yes | |||||
| Actin | yes | |||||
| 5 | ARV7 | yes | 2 | 2/2 | ||
| K19 | yes | |||||
| Actin | yes | |||||
| 3 | ARV7 | yes | 4 | 1/4 | ||
| K19 | yes | 3/4 | ||||
| Actin | yes | 4/4 | ||||
| 1 | ARV7 | yes | 3 | 1/3 | ||
| K19 | yes | 2/3 | ||||
| Actin | yes | 3/3 | ||||
| VCaP | + | 10 | ARV7 | yes | 2 | 2/2 |
| K19 | yes | |||||
| Actin | yes | |||||
| 5 | ARV7 | yes | 1 | 1/1 | ||
| K19 | N/D | |||||
| Actin | yes |
| Tube | Cell Line | ARV7 Status | Cell Amount | Target | Detection | N | Ratio [detection/N] |
|---|---|---|---|---|---|---|---|
| EDTA | HD | − | 0 | ARV7 | N/D | 3 | 3/3 |
| K19 | N/D | ||||||
| Actin | yes | ||||||
| 22RV1 | + | 10 | ARV7 | N/D | 1 | 1/1 | |
| K19 | yes | ||||||
| Actin | yes | ||||||
| 5 | ARV7 | yes | 3 | 2/3 | |||
| K19 | yes | 2/3 | |||||
| Actin | yes | 3/3 | |||||
| Adnagen | HD | − | 0 | ARV7 | yes | 3 | 1/3 |
| K19 | yes | 2/3 | |||||
| Actin | yes | 3/3 | |||||
| 22RV1 | + | 10 | ARV7 | N/D | 1 | 1/1 | |
| K19 | N/D | ||||||
| Actin | yes | ||||||
| 5 | ARV7 | yes | 3 | 3/3 | |||
| K19 | yes | 1/3 | |||||
| Actin | yes | 3/3 |
| CellSearch | qPCR | |||||
|---|---|---|---|---|---|---|
| Sample | CTC Count | AR (nucl.) | AR (cytopl.) | ARV7 | K19 | Actin |
| UKE-1 | - | - | - | yes | yes | yes |
| UKE-2 | - | - | - | N/D | N/D | yes |
| UKE-3 | - | - | - | N/D | N/D | yes |
| UKE-4 | 0 | - | - | N/D | N/D | yes |
| UKE-5 | 0 | - | - | N/D | N/D | yes |
| UKE-6 | 0 | - | - | N/D | N/D | yes |
| UKE-7 | 0 | 0 | 0 | N/D | N/D | yes |
| UKE-8 | 1 | 0 | yes (1) | N/D | N/D | yes |
| UKE-9 | 1 | yes (1) | 0 | N/D | N/D | yes |
| UKE-10 | 1 | 0 | 0 | N/D | yes | yes |
| UKE-11 | 1 | 0 | yes (1) | N/D | yes | yes |
| UKE-12 | 1 | - | - | yes | yes | yes |
| UKE-13 | 1 | 0 | 0 | yes | yes | yes |
| UKE-14 | 2 | yes (1) | yes (1) | N/D | yes | yes |
| UKE-15 | 6 | 0 | 0 | N/D | N/D | yes |
| UKE-16 | 6 | - | - | N/D | N/D | yes |
| UKE-17 | 8 | yes (3) | yes (4) | yes | yes | yes |
| UKE-18 | 9 | 0 | yes (9) | yes | yes | yes |
| UKE-19 | 11 | 0 | yes (11) | yes | yes | yes |
| UKE-20 | 11 | 0 | yes (11) | yes | yes | yes |
| UKE-21 | 14 | - | - | N/D | yes | yes |
| UKE-22 | 16 | - | - | yes | yes | yes |
| UKE-23 | 22 | - | - | yes | N/D | yes |
| UKE-24 | 80 | - | - | yes | yes | yes |
| UKE-25 | 156 | - | - | yes | yes | yes |
| UKE-26 | 398 | - | - | yes | yes | yes |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hille, C.; Gorges, T.M.; Riethdorf, S.; Mazel, M.; Steuber, T.; Von Amsberg, G.; König, F.; Peine, S.; Alix-Panabières, C.; Pantel, K. Detection of Androgen Receptor Variant 7 (ARV7) mRNA Levels in EpCAM-Enriched CTC Fractions for Monitoring Response to Androgen Targeting Therapies in Prostate Cancer. Cells 2019, 8, 1067. https://doi.org/10.3390/cells8091067
Hille C, Gorges TM, Riethdorf S, Mazel M, Steuber T, Von Amsberg G, König F, Peine S, Alix-Panabières C, Pantel K. Detection of Androgen Receptor Variant 7 (ARV7) mRNA Levels in EpCAM-Enriched CTC Fractions for Monitoring Response to Androgen Targeting Therapies in Prostate Cancer. Cells. 2019; 8(9):1067. https://doi.org/10.3390/cells8091067
Chicago/Turabian StyleHille, Claudia, Tobias M. Gorges, Sabine Riethdorf, Martine Mazel, Thomas Steuber, Gunhild Von Amsberg, Frank König, Sven Peine, Catherine Alix-Panabières, and Klaus Pantel. 2019. "Detection of Androgen Receptor Variant 7 (ARV7) mRNA Levels in EpCAM-Enriched CTC Fractions for Monitoring Response to Androgen Targeting Therapies in Prostate Cancer" Cells 8, no. 9: 1067. https://doi.org/10.3390/cells8091067
APA StyleHille, C., Gorges, T. M., Riethdorf, S., Mazel, M., Steuber, T., Von Amsberg, G., König, F., Peine, S., Alix-Panabières, C., & Pantel, K. (2019). Detection of Androgen Receptor Variant 7 (ARV7) mRNA Levels in EpCAM-Enriched CTC Fractions for Monitoring Response to Androgen Targeting Therapies in Prostate Cancer. Cells, 8(9), 1067. https://doi.org/10.3390/cells8091067






