miR-263b Controls Circadian Behavior and the Structural Plasticity of Pacemaker Neurons by Regulating the LIM-Only Protein Beadex
Abstract
:1. Introduction
2. Results
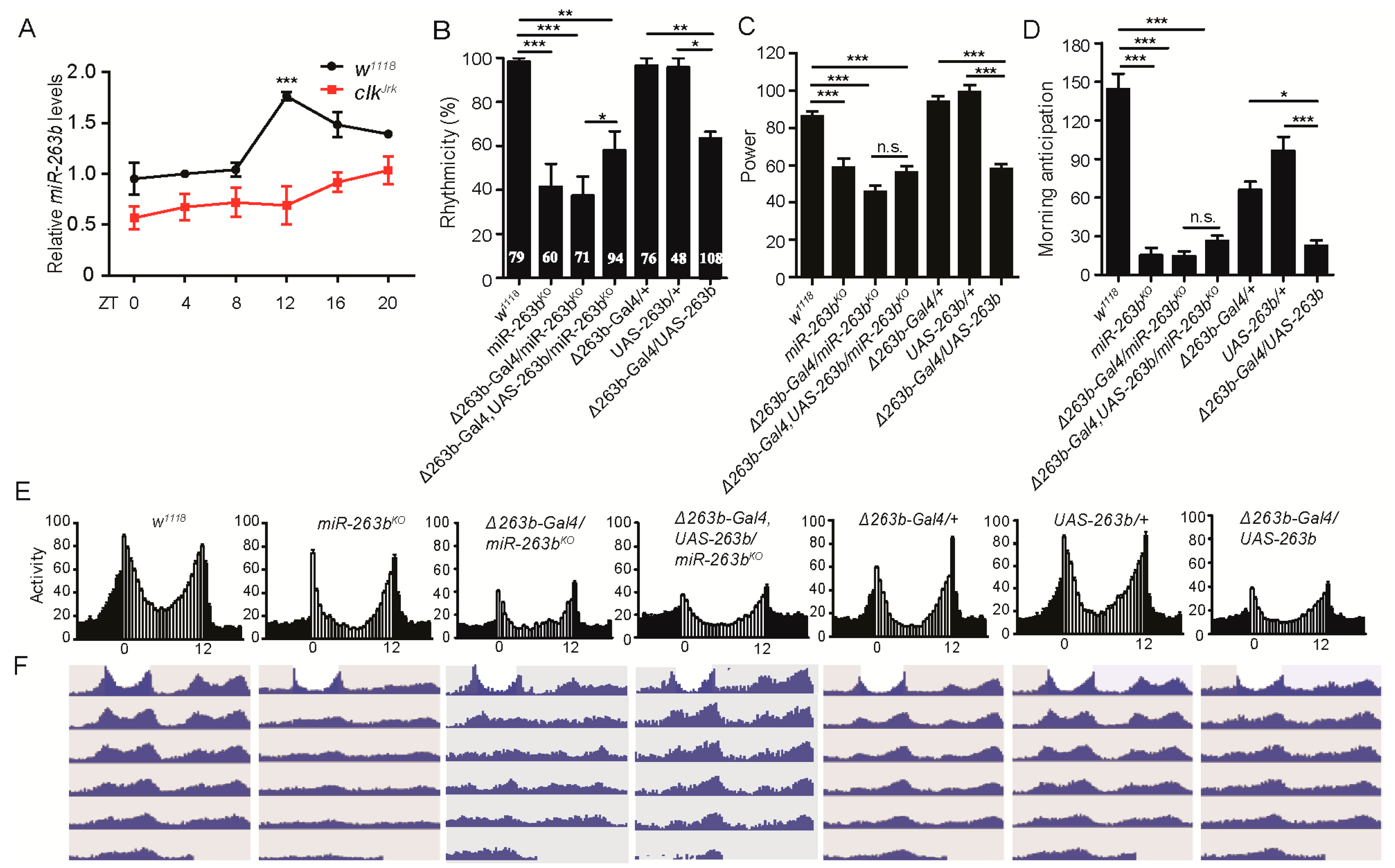
2.1. miR-263b Regulates Circadian Behavior
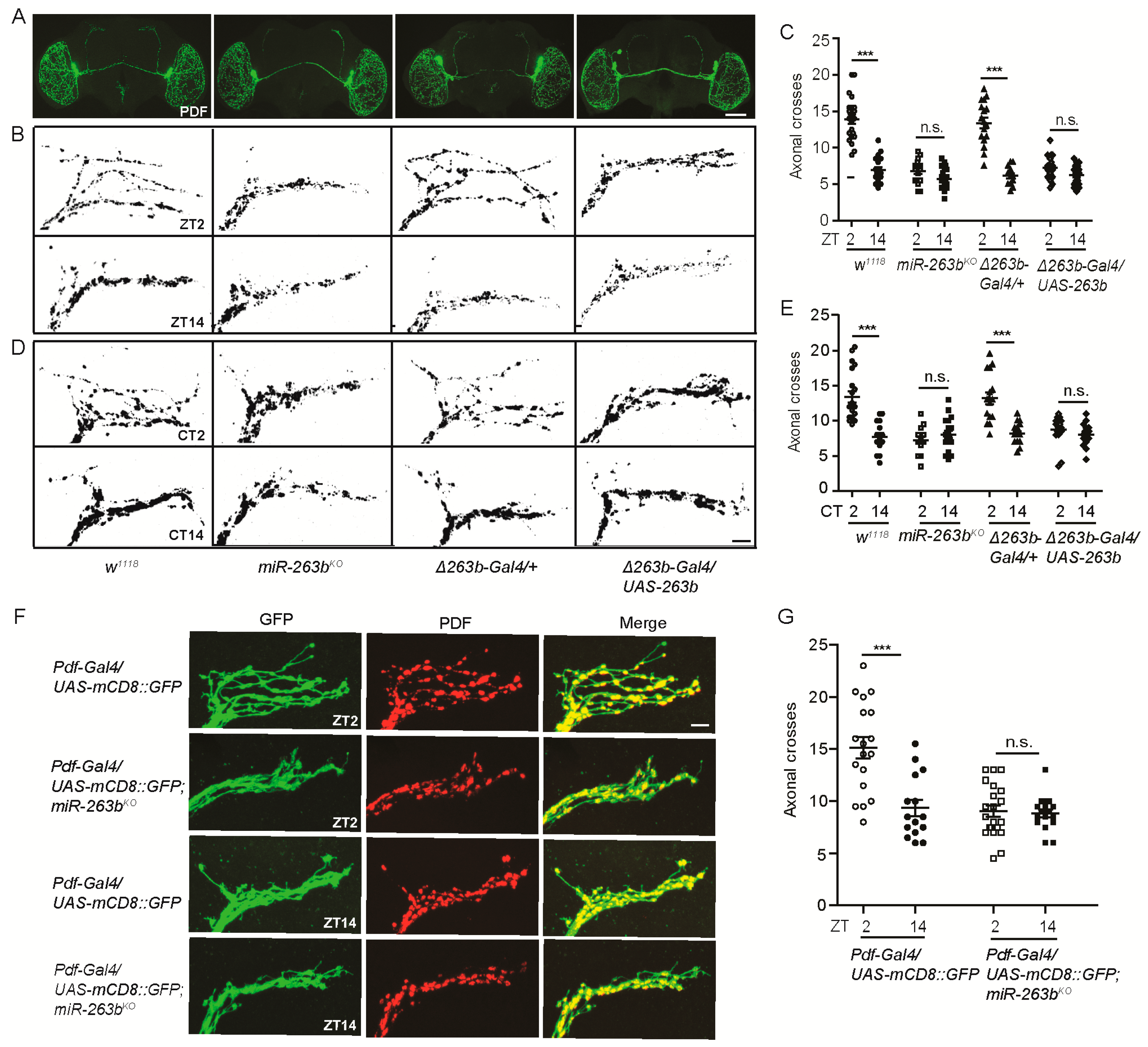
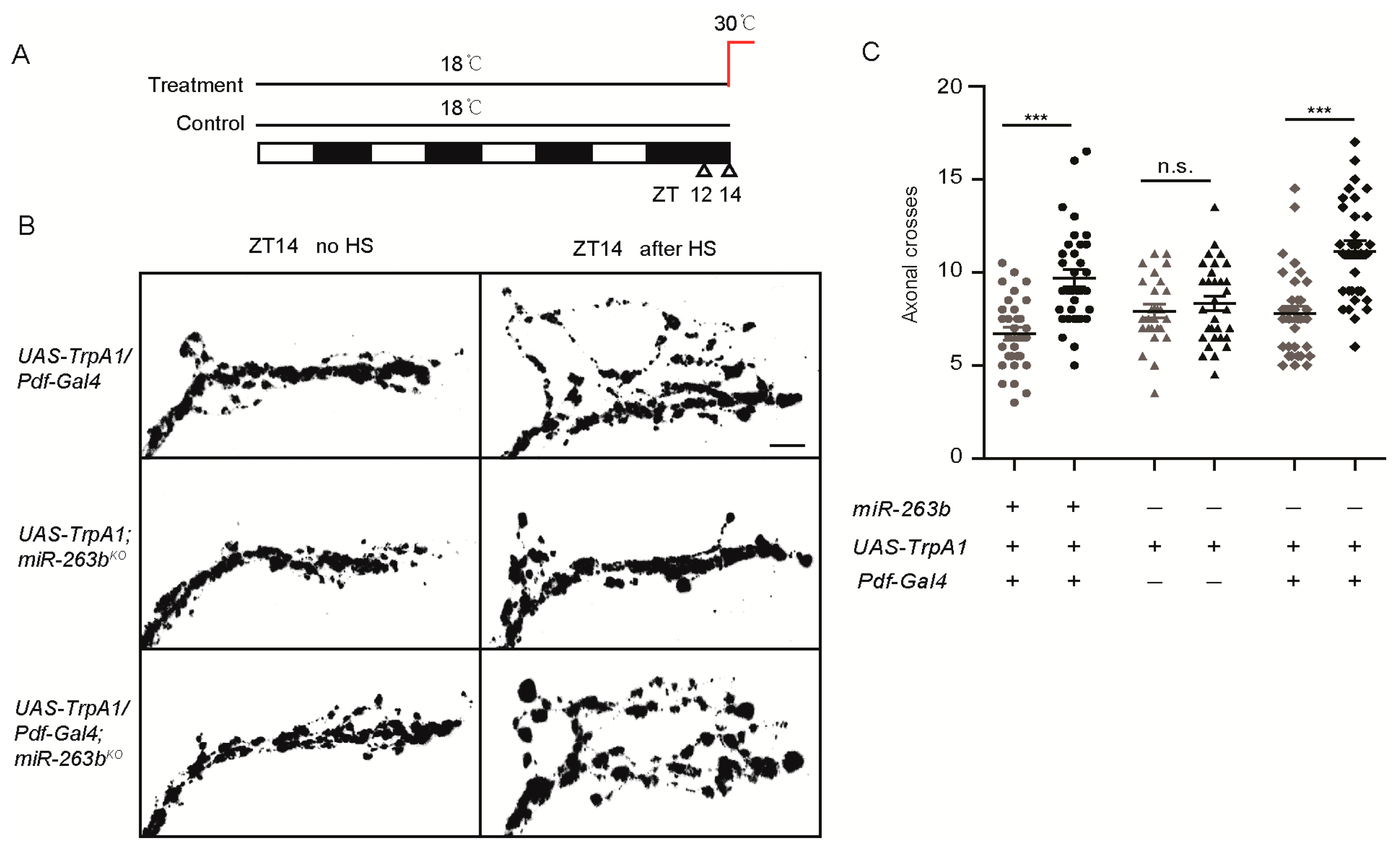
2.2. miR-263b Is Required for the Circadian Structural Plasticity of sLNvs Axonal Projections
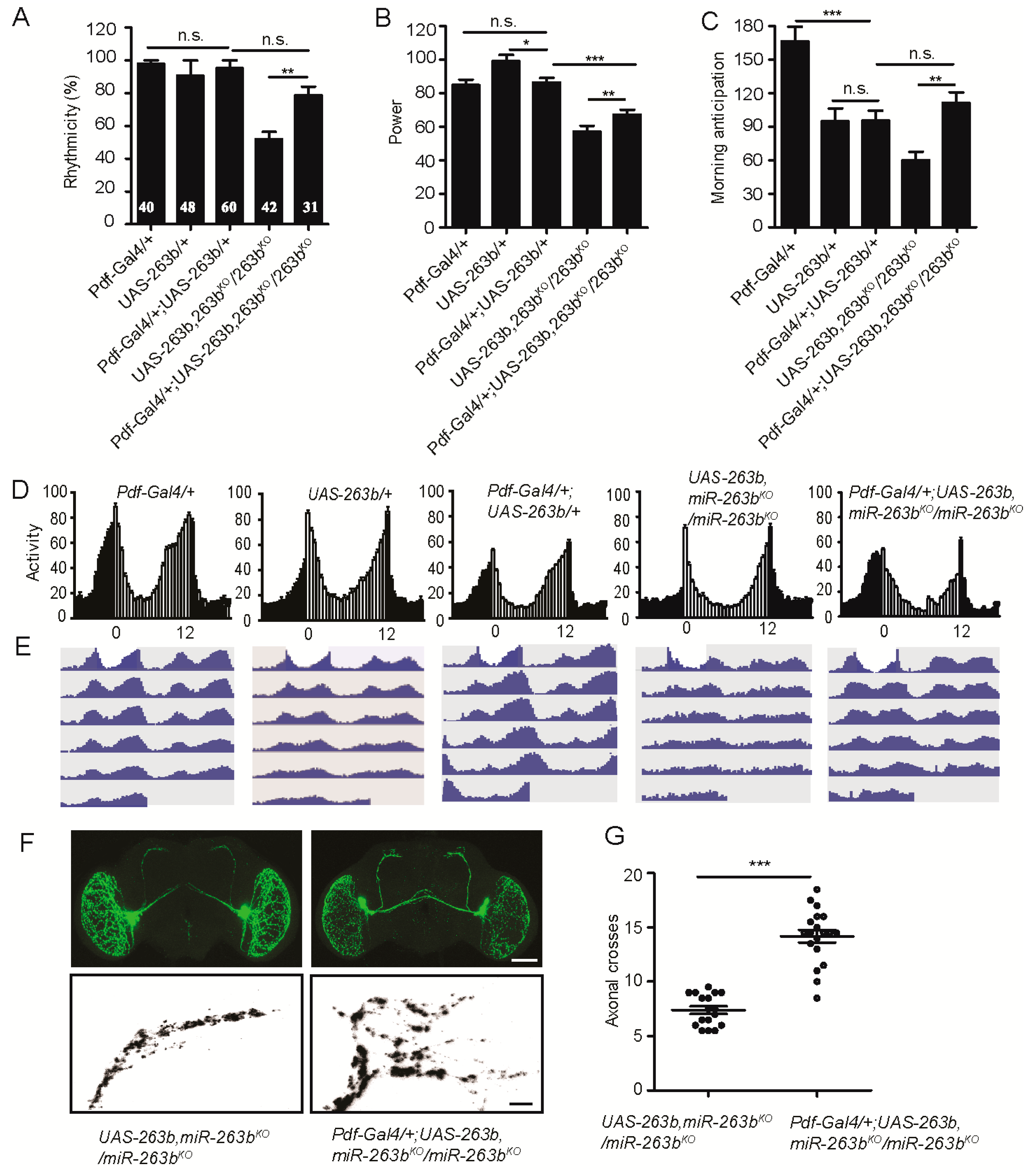
2.3. miR-263b Is Required in the PDF-Positive sLNvs for Circadian Behavior
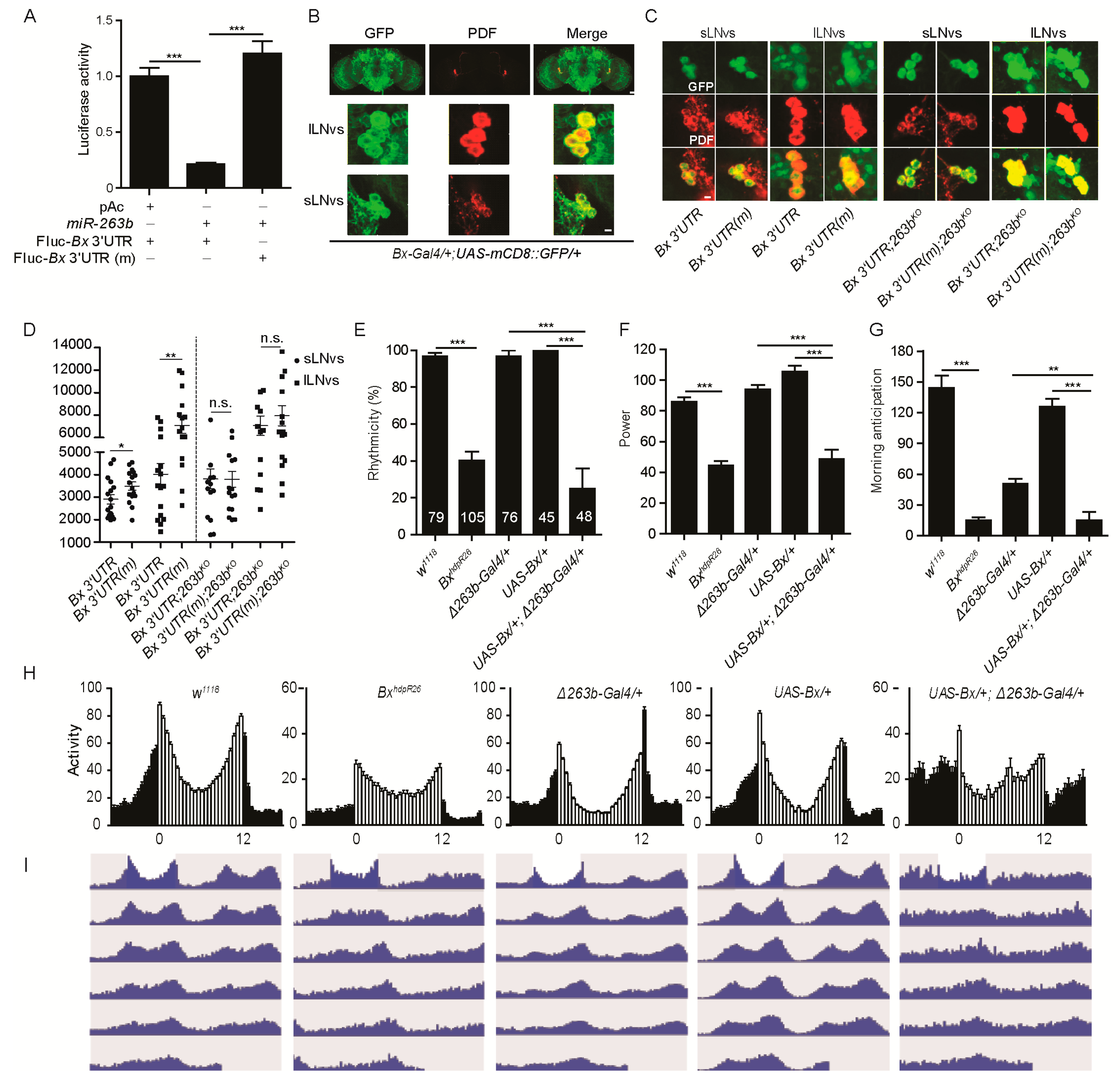
2.4. Bx Regulates Circadian Rhythms, and Its Expression Is Suppressed by miR-263b
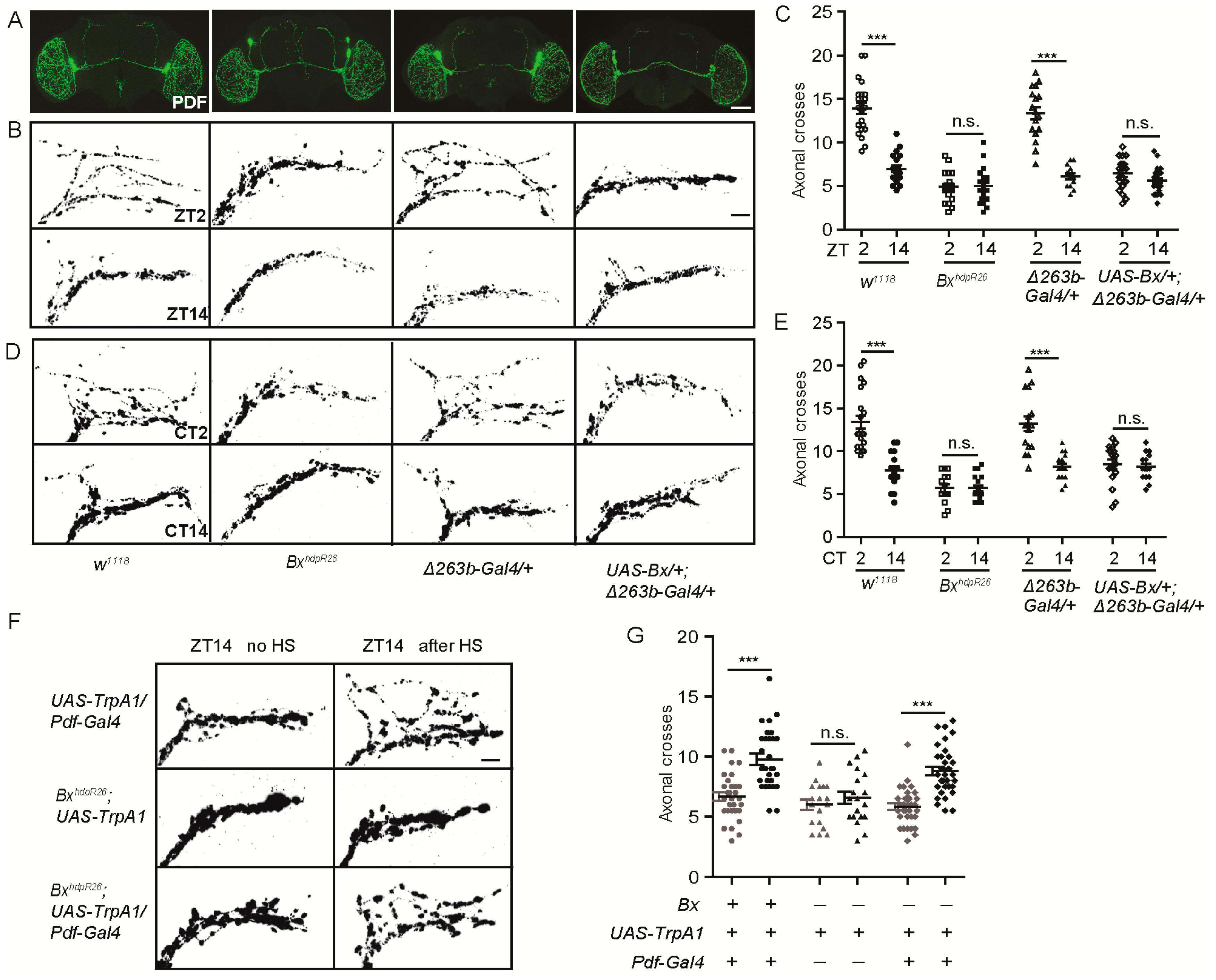
2.5. Bx Regulates the Arborization Rhythms of sLNv Dorsal Projections
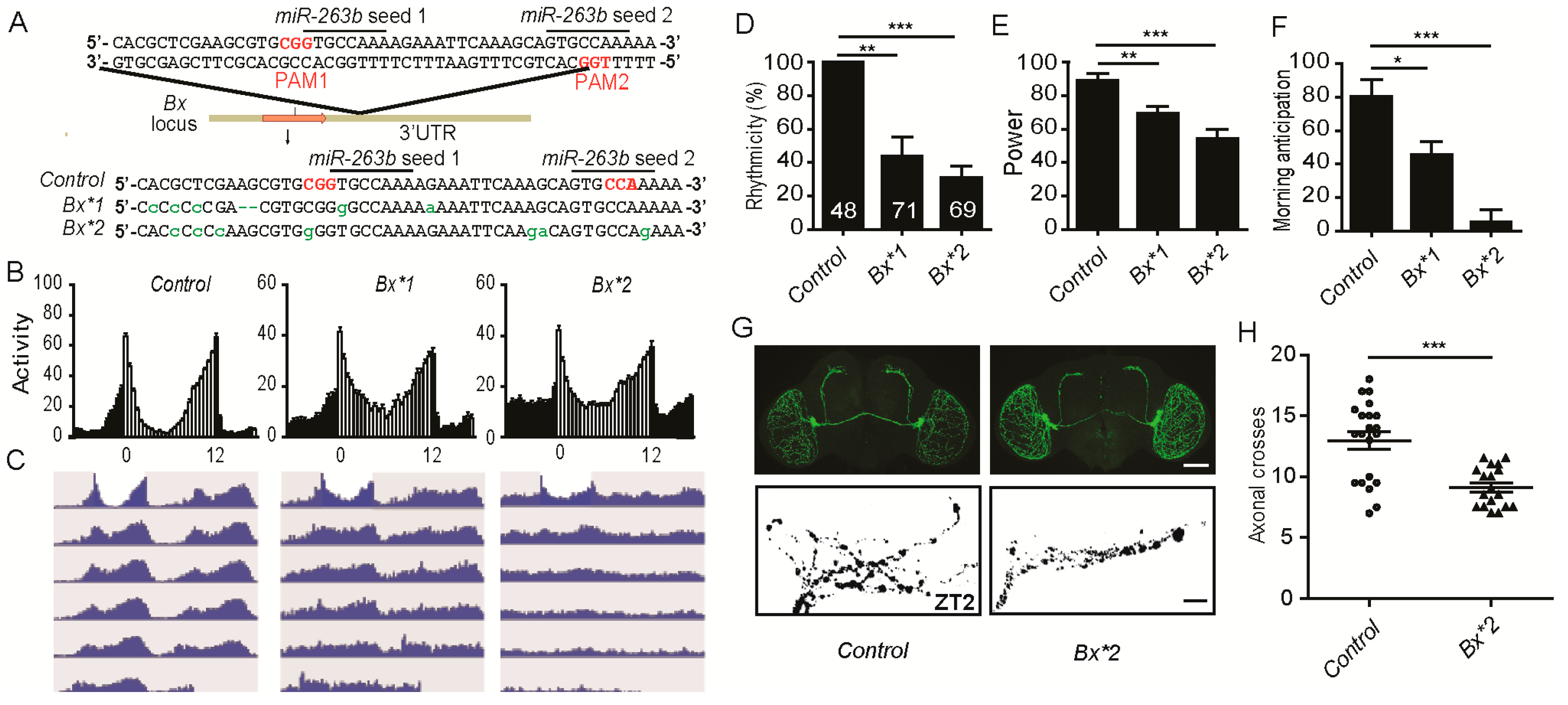
2.6. miR-263b Binding Sites in the Bx 3′ UTR Are Essential for Circadian Function of Bx
3. Discussion
4. Materials and Methods
4.1. Fly Stocks
4.2. Behavioral Experiments and Analysis
4.3. Whole-Mount Immunohistochemistry and Quantification
4.4. Quantitative Real-Time PCR
4.5. S2 Cell Luciferase Assay
4.6. Analysis of Axonal Morphology by Modified Sholl’s Method
4.7. Statistics Analysis
5. Significance Statement
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Crane, B.R.; Young, M.W. Interactive features of proteins composing eukaryotic circadian clocks. Annu. Rev. Biochem. 2014, 83, 191–219. [Google Scholar] [CrossRef] [PubMed]
- Hardin, P.E.; Panda, S. Circadian timekeeping and output mechanisms in animals. Curr. Opin. Neurobiol. 2013, 23, 724–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tataroglu, O.; Emery, P. The molecular ticks of the Drosophila circadian clock. Curr. Opin. Insect Sci. 2015, 7, 51–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubowy, C.; Sehgal, A. Circadian Rhythms and Sleep in Drosophila melanogaster. Genetics 2017, 205, 1373–1397. [Google Scholar] [CrossRef] [PubMed]
- Chiu, J.C.; Ko, H.W.; Edery, I. NEMO/NLK Phosphorylates PERIOD to Initiate a Time-Delay Phosphorylation Circuit that Sets Circadian Clock Speed. Cell 2011, 145, 357–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grima, B.; Dognon, A.; Lamouroux, A.; Chelot, E.; Rouyer, F. CULLIN-3 Controls TIMELESS Oscillations in the Drosophila Circadian Clock. PLoS Biol. 2012, 10, e10011367. