The Prospect and Challenges to the Flow of Liquid Biopsy in Africa
Abstract
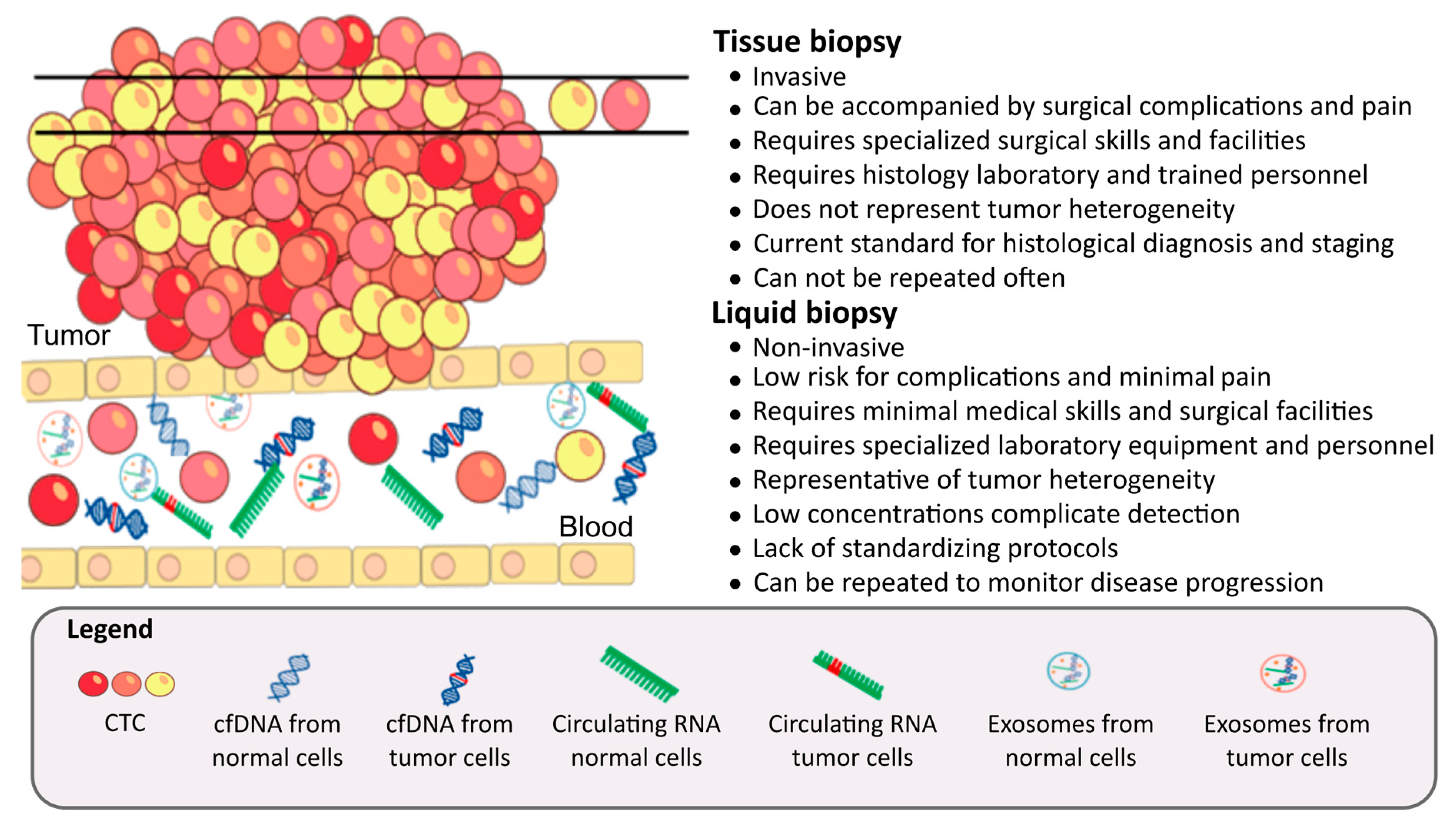
1. Introduction
2. Component of Liquid Biopsy
2.1. Circulating Tumor Cells
2.2. Circulating Tumor DNA
2.3. Circulating Tumor RNAs
2.3.1. Circulating Coding RNA
2.3.2. Long Non-Coding RNAs
2.3.3. Circulating microRNAs
2.4. Other Circulating Molecules
2.4.1. Exosomes
2.4.2. Circulating Proteins and Peptides
3. Challenges to Implementation of Liquid Biopsy Technology in Africa
4. The Prospect for Liquid Biopsy in Africa
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Jemal, A.; Center, M.M.; DeSantis, C.; Ward, E.M. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidem. Biomar. 2010, 19, 1893–1907. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.R.; Boniol, M.; Joubert, C.; Hery, C.; Haukka, J.; Autier, P.; Nishino, Y.; Sobue, T.; Chen, C.J.; You, S.L.; et al. Secular trends in breast cancer mortality in five east asian populations: Hong kong, japan, korea, singapore and taiwan. Cancer Sci. 2010, 101, 1241–1246. [Google Scholar] [CrossRef] [PubMed]
- Youlden, D.R.; Cramb, S.M.; Dunn, N.A.; Muller, J.M.; Pyke, C.M.; Baade, P.D. The descriptive epidemiology of female breast cancer: An international comparison of screening, incidence, survival and mortality. Cancer Epidemiol. 2012, 36, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Vorobiof, D.A.; Abratt, R. The cancer burden in africa. S. Afr. Med. J. 2007, 97, 937–939. [Google Scholar] [PubMed]
- Ibrahim, N.A.; Oludara, M.A. Socio-demographic factors and reasons associated with delay in breast cancer presentation: A study in nigerian women. Breast 2012, 21, 416–418. [Google Scholar] [CrossRef] [PubMed]
- Ouasmani, F.; Hanchi, Z.; Haddou Rahou, B.; Bekkali, R.; Ahid, S.; Mesfioui, A. Determinants of patient delay in seeking diagnosis and treatment among moroccan women with cervical cancer. Obstet. Gynecol. Int. 2016, 2016, 4840762. [Google Scholar] [CrossRef]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef]
- Cheung, A.H.; Chow, C.; To, K.F. Latest development of liquid biopsy. J. Thorac. Dis. 2018, 10, S1645–S1651. [Google Scholar] [CrossRef]
- Wong, A.I.; Lo, Y.D. Noninvasive fetal genomic, methylomic, and transcriptomic analyses using maternal plasma and clinical implications. Trends Mol. Med. 2015, 21, 98–108. [Google Scholar] [CrossRef]
- Esteller, M.; Sanchez-Cespedes, M.; Rosell, R.; Sidransky, D.; Baylin, S.B.; Herman, J.G. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res. 1999, 59, 67–70. [Google Scholar] [PubMed]
