Binding of SU(VAR)3-9 Partially Depends on SETDB1 in the Chromosomes of Drosophila melanogaster
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Constructs
2.2. Fly Stocks
- y w attP18[mini-hsp70-Δt-Dam]; egg1473/TSTL
- y w attP18[mini-hsp70-Δt-Su(var)3-9-Dam]; egg1473/TSTL
- y w attP18[full-hsp70-t-Dam]; Df(2R)Dll-MP/Cy
- y w attP18[full-hsp70-t-Su(var)3-9-Dam]; Df(2R)Dll-MP/Cy
- y w; nanos-CRE egg1473/Cy
- y w attP18[full-hsp70-t-Dam]; +; +
- y w attP18[full-hsp70-t-Su(var)3-9-Dam]; +; +
- y w; nanos-CRE; +
2.3. Tissue-Specific DamID-Seq Technique
2.4. Availability of Data
3. Results
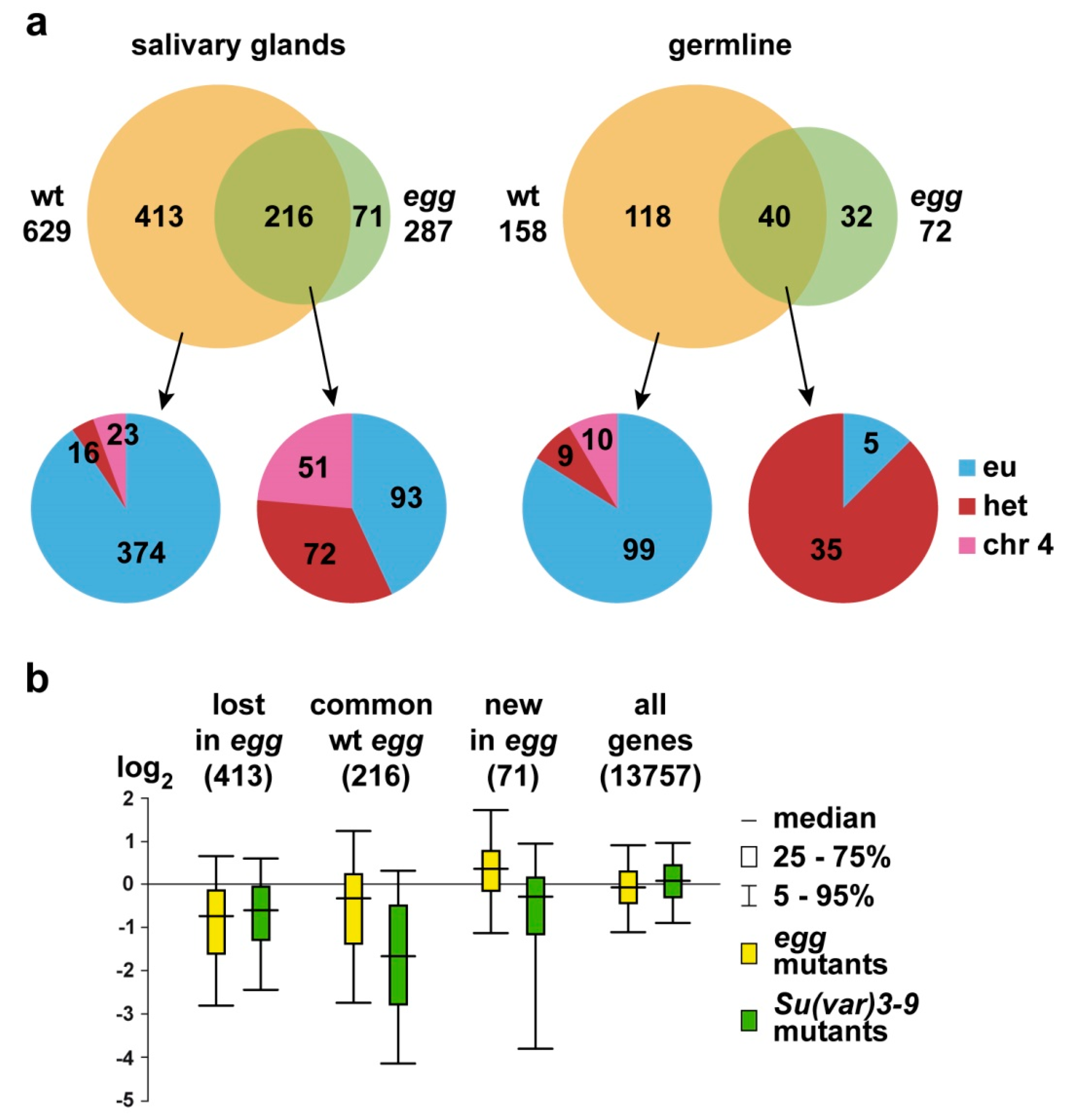
3.1. SU(VAR)3-9 Binding with Single Copy Genes is Mostly SETDB1-Dependent
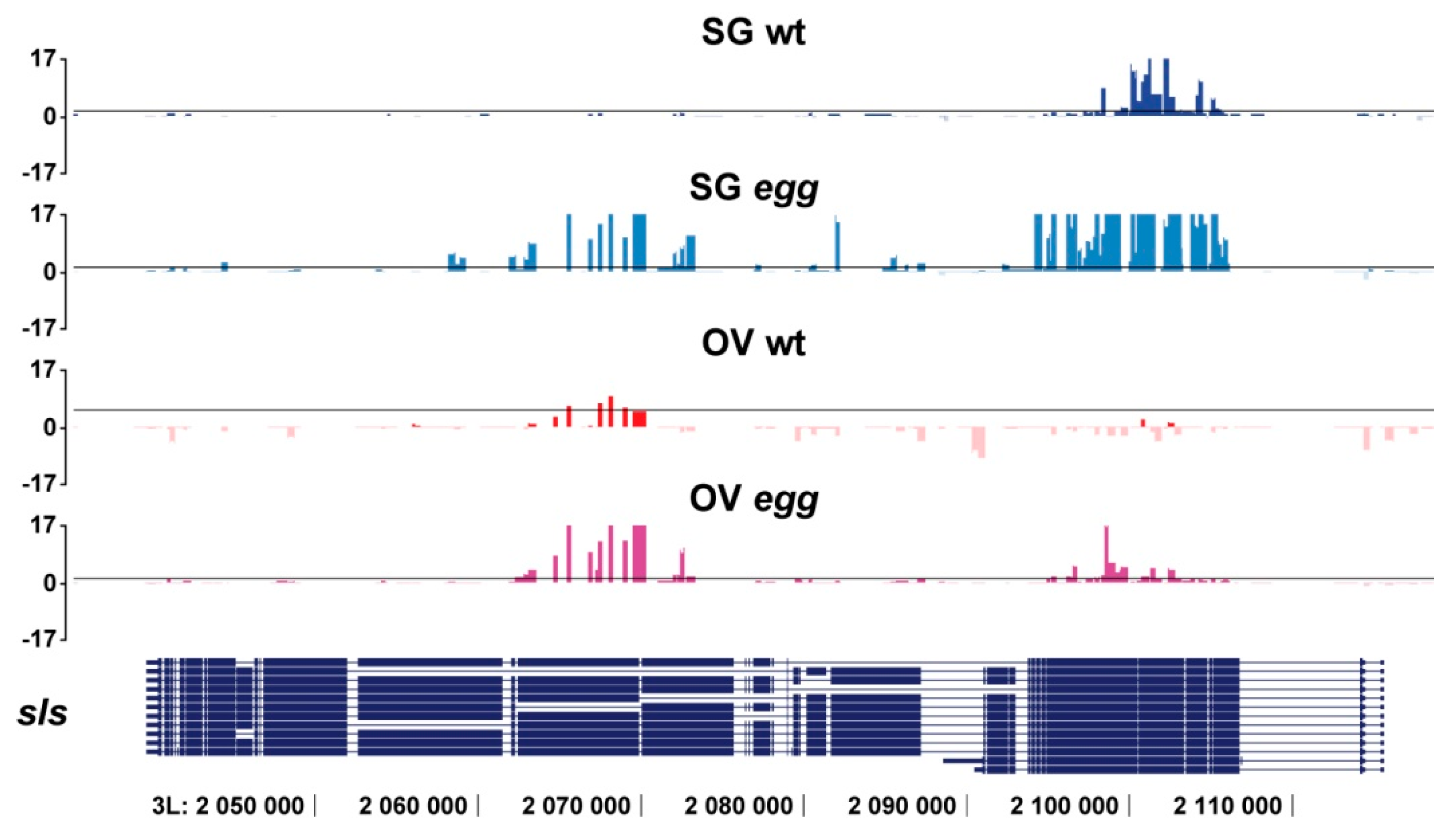
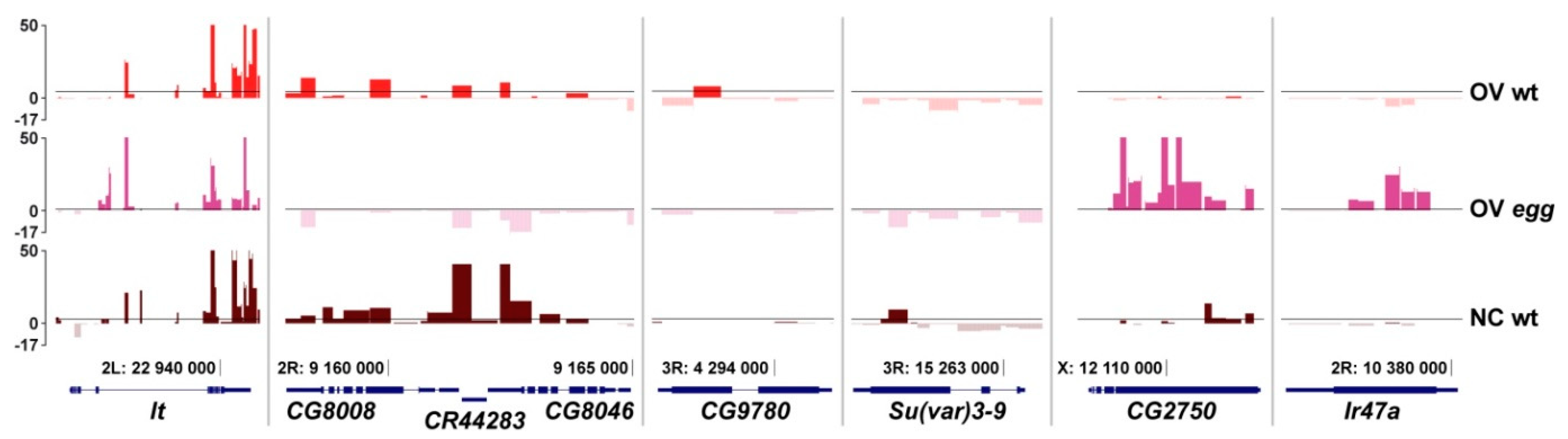
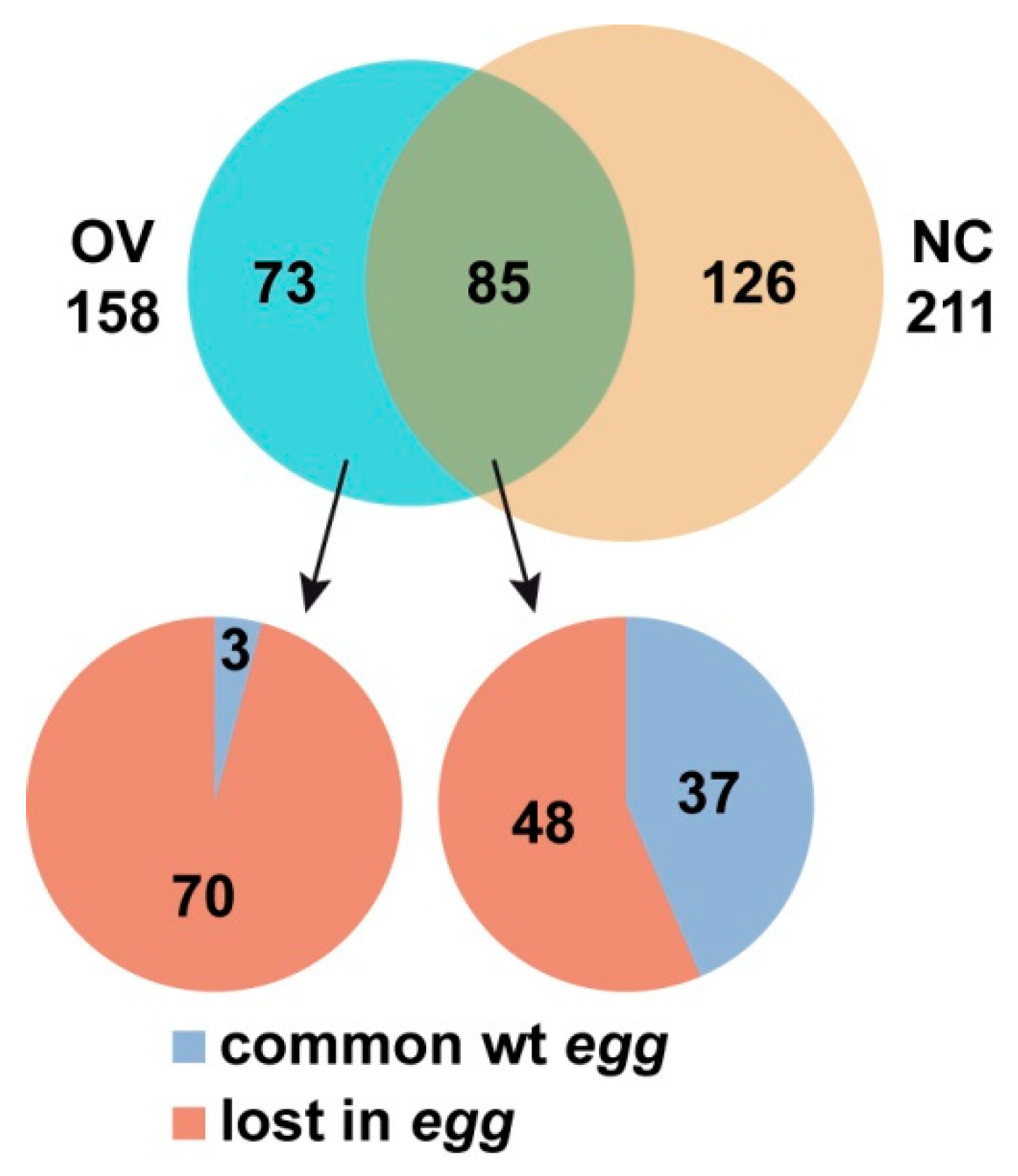
3.2. SU(VAR)3-9 Binding to the Chromosomes in the Female Germline is Dynamic throughout Development
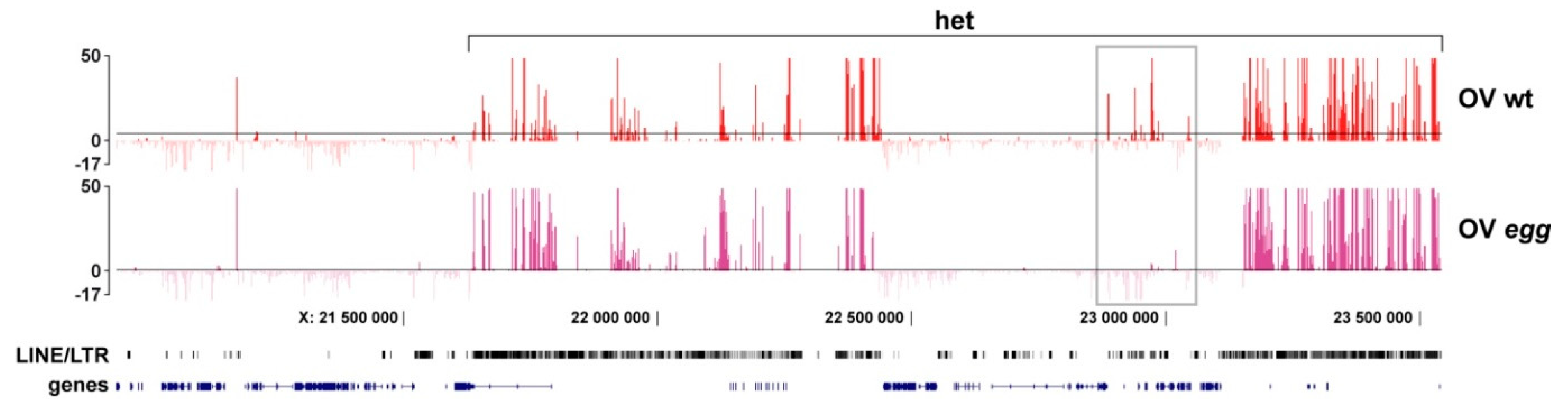
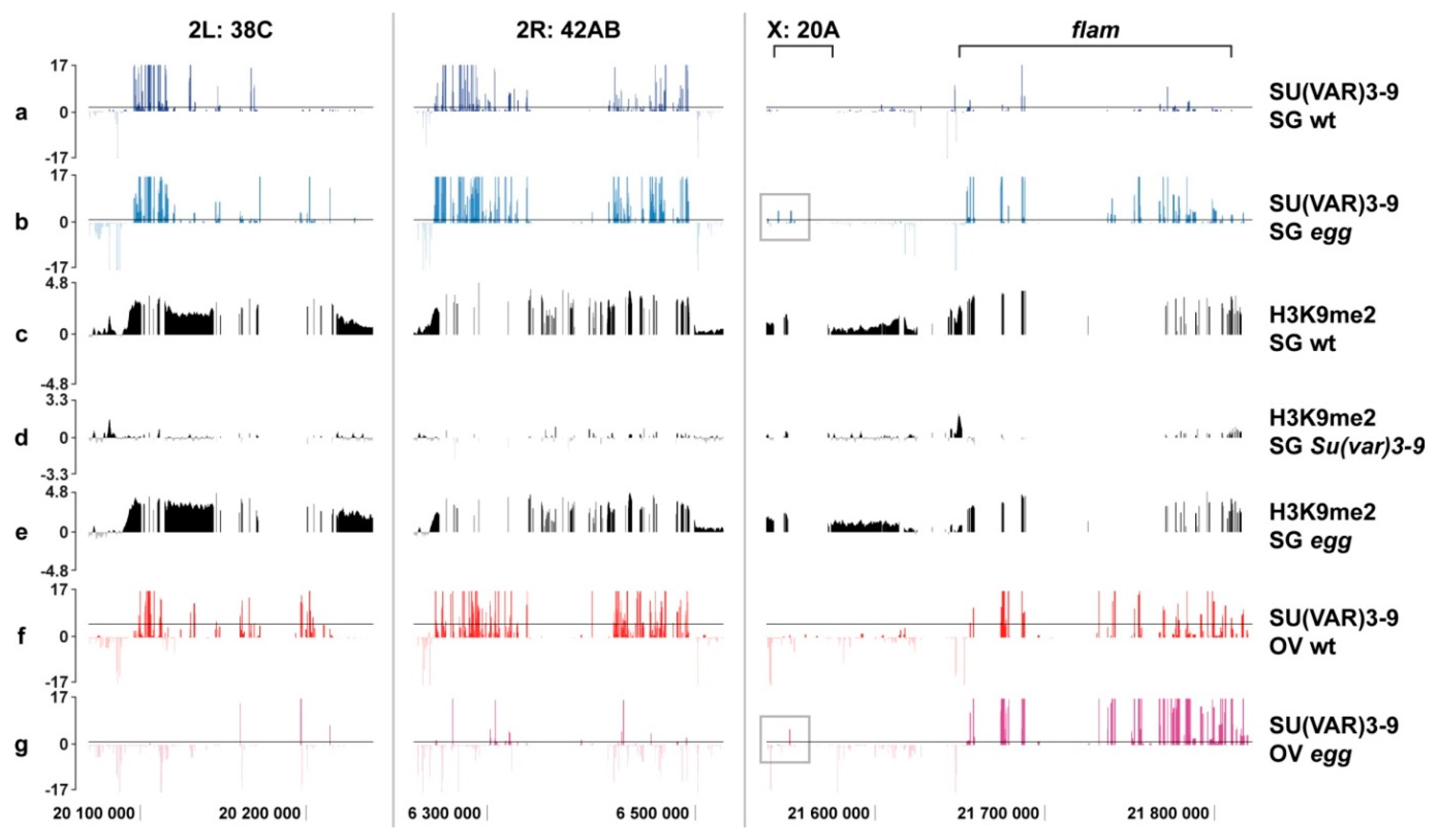
3.3. Binding of SU(VAR)3-9 to piRNA Clusters Proceeds via Distinct Mechanisms in the Somatic and Germline Cells
4. Discussion
4.1. Interplay between SU(VAR)3-9 and SETDB1
4.2. SU(VAR)3-9 and SETDB1 in the Female Germline Cells
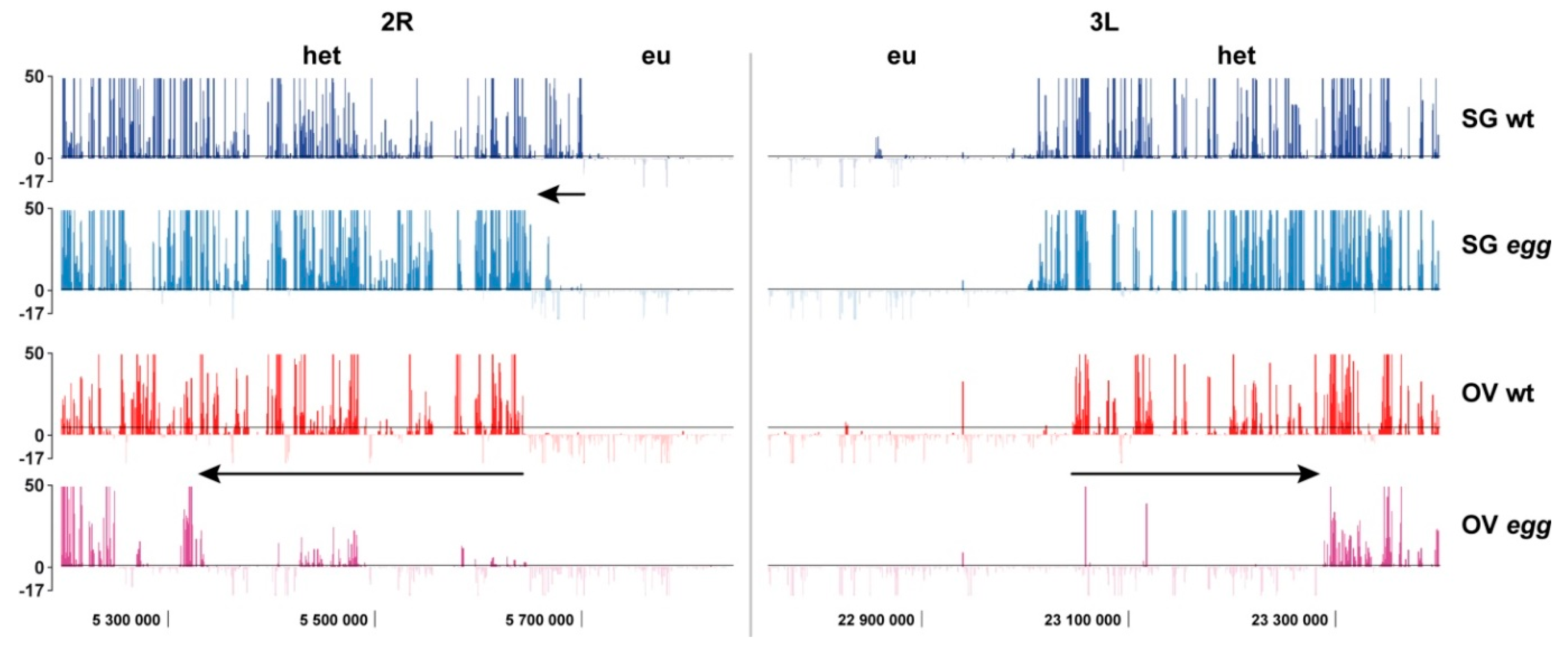
4.3. Border between Eu- and Heterochromatin
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Iyer, L.M.; Abhiman, S.; Aravind, L. Natural History of Eukaryotic DNA Methylation Systems. Prog. Mol. Biol. Transl. Sci. 2011, 101, 25–104. [Google Scholar]