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.Y.; Jeong, E.H.; Park, S.; Jeong, H.J.; Edery, I.; Cho, J.W. A role for O-GlcNAcylation in setting circadian clock speed. Genes Dev. 2012, 26, 490–502. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.F.; Li, Y.; Tang, C.H.A.; Abruzzi, K.C.; Rodriguez, J.; Pescatore, S.; Roshbash, M. CLOCK deubiquitylation by USP8 inhibits CLK/CYC transcription in Drosophila. Genes Dev. 2012, 26, 2536–2549. [Google Scholar] [CrossRef] [PubMed]
- Ko, H.W.; Jiang, J.; Edery, I. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature 2002, 420, 673–678. [Google Scholar] [CrossRef] [PubMed]
- Beckwith, E.J.; Ceriani, M.F. Experimental assessment of the network properties of the Drosophila circadian clock. J Comp. Neurol. 2015, 523, 982–996. [Google Scholar] [CrossRef] [PubMed]
- Johard, H.A.D.; Yoishii, T.; Dircksen, H.; Cusumano, P.; Rouyer, F.; Helfrich-Forster, C.; Nassel, D.R. Peptidergic Clock Neurons in Drosophila: Ion Transport Peptide and Short Neuropeptide F in Subsets of Dorsal and Ventral Lateral Neurons. J. Comp. Neurol. 2009, 516, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Nitabach, M.N.; Taghert, P.H. Organization of the Drosophila circadian control circuit. Curr. Biol. 2008, 18, R84–R93. [Google Scholar] [CrossRef] [PubMed]
- Grima, B.; Chelot, E.; Xia, R.; Rouyer, F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 2004, 431, 869–873. [Google Scholar] [CrossRef] [PubMed]
- Stoleru, D.; Peng, Y.; Agosto, J.; Rosbash, M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 2004, 431, 862–868. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Yu, J.W.; Jung, H.J.; Abruzzi, K.C.; Luo, W.F.; Griffith, L.C.; Rosbash, M. Circadian neuron feedback controls the Drosophila sleep-activity profile. Nature 2016, 536, 292. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.X.; Bilodeau-Wentworth, D.; Hardin, P.E.; Emery, P. Light and Temperature Control the Contribution of Specific DN1 Neurons to Drosophila Circadian Behavior. Curr. Biol. 2010, 20, 600–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadlapalli, S.; Jiang, C.; Bahle, A.; Reddy, P.; Eyhofer, E.M.; Shafer, O.T. Circadian clock neurons constantly monitor environmental temperature to set sleep timing. Nature 2018, 555, 98. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, H.; Head, L.M.; Ling, J.L.; Tang, X.; Liu, Y.L.; Hardin, P.E.; Emery, P.; Hamada, F.N. Circadian Rhythm of Temperature Preference and Its Neural Control in Drosophila. Curr. Biol. 2012, 22, 1851–1857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohawk, J.A.; Green, C.B.; Takahashi, J.S. Central and Peripheral Circadian Clocks in Mammals. Annu. Rev. Neurosci. 2012, 35, 445–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reppert, S.M.; Weaver, D.R. Coordination of circadian timing in mammals. Nature 2002, 418, 935–941. [Google Scholar] [CrossRef] [PubMed]
- Renn, S.C.P.; Park, J.H.; Rosbash, M.; Hall, J.C.; Taghert, P.H. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 1999, 99, 791–802. [Google Scholar] [CrossRef]
- Mertens, I.; Vandingenen, A.; Johnson, E.C.; Shafer, O.T.; Li, W.; Trigg, J.S.; Loof, A.D.; Schoofs, L.; Taghert, P.H. PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron 2005, 48, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Helfrichforster, C. The Period Clock Gene Is Expressed in Central-Nervous-System Neurons Which Also Produce a Neuropeptide That Reveals the Projections of Circadian Pacemaker Cells within the Brain of Drosophila-Melanogaster. Proc. Natl. Acad. Sci. USA 1995, 92, 612–616. [Google Scholar] [CrossRef] [PubMed]
- Helfrichforster, C.; Homberg, U. Pigment-Dispersing Hormone-Immunoreactive Neurons in the Nervous-System of Wild-Type Drosophila-Melanogaster and of Several Mutants with Altered Circadian Rhythmicity. J. Comp. Neurol. 1993, 337, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Helfrich-Forster, C.; Lee, G.; Liu, L.; Rosbash, M.; Hall, J.C. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc. Natl. Acad. Sci. USA 2000, 97, 3608–3613. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, M.P.; Berni, J.; Ceriani, M.F. Circadian remodeling of neuronal circuits involved in rhythmic behavior. PLoS Biol. 2008, 6, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Depetris-Chauvin, A.; Berni, J.; Aranovich, E.J.; Muraro, N.I.; Beckwith, E.J.; Ceriani, M.F. Adult-Specific Electrical Silencing of Pacemaker Neurons Uncouples Molecular Clock from Circadian Outputs. Curr. Biol. 2011, 21, 1783–1793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sivachenko, A.; Li, Y.; Abruzzi, K.C.; Rosbash, M. The Transcription Factor Mef2 Links the Drosophila Core Clock to Fas2, Neuronal Morphology, and Circadian Behavior. Neuron 2013, 79, 281–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Depetris-Chauvin, A.; Fernandez-Gamba, A.; Gorostiza, E.A.; Herrero, A.; Castano, E.M.; Ceriani, M.F. Mmp1 Processing of the PDF Neuropeptide Regulates Circadian Structural Plasticity of Pacemaker Neurons. PLoS Genet. 2014, 10, e1004700. [Google Scholar] [CrossRef]