- Kopreski, M.S.; Benko, F.A.; Kwee, C.; Leitzel, K.E.; Eskander, E.; Lipton, A.; Gocke, C.D. Detection of mutant k-ras DNA in plasma or serum of patients with colorectal cancer. Br. J. Cancer 1997, 76, 1293–1299. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Cespedes, M.; Monzo, M.; Rosell, R.; Pifarre, A.; Calvo, R.; Lopez-Cabrerizo, M.P.; Astudillo, J. Detection of chromosome 3p alterations in serum DNA of non-small-cell lung cancer patients. Ann. Oncol. 1998, 9, 113–116. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Nakamori, S.; Ohzato, H.; Oshima, S.; Aoki, T.; Higaki, N.; Sugimoto, K.; Akagi, K.; Fujiwara, Y.; Nishisho, I.; et al. Detection of k-ras gene mutations in plasma DNA of patients with pancreatic adenocarcinoma: Correlation with clinicopathological features. Clin. Cancer Res. 1998, 4, 1527–1532. [Google Scholar] [PubMed]
- Adeola, H.A.; Soares, N.C.; Paccez, J.D.; Kaestner, L.; Blackburn, J.M.; Zerbini, L.F. Discovery of novel candidate urinary protein biomarkers for prostate cancer in a multiethnic cohort of south african patients via label-free mass spectrometry. Proteomics Clin. Appl. 2015, 9, 597–609. [Google Scholar] [CrossRef] [PubMed]
- Montaser, L.M.; Sonbol, A.A.E.; EL-Gammal, A.S.; El-Yazeed, A.S.A. Role of circulating endothelial progenitor cells in patients with breast cancer. Int. J. Curr. Res. 2018, 10, 65250–65256. [Google Scholar]
- Elnagdy, M.H.; Farouk, O.; Seleem, A.K.; Nada, H.A. Tff1 and tff3 mrnas are higher in blood from breast cancer patients with metastatic disease than those without. J. Oncol. 2018, 2018, 4793498. [Google Scholar] [CrossRef]
- Sayed, M.; Zahran, A.M.; Hassan, M.S.F.; Mohamed, D.O. Circulating tumor cells and cancer stem cells: Clinical implications in nonmetastatic breast cancer. Breast Cancer Basic Clin. Res. 2016, 10, 197. [Google Scholar] [CrossRef]
- Bahnassy, A.A.; Zekri, A.R.; El-Bastawisy, A.; Fawzy, A.; Shetta, M.; Hussein, N.; Omran, D.; Ahmed, A.A.; El-Labbody, S.S. Circulating tumor and cancer stem cells in hepatitis c virus-associated liver disease. World J. Gastroenterol. 2014, 20, 18240–18248. [Google Scholar] [CrossRef]
- Mansour, A.H.; Elkhodary, T.R.; Anwar, R.; Habeeb, M.R.; Mohammed, M.A. Regulation of cancer stem cell marker (cd133) by transforming growth factor beta in hepatocellular carcinoma. Int. J. Cancer 2014, 10, 65–73. [Google Scholar]
- Zedan, A.; Zahran, A.M.; Maximos, D.W.; Hassan, M.F.S. The role of circulating tumor cells (ctcs) in predicting the response of primary (neoadjuvant) chemotherapy and its impact as a prognostic factor in early breast cancer. SECI Oncol. 2014, 2, 68–75. [Google Scholar] [CrossRef]
- Ebeed, S.A.; El-Moneim, N.; Saad, A.; Zaher, E.R.; Yassin, O.G.; Khamis, S.A. Diagnostic and prognostic value of circulating tumor cells in female breast cancer patients. Alexandria Med. J. 2012, 48, 197–206. [Google Scholar] [CrossRef][Green Version]