- Lyko, F.; Ramsahoye, B.H.; Jaenisch, R. DNA Methylation in Drosophila melanogaster. Nature 2000, 408, 538–540. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of Chromatin by Histone Modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Copur, O.; Gorchakov, A.; Finkl, K.; Kuroda, M.I.; Muller, J. Sex-Specific Phenotypes of Histone H4 Point Mutants Establish Dosage Compensation as the Critical Function of H4k16 Acetylation in Drosophila. Proc. Natl. Acad. Sci. USA 2018, 115, 13336–13341. [Google Scholar] [CrossRef]
- Taylor, G.C.; Eskeland, R.; Hekimoglu-Balkan, B.; Pradeepa, M.M.; Bickmore, W.A. H4K16 Acetylation Marks Active Genes and Enhancers of Embryonic Stem Cells, but Does not Alter Chromatin Compaction. Genome Res. 2013, 23, 2053–2065. [Google Scholar] [CrossRef]
- Krauss, V. Glimpses of Evolution: Heterochromatic Histone H3K9 Methyltransferases Left Its Marks Behind. Genetica 2008, 133, 93–106. [Google Scholar] [CrossRef]
- Du, J.; Johnson, L.M.; Jacobsen, S.E.; Patel, D.J. DNA Methylation Pathways and Their Crosstalk with Histone Methylation. Nat. Rev. Mol. Cell. Biol. 2015, 16, 519–532. [Google Scholar] [CrossRef]
- Taverna, S.D.; Coyne, R.S.; Allis, C.D. Methylation of Histone H3 at Lysine 9 Targets Programmed DNA Elimination in Tetrahymena. Cell 2002, 110, 701–711. [Google Scholar] [CrossRef]
- Cowell, I.G.; Aucott, R.; Mahadevaiah, S.K.; Burgoyne, P.S.; Huskisson, N.; Bongiorni, S.; Prantera, G.; Fanti, L.; Pimpinelli, S.; Wu, R.; et al. Heterochromatin, HP1 and Methylation at Lysine 9 of Histone H3 in Animals. Chromosoma 2002, 111, 22–36. [Google Scholar] [CrossRef]