- Petsakou, A.; Sapsis, T.P.; Blau, J. Circadian Rhythms in Rho1 Activity Regulate Neuronal Plasticity and Network Hierarchy. Cell 2015, 162, 823–835. [Google Scholar] [CrossRef] [Green Version]
- Gunawardhana, K.L.; Hardin, P.E. VRILLE Controls PDF Neuropeptide Accumulation and Arborization Rhythms in Small Ventrolateral Neurons to Drive Rhythmic Behavior in Drosophila. Curr. Biol. 2017, 27, 3442–3453. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Colpaert, R.M.W.; Calore, M. MicroRNAs in Cardiac Diseases. Cells 2019, 8, 737. [Google Scholar] [CrossRef] [PubMed]
- Falzone, L.; Lupo, G.; Rosa, G.R.M.L.; Crimi, S.; Anfuso, C.D.; Salemi, R.; Candido, S.; Libra, M.; Candido, S. Identification of Novel MicroRNAs and Their Diagnostic and Prognostic Significance in Oral Cancer. Cancers 2019, 11, 610. [Google Scholar] [CrossRef] [PubMed]
- Tomankova, T.; Petrek, M.; Kriegova, E. Involvement of microRNAs in physiological and pathological processes in the lung. Respir. Res. 2010, 11, 159. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Viveros, L.; Chiang, C.K.; Ong, J.L.K.; Hegazi, S.; Cheng, A.H.; Bouchard-Cannon, P.; Fana, M.; Lowden, C.; Zhang, P.; Bothorel, B. miR-132/212 Modulates Seasonal Adaptation and Dendritic Morphology of the Central Circadian Clock. Cell Rep. 2017, 19, 505–520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, Y.B.; Zhang, Y. Emerging roles for microRNA in the regulation of Drosophila circadian clock. BMC Neurosci. 2018, 19, 1. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.Y.M.; Papp, J.W.; Varlamova, O.; Dziema, H.; Russell, B.; Curfman, J.P.; Nakazawa, T.; Shimizu, K.; Okamura, H.; Impey, S. microRNA modulation of circadian-clock period and entrainment. Neuron 2007, 54, 813–829. [Google Scholar] [CrossRef]