- Teama, S.H.; Agwa, S.H.A. Detection of circulating tumor cells by nested rt-pcr targeting egfr/cea/ck20mrnas in colorectal carcinoma patients. Egypt. J. Med. Hum. Genet. 2010, 11, 173–180. [Google Scholar] [CrossRef][Green Version]
- Zehentner, B.K.; Persing, D.H.; Deme, A.; Toure, P.; Hawes, S.E.; Brooks, L.; Feng, Q.; Hayes, D.C.; Critichlow, C.W.; Houghton, R.L.; et al. Mammaglobin as a novel breast cancer biomarker: Multigene reverse transcription-pcr assay and sandwich elisa. Clin. Chem. 2004, 50, 2069–2076. [Google Scholar] [CrossRef] [PubMed]
- Hanekom, G.; Stubbings, H.; Johnson, C.; Kidson, S. The detection of circulating melanoma cells correlates with tumour thickness and ulceration but is not predictive of metastasis for patients with primary melanoma. Melanoma Res. 1999, 9, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Marchio, A.; Amougou Atsama, M.; Bere, A.; Komas, N.P.; Noah Noah, D.; Atangana, P.J.A.; Camengo-Police, S.M.; Njouom, R.; Bekondi, C.; Pineau, P. Droplet digital pcr detects high rate of tp53 r249s mutants in cell-free DNA of middle african patients with hepatocellular carcinoma. Clin. Exp. Med. 2018, 18, 421–431. [Google Scholar] [CrossRef]
- Hussein, N.A.; Mohamed, S.N.; Ahmed, M.A. Plasma alu-247, alu-115, and cfdna integrity as diagnostic and prognostic biomarkers for breast cancer. Appl. Biochem. Biotech. 2018, 187, 1028–1045. [Google Scholar] [CrossRef]
- Soliman, S.E.-S.; Alhanafy, A.M.; Habib, M.S.E.d.; Hagag, M.; Ibrahem, R.A.L. Serum circulating cell free DNA as potential diagnostic and prognostic biomarker in non small cell lung cancer. BB Rep. 2018, 15, 45–51. [Google Scholar] [CrossRef]
- Fawzy, A.; Sweify, K.M.; El-Fayoumy, H.M.; Nofal, N. Quantitative analysis of plasma cell-free DNA and its DNA integrity in patients with metastatic prostate cancer using alu sequence. J. Egypt. Natl. Canc. Inst. 2016, 28, 235–242. [Google Scholar] [CrossRef]
- Ibrahim, I.H.; Kamel, M.M.; Ghareeb, M. Circulating DNA in egyptian women with breast cancer. Asian Pac. J. Cancer Prev. 2016, 17, 2989–2993. [Google Scholar]
- El-Gayar, D.; El-Abd, N.; Hassan, N.; Ali, R. Increased free circulating DNA integrity index as a serum biomarker in patients with colorectal carcinoma. Asian Pac. J. Cancer Prev. 2016, 17, 939–944. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, E.H.; Fawzy, A.; Ahmad, O.K.; Ali, A.M. Plasma circulating cell-free nuclear and mitochondrial DNA as potential biomarkers in the peripheral blood of breast cancer patients. Asian Pac. J. Cancer Prev. 2015, 16, 8299–8305. [Google Scholar] [CrossRef] [PubMed]