- Armstrong, R.L.; Penke, T.J.R.; Chao, S.K.; Gentile, G.M.; Strahl, B.D.; Matera, A.G.; McKay, D.J.; Duronio, R.J. H3K9 Promotes Under-Replication of Pericentromeric Heterochromatin in Drosophila Salivary Gland Polytene Chromosomes. Genes 2019, 10, 93. [Google Scholar] [CrossRef]
- Vakoc, C.R.; Mandat, S.A.; Olenchock, B.A.; Blobel, G.A. Histone H3 Lysine 9 Methylation and HP1gamma Are Associated with Transcription Elongation through Mammalian Chromatin. Mol. Cell 2005, 19, 381–391. [Google Scholar] [CrossRef]
- Saint-André, V.; Batsché, E.; Rachez, C.; Muchardt, C. Histone H3 Lysine 9 Trimethylation and HP1γ Favor Inclusion of Alternative Exons. Nat. Struct. Mol. Biol. 2011, 18, 337–344. [Google Scholar] [CrossRef]
- Bieberstein, N.I.; Kozáková, E.; Huranová, M.; Thakur, P.K.; Krchňáková, Z.; Krausová, M.; Oesterreich, F.C.; Staněk, D. TALE-Directed Local Modulation of H3K9 Methylation Shapes Exon Recognition. Sci. Rep. 2016, 6, 29961. [Google Scholar] [CrossRef]
- Rea, S.; Eisenhaber, F.; O’Carroll, D.; Strahl, B.D.; Sun, Z.W.; Schmid, M.; Opravil, S.; Mechtler, K.; Ponting, C.P.; Allis, C.D.; et al. Regulation of Chromatin Structure by Site-Specific Histone H3 Methyltransferases. Nature 2000, 406, 593–599. [Google Scholar] [CrossRef]
- O’Carroll, D.; Scherthan, H.; Peters, A.H.; Opravil, S.; Haynes, A.R.; Laible, G.; Rea, S.; Schmid, M.; Lebersorger, A.; Jerratsch, M.; et al. Isolation and Characterization of Suv39h2, a Second Histone H3 Methyltransferase Gene That Displays Testis-Specific Expression. Mol. Cell. Biol. 2000, 20, 9423–9433. [Google Scholar] [CrossRef]