- Yoo, S.H.; Kojima, S.; Shimomura, K.; Koike, N.; Buhr, E.D.; Furukawa, T.; Ko, C.H.; Gloston, G.; Ayoub, C.; Nohara, K.; et al. Period2 3 ‘-UTR and microRNA-24 regulate circadian rhythms by repressing PERIOD2 protein accumulation. Proc. Natl. Acad. Sci. USA 2017, 114, E8855–E8864. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.F.; Liu, Z.X.; Li, T.J.; Zhang, R.F.; Xue, Y.B.; Zhong, Y.; Bai, W.W.; Zhou, D.S.; Zhao, Z.W. Regulation of Drosophila circadian rhythms by miRNA let-7 is mediated by a regulatory cycle. Nat. Commun. 2014, 5, 5549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Rosbash, M. mir-276a strengthens Drosophila circadian rhythms by regulating timeless expression. Proc. Natl. Acad. Sci. USA 2016, 113, E2965–E2972. [Google Scholar] [CrossRef] [PubMed]
- Kadener, S.; Menet, J.S.; Sugino, K.; Horwich, M.D.; Weissbein, U.; Nawathean, P.; Vagin, V.V.; Zamore, P.D.; Nelson, S.B.; Rosbash, M. A role for microRNAs in the Drosophila circadian clock. Genes Dev. 2009, 23, 2179–2191. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Emery, P. GW182 Controls Drosophila Circadian Behavior and PDF-Receptor Signaling. Neuron 2013, 78, 152–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, W.Y.; Sehgal, A. Regulation of Circadian Behavioral Output via a MicroRNA-JAK/STAT Circuit. Cell 2012, 148, 765–779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vodala, S.; Pescatore, S.; Rodriguez, J.; Buescher, M.; Chen, Y.W.; Weng, R.F.; Cohen, S.M.; Rosbash, M. The Oscillating miRNA 959-964 Cluster Impacts Drosophila Feeding Time and Other Circadian Outputs. Cell Metab. 2012, 16, 601–612. [Google Scholar] [CrossRef] [Green Version]
- Garaulet, D.L.; Sun, K.L.; Li, W.H.; Wen, J.Y.; Panzarino, A.M.; O’Neil, J.L.; Hiesinger, P.R.; Young, M.W.; Lai, E.C. miR-124 Regulates Diverse Aspects of Rhythmic Behavior in Drosophila. J. Neurosci. 2016, 36, 3414–3421. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lamba, P.; Guo, P.Y.; Emery, P. miR-124 Regulates the Phase of Drosophila Circadian Locomotor Behavior. J. Neurosci. 2016, 36, 2007–2013. [Google Scholar] [CrossRef]
- Niu, Y.; Liu, Z.; Nian, X.; Xu, X.; Zhang, Y. miR-210 controls the evening phase of circadian locomotor rhythms through repression of Fasciclin 2. PLoS Genet. 2019, 15, e1007655. [Google Scholar] [CrossRef]
- Yang, M.; Lee, J.E.; Padgett, R.W.; Edery, I. Circadian regulation of a limited set of conserved microRNAs in Drosophila. BMC Genomics 2008, 9, 83. [Google Scholar] [CrossRef]
- Hilgers, V.; Bushati, N.; Cohen, S.M. Drosophila microRNAs 263a/b Confer Robustness during Development by Protecting Nascent Sense Organs from Apoptosis. PLoS Biol. 2010, 8, e1000396. [Google Scholar] [CrossRef]