- Baumbusch, L.O.; Thorstensen, T.; Krauss, V.; Fischer, A.; Naumann, K.; Assalkhou, R.; Schulz, I.; Reuter, G.; Aalen, R.B. The Arabidopsis thaliana Genome Contains at Least 29 Active Genes Encoding SET Domain Proteins that Can Be Assigned to Four Evolutionarily Conserved Classes. Nucleic Acids Res. 2001, 29, 4319–4333. [Google Scholar] [CrossRef]
- Schotta, G.; Ebert, A.; Krauss, V.; Fischer, A.; Hoffmann, J.; Rea, S.; Jenuwein, T.; Dorn, R.; Reuter, G. Central Role of Drosophila SU(VAR)3-9 in Histone H3-K9 Methylation and Heterochromatic Gene Silencing. EMBO J. 2002, 21, 1121–1131. [Google Scholar] [CrossRef]
- Ebert, A.; Schotta, G.; Lein, S.; Kubicek, S.; Krauss, V.; Jenuwein, T.; Reuter, G. Su(var) Genes Regulate the Balance between Euchromatin and Heterochromatin in Drosophila. Genes Dev. 2004, 18, 2973–2983. [Google Scholar] [CrossRef]
- Seum, C.; Reo, E.; Peng, H.; Rauscher, F.J., III; Spierer, P.; Bontron, S. Drosophila SETDB1 Is Required for Chromosome 4 Silencing. PLoS Genet. 2007, 3, e76. [Google Scholar] [CrossRef]
- Tzeng, T.Y.; Lee, C.H.; Chan, L.W.; Shen, C.K. Epigenetic Regulation of the Drosophila Chromosome 4 by the Histone H3K9 Methyltransferase dSETDB1. Proc. Natl. Acad. Sci. USA 2007, 104, 12691–12696. [Google Scholar] [CrossRef]
- Yoon, J.; Lee, K.S.; Park, J.S.; Yu, K.; Paik, S.G.; Kang, Y.K. dSETDB1 and SU(VAR)3-9 Sequentially Function During Germline-Stem Cell Differentiation in Drosophila melanogaster. PLoS ONE 2008, 3, e2234. [Google Scholar] [CrossRef]
- Clough, E.; Moon, W.; Wang, S.; Smith, K.; Hazelrigg, T. Histone Methylation Is Required for Oogenesis in Drosophila. Development 2007, 134, 157–165. [Google Scholar] [CrossRef]
- Wang, X.; Pan, L.; Wang, S.; Zhou, J.; McDowell, W.; Park, J.; Haug, J.; Staehling, K.; Tang, H.; Xie, T. Histone H3K9 Trimethylase Eggless Controls Germline Stem Cell Maintenance and Differentiation. PLoS Genet. 2011, 7, e1002426. [Google Scholar] [CrossRef]
- Clough, E.; Tedeschi, T.; Hazelrigg, T. Epigenetic Regulation of Oogenesis and Germ Stem Cell Maintenance by the Drosophila Histone Methyltransferase Eggless/dSetDB1. Dev. Biol. 2014, 388, 181–191. [Google Scholar] [CrossRef]
- Smolko, A.E.; Shapiro-Kulnane, L.; Salz, H.K. The H3K9 Methyltransferase SETDB1 Maintains Female Identity in Drosophila Germ Cells. Nat. Commun. 2018, 9, 4155. [Google Scholar] [CrossRef]
- Tschiersch, B.; Hofmann, A.; Krauss, V.; Dorn, R.; Korge, G.; Reuter, G. The Protein Encoded by the Drosophila Position-Effect Variegation Suppressor Gene Su(var)3-9 Combines Domains of Antagonistic Regulators of Homeotic Gene Complexes. EMBO J. 1994, 13, 3822–3831. [Google Scholar] [CrossRef]
- Mis, J.; Ner, S.S.; Grigliatti, T.A. Identification of Three Histone Methyltransferases in Drosophila: dG9a Is a Suppressor of PEV and Is Required for Gene Silencing. Mol. Gen. Genom. 2006, 275, 513–526. [Google Scholar] [CrossRef]
- Maksimov, D.A.; Laktionov, P.P.; Posukh, O.V.; Belyakin, S.N.; Koryakov, D.E. Genome-Wide Analysis of SU(VAR)3-9 Distribution in Chromosomes of Drosophila melanogaster. Chromosoma 2018, 127, 85–102. [Google Scholar] [CrossRef]