- Gorostiza, E.A.; Depetris-Chauvin, A.; Frenkel, L.; Pirez, N.; Ceriani, M.F. Circadian Pacemaker Neurons Change Synaptic Contacts across the Day. Curr. Biol. 2014, 24, 2161–2167. [Google Scholar] [CrossRef] [Green Version]
- Hamada, F.N.; Rosenzweig, M.; Kang, K.; Pulver, S.R.; Ghezzi, A.; Jegla, T.J.; Garrity, P.A. An internal thermal sensor controlling temperature preference in Drosophila. Nature 2008, 454, 217. [Google Scholar] [CrossRef]
- Tsai, L.T.Y.; Bainton, R.J.; Blau, J.; Heberlein, U. Lmo mutants reveal a novel role for circadian pacemaker neurons in cocaine-induced behaviors. PLoS Biol. 2004, 2, 2122–2134. [Google Scholar] [CrossRef]
- Brennecke, J.; Stark, A.; Russell, R.B.; Cohen, S.M. Principles of microRNA-target recognition. PLoS Biol. 2005, 3, e85. [Google Scholar] [CrossRef]
- You, S.; Fulga, T.A.; Van Vactor, D.; Jackson, F.R. Regulation of Circadian Behavior by Astroglial MicroRNAs in Drosophila. Genetics 2018, 208, 1195–1207. [Google Scholar] [CrossRef]
- Girardet, C.; Blanchard, M.P.; Ferracci, G.; Leveque, C.; Moreno, M.; Francois-Bellan, A.M.; Becquet, D.; Bosler, O. Daily changes in synaptic innervation of VIP neurons in the rat suprachiasmatic nucleus: Contribution of glutamatergic afferents. Eur. J. Neurosci. 2010, 31, 359–370. [Google Scholar] [CrossRef]
- Clokie, S.J.H.; Lau, P.; Kim, H.H.; Coon, S.L.; Klein, D.C. MicroRNAs in the Pineal Gland miR-483 regulates melatonin synthesis by targeting arylalkylamine N-acetyltransferase. J. Biol. Chem. 2012, 287, 25312–25324. [Google Scholar] [CrossRef]
- Xu, S.B.; Witmer, P.D.; Lumayag, S.; Kovacs, B.; Valle, D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J. Biol. Chem. 2007, 282, 25053–25066. [Google Scholar] [CrossRef]
- Dambal, S.; Shah, M.; Mihelich, B.; Nonn, L. The microRNA-183 cluster: The family that plays together stays together. Nucleic Acids Res. 2015, 43, 7173–7188. [Google Scholar] [CrossRef]
- Bejarano, F.; Smibert, P.; Lai, E.C. miR-9a prevents apoptosis during wing development by repressing Drosophila LIM-only. Dev. Biol. 2010, 338, 63–73. [Google Scholar] [CrossRef] [Green Version]
- Grima, B.; Lamouroux, A.; Chelot, E.; Papin, C.; Limbourg-Bouchon, B.; Rouyer, F. The F-box protein Slimb controls the levels of clock proteins Period and Timeless. Nature 2002, 420, 178–182. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
| Genotype | N | % Rhythmic | Period (h) ± SEM | Power * ± SEM |