- Lundberg, L.E.; Stenberg, P.; Larsson, J. HP1a, Su(var)3-9, SETDB1 and POF Stimulate or Repress Gene Expression Depending on Genomic Position, Gene Length and Expression Pattern in Drosophila melanogaster. Nucleic Acids Res. 2013, 41, 4481–4494. [Google Scholar] [CrossRef]
- Fritsch, L.; Robin, P.; Mathieu, J.R.R.; Souidi, M.; Hinaux, H.; Rougeulle, C.; Harel-Bellan, A.; Ameyar-Zazoua, M.; Ait-Si-Ali, S. A Subset of the Histone H3 Lysine 9 Methyltransferases Suv39h1, G9a, GLP, and SETDB1 Participate in a Multimeric Complex. Mol. Cell 2010, 37, 46–56. [Google Scholar] [CrossRef]
- Rørth, P. Gal4 in the Drosophila Female Germline. Mech. Dev. 1998, 78, 113–118. [Google Scholar] [CrossRef]
- Maksimov, D.A.; Laktionov, P.P.; Belyakin, S.N. Data Analysis Algorithm for DamID-seq Profiling of Chromatin Proteins in Drosophila melanogaster. Chromosome Res. 2016, 24, 481–494. [Google Scholar] [CrossRef]
- Lyko, F.; Maleszka, R. Insects as Innovative Models for Functional Studies of DNA Methylation. Trends Genet. 2011, 27, 127–131. [Google Scholar] [CrossRef]
- Zhang, G.; Huang, H.; Liu, D.; Cheng, Y.; Liu, X.; Zhang, W.; Yin, R.; Zhang, D.; Zhang, P.; Liu, J.; et al. N6-methyladenine DNA Modification in Drosophila. Cell 2015, 161, 893–906. [Google Scholar] [CrossRef]
- Figueiredo, M.L.A.; Philip, P.; Stenberg, P.; Larsson, J. HP1a Recruitment to Promoters Is Independent of H3K9 Methylation in Drosophila melanogaster. PLoS Genet. 2012, 8, e1003061. [Google Scholar] [CrossRef]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete Small RNA-Generating Loci as Master Regulators of Transposon Activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef]
- Rangan, P.; Malone, C.D.; Navarro, C.; Newbold, S.P.; Hayes, P.S.; Sachidanandam, R.; Hannon, G.J.; Lehmann, R. piRNA Production Requires Heterochromatin Formation in Drosophila. Curr. Biol. 2011, 21, 1373–1379. [Google Scholar] [CrossRef]
- Sienski, G.; Batki, J.; Senti, K.A.; Dönertas, D.; Tirian, L.; Meixner, K.; Brennecke, J. Silencio/CG9754 Connects the Piwi-piRNA Complex to the Cellular Heterochromatin Machinery. Genes Dev. 2015, 29, 2258–2271. [Google Scholar] [CrossRef]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-Interacting RNAs: Small RNAs with Big Functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef]
- Padeken, J.; Zeller, P.; Towbin, B.; Katic, I.; Kalck, V.; Methot, S.P.; Gasser, S.M. Synergistic Lethality between BRCA1 and H3K9me2 Loss Reflects Satellite Derepression. Genes Dev. 2019, 33, 436–451. [Google Scholar] [CrossRef]
- Seller, C.A.; Cho, C.-Y.; O’Farrell, P.H. Rapid Embryonic Cell Cycles Defer the Establishment of Heterochromatin by Eggless/SetDB1 in Drosophila. Genes Dev. 2019, 33, 403–417. [Google Scholar] [CrossRef]
- Johansson, A.M.; Stenberg, P.; Pettersson, F.; Larsson, J. POF and HP1 Bind Expressed Exons, Suggesting a Balancing Mechanism for Gene Regulation. PLoS Genet. 2007, 3, e209. [Google Scholar] [CrossRef]
- O’Geen, H.; Squazzo, S.L.; Iyengar, S.; Blahnik, K.; Rinn, J.L.; Chang, H.Y.; Green, R.; Farnham, P.J. Genome-Wide Analysis of KAP1 Binding Suggests Autoregulation of KRAB-ZNFs. PLoS Genet. 2007, 3, e89. [Google Scholar] [CrossRef]