|---|---|---|---|---|
| w1118 | 79 | 98.5 ± 1.5 | 24.4 ± 0.1 | 86.0 ± 2.7 |
| miR-263bKO | 60 | 41.5 ± 10.4 | 24.3 ± 0.1 | 58.7 ± 4.8 |
| Δ263b-Gal4/miR-263bKO | 71 | 39.6 ± 9.0 | 24.5 ± 0.1 | 45.7 ± 3.4 |
| Δ263b-Gal4, UAS-263b/miR-263bKO | 94 | 58.0 ± 8.7 | 24.2 ± 0.1 | 56.2 ± 3.5 |
| Δ263b-Gal4/+ | 76 | 96.6 ± 3.3 | 24.5 ± 0.0 | 94.1 ± 3.7 |
| UAS-263b/+ | 48 | 95.8 ± 4.2 | 23.8 ± 0.1 | 99.2 ± 3.7 |
| Δ263b-Gal4/UAS-263b | 45 | 68.8 ± 2.6 | 24.2 ± 0.2 | 61.4 ± 4.5 |
| UAS-Bx/+ | 45 | 100.0 ± 0.0 | 24.0 ± 0.1 | 105.7 ± 3.6 |
| BxhdpR26 | 105 | 40.1 ± 4.8 | 25.7 ± 0.2 | 44.7 ± 2.8 |
| UAS-Bx/+; Δ263b-Gal4/+ | 48 | 25.1 ± 10.9 | 24.4 ± 0.4 | 48.6 ± 6.3 |
| yw, Act5C-Cas9 | 48 | 100.0 ± 0.0 | 24.4 ± 0.2 | 89.1 ± 4.0 |
| Bx*1 | 71 | 43.6 ± 11.7 | 24.1 ± 0.1 | 69.5 ± 4.3 |
| Bx*2 | 69 | 31.1 ± 6.7 | 24.3 ± 0.1 | 54.3 ± 5.4 |
| Pdf-Gal4/+ | 40 | 98.0 ± 2.0 | 24.4 ± 0.1 | 84.9 ± 3.3 |
| Pdf-Gal4/+; UAS-263b/+ | 60 | 95.2 ± 4.8 | 26.1 ± 0.1 | 86.6 ± 2.5 |
| UAS-263b, miR-263bKO/miR-263bKO | 42 | 52.1 ± 4.4 | 23.9 ± 0.1 | 57.2 ± 3.5 |
| Pdf-Gal4/+; miR-263bKO/UAS-263b, miR-263bKO | 57 | 78.8 ± 5.3 | 25.5 ± 0.1 | 67.3 ± 3.0 |
| tubulin-Gal80ts/+; Δ263b-Gal4/+ (29 °C) | 70 | 91.8 ± 3.4 | 24.0 ± 0.1 | 78.0 ± 3.5 |
| UAS-263b/+ (29 °C) | 48 | 89.9 ± 1.7 | 23.8 ± 0.1 | 90.6 ± 4.3 |
| UAS-Bx/+ (29 °C) | 31 | 85.6 ± 8.9 | 23.9 ± 0.0 | 88.3 ± 3.5 |
| tubulin-Gal80ts/+; Δ263b-Gal4, UAS-263b (29 °C) | 70 | 50.0 ± 7.4 | 24.3 ± 0.1 | 56.3 ± 4.1 |
| tubulin-Gal80ts/UAS-Bx; Δ263b-Gal4/+ (29 °C) | 24 | 41.6 ± 8.3 | 24.5 ± 0.2 | 48.6 ± 6.7 |
| tubulin-Gal80ts/+; Δ263b-Gal4/+ (18 °C) | 62 | 90.4 ± 6.6 | 23.6 ± 0.1 | 103.9 ± 6.5 |
| UAS-263b/+ (18 °C) | 68 | 83.5 ± 4.9 | 23.7 ± 0.1 | 72.4 ± 4.9 |
| UAS-Bx/+ (18 °C) | 25 | 85.4 ± 1.4 | 24.1 ± 0.2 | 57.4 ± 5.7 |
| tubulin-Gal80ts/+; Δ263b-Gal4, UAS-263b (18 °C) | 50 | 66.9 ± 3.5 | 24.5 ± 0.4 | 55.5 ± 3.9 |
| tubulin-Gal80ts/UAS-Bx; Δ263b-Gal4/+ (18 °C) | 25 | 68.3 ± 4.3 | 23.7 ± 0.2 | 41.3 ± 4.9 |
| Bereft24 | 70 | 53.2 ± 2.3 | 23.8 ± 0.1 | 41.8 ± 2.0 |
| Δ263a-Gal4/+ | 68 | 96.7 ± 3.3 | 24.2 ± 0.1 | 100.8 ± 5.5 |
| Δ263a-Gal4/UAS-263a | 86 | 79.7 ± 4.9 | 23.8 ± 0.1 | 69.6 ± 4.5 |
| Genotype (Time Point) | N | Genotype (Time Point) | N |
|---|---|---|---|
| w1118 (ZT2) | 22 | w1118 (ZT14) | 20 |
| w1118 (CT2) | 21 | w1118 (CT14) | 15 |
| miR-263bKO (ZT2) | 18 | miR-263bKO (ZT14) | 21 |