- Frietze, S.; O’Geen, H.; Blahnik, K.R.; Jin, V.X.; Farnham, P.J. ZNF274 Recruits the Histone Methyltransferase SETDB1 to the 39 Ends of ZNF Genes. PLoS ONE 2010, 5, e15082. [Google Scholar] [CrossRef]
- Yasuhara, J.C.; Wakimoto, B.T. Molecular Landscape of Modified Histones in Drosophila Heterochromatic Genes and Euchromatin-Heterochromatin Transition Zones. PLoS Genet. 2008, 4, e16. [Google Scholar] [CrossRef]
- Riddle, N.C.; Minoda, A.; Kharchenko, P.V.; Alekseyenko, A.A.; Schwartz, Y.B.; Tolstorukov, M.Y.; Gorchakov, A.A.; Jaffe, J.D.; Kennedy, C.; Linder-Basso, D.; et al. Plasticity in Patterns of Histone Modifications and Chromosomal Proteins in Drosophila Heterochromatin. Genome Res. 2011, 21, 147–163. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maksimov, D.A.; Koryakov, D.E. Binding of SU(VAR)3-9 Partially Depends on SETDB1 in the Chromosomes of Drosophila melanogaster. Cells 2019, 8, 1030. https://doi.org/10.3390/cells8091030
Maksimov DA, Koryakov DE. Binding of SU(VAR)3-9 Partially Depends on SETDB1 in the Chromosomes of Drosophila melanogaster. Cells. 2019; 8(9):1030. https://doi.org/10.3390/cells8091030
Chicago/Turabian StyleMaksimov, Daniil A., and Dmitry E. Koryakov. 2019. "Binding of SU(VAR)3-9 Partially Depends on SETDB1 in the Chromosomes of Drosophila melanogaster" Cells 8, no. 9: 1030. https://doi.org/10.3390/cells8091030
APA StyleMaksimov, D. A., & Koryakov, D. E. (2019). Binding of SU(VAR)3-9 Partially Depends on SETDB1 in the Chromosomes of Drosophila melanogaster. Cells, 8(9), 1030. https://doi.org/10.3390/cells8091030