| miR-263bKO (CT2) | 15 | miR-263bKO (CT14) | 18 |
| BxhdpR26 (ZT2) | 21 | BxhdpR26 (ZT14) | 20 |
| BxhdpR26 (CT2) | 15 | BxhdpR26 (CT14) | 18 |
| Δ263b-Gal4/+ (ZT2) | 17 | Δ263b-Gal4/+ (ZT14) | 14 |
| Δ263b-Gal4/+ (CT2) | 16 | Δ263b-Gal4/+ (CT14) | 17 |
| Δ263b-Gal4/UAS-263b (ZT2) | 19 | Δ263b-Gal4/UAS-263b (ZT14) | 20 |
| Δ263b-Gal4/UAS-263b (CT2) | 16 | Δ263b-Gal4/UAS-263b (CT14) | 15 |
| UAS-Bx/+; Δ263b-Gal4/+ (ZT2) | 22 | UAS-Bx/+; Δ263b-Gal4/+ (ZT14) | 23 |
| UAS-Bx/+; Δ263b-Gal4/+ (CT2) | 17 | UAS-Bx/+; Δ263b-Gal4/+ (CT14) | 17 |
| Pdf-Gal4/UAS-mCD8::GFP (ZT2) | 18 | Pdf-Gal4/UAS-mCD8::GFP (ZT14) | 15 |
| Pdf-Gal4/UAS-mCD8::GFP;miR-263bKO (ZT2) | 21 | Pdf-Gal4/UAS-mCD8::GFP;miR-263bKO (ZT14) | 17 |
| yw, Act5C-Cas9 (ZT2) | 21 | Bx*2 (ZT2) | 18 |
| UAS-263b, miR-263bKO/miR-263bKO (ZT2) | 17 | Pdf-Gal4/+; miR-263bKO,UAS-263b/miR-263bKO(ZT2) | 19 |
| Pdf-Gal4/UAS-TrpA1 (ZT14 no HS) | 31 | Pdf-Gal4/UAS-TrpA1 (ZT14 after HS) | 34 |
| UAS-TrpA1; miR-263bKO (ZT14 no HS) | 26 | UAS-TrpA1; miR-263bKO (ZT14 after HS) | 31 |
| Pdf-Gal4/UAS-TrpA1; miR-263bKO (ZT14 no HS) | 33 | Pdf-Gal4/UAS-TrpA1; miR-263bKO (ZT14 after HS) | 33 |
| BxhdpR26; UAS-TrpA1 (ZT14 no HS) | 18 | BxhdpR26; UAS-TrpA1 (ZT14 after HS) | 19 |
| BxhdpR26; Pdf-Gal4/UAS-TrpA1 (ZT14 no HS) | 34 | BxhdpR26; Pdf-Gal4/UAS-TrpA1 (ZT14 after HS) | 33 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nian, X.; Chen, W.; Bai, W.; Zhao, Z.; Zhang, Y. miR-263b Controls Circadian Behavior and the Structural Plasticity of Pacemaker Neurons by Regulating the LIM-Only Protein Beadex. Cells 2019, 8, 923. https://doi.org/10.3390/cells8080923
Nian X, Chen W, Bai W, Zhao Z, Zhang Y. miR-263b Controls Circadian Behavior and the Structural Plasticity of Pacemaker Neurons by Regulating the LIM-Only Protein Beadex. Cells. 2019; 8(8):923. https://doi.org/10.3390/cells8080923
Chicago/Turabian StyleNian, Xiaoge, Wenfeng Chen, Weiwei Bai, Zhangwu Zhao, and Yong Zhang. 2019. "miR-263b Controls Circadian Behavior and the Structural Plasticity of Pacemaker Neurons by Regulating the LIM-Only Protein Beadex" Cells 8, no. 8: 923. https://doi.org/10.3390/cells8080923
APA StyleNian, X., Chen, W., Bai, W., Zhao, Z., & Zhang, Y. (2019). miR-263b Controls Circadian Behavior and the Structural Plasticity of Pacemaker Neurons by Regulating the LIM-Only Protein Beadex. Cells, 8(8), 923. https://doi.org/10.3390/cells8080923