- Zaher, E.R.; Anwar, M.M.; Kohail, H.M.; El-Zoghby, S.M.; Abo-El-Eneen, M.S. Cell-free DNA concentration and integrity as a screening tool for cancer. Indian J. Cancer 2013, 50, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Hashad, D.; Sorour, A.; Ghazal, A.; Talaat, I. Free circulating tumor DNA as a diagnostic marker for breast cancer. J. Clin. Lab. Anal. 2012, 26, 467–472. [Google Scholar] [CrossRef] [PubMed]
- El-Shazly, S.F.; Eid, M.A.; El-Sourogy, H.A.; Attia, G.F.; Ezzat, S.A. Evaluation of serum DNA integrity as a screening and prognostic tool in patients with hepatitis c virus-related hepatocellular carcinoma. Int. J. Biol. Markers 2010, 25, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Iyer, P.; Zekri, A.R.; Hung, C.W.; Schiefelbein, E.; Ismail, K.; Hablas, A.; Seifeldin, I.A.; Soliman, A.S. Concordance of DNA methylation pattern in plasma and tumor DNA of egyptian hepatocellular carcinoma patients. Exp. Mol. Pathol. 2010, 88, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Hosny, G.; Farahat, N.; Hainaut, P. Tp53 mutations in circulating free DNA from egyptian patients with non-hodgkin’s lymphoma. Cancer Lett. 2009, 275, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Hosny, G.; Farahat, N.; Tayel, H.; Hainaut, P. Ser-249 tp53 and ctnnb1 mutations in circulating free DNA of egyptian patients with hepatocellular carcinoma versus chronic liver diseases. Cancer Lett. 2008, 264, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Kirk, G.D.; Lesi, O.A.; Mendy, M.; Szymanska, K.; Whittle, H.; Goedert, J.J.; Hainaut, P.; Montesano, R. 249(ser) tp53 mutation in plasma DNA, hepatitis b viral infection, and risk of hepatocellular carcinoma. Oncogene 2005, 24, 5858–5867. [Google Scholar] [CrossRef]
- Szymanska, K.; Lesi, O.A.; Kirk, G.D.; Sam, O.; Taniere, P.; Scoazec, J.Y.; Mendy, M.; Friesen, M.D.; Whittle, H.; Montesano, R.; et al. Ser-249tp53 mutation in tumour and plasma DNA of hepatocellular carcinoma patients from a high incidence area in the gambia, west Africa. Int. J. Cancer 2004, 110, 374–379. [Google Scholar] [CrossRef]
- Van der Vaart, M.; Semenov, D.V.; Kuligina, E.V.; Richter, V.A.; Pretorius, P.J. Characterisation of circulating DNA by parallel tagged sequencing on the 454 platform. Clin. Chim. Acta 2009, 409, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Van der Vaart, M.; Pretorius, P.J. A method for characterization of total circulating DNA. Ann. N. Y. Acad. Sci. 2008, 1137, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Abd-El-Fattah, A.A.; Sadik, N.A.; Shaker, O.G.; Aboulftouh, M.L. Differential micrornas expression in serum of patients with lung cancer, pulmonary tuberculosis, and pneumonia. Cell Biochem. Biophys. 2013, 67, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Ali, L.H.; Higazi, A.M.; Moness, H.M.; Farag, N.M.; Saad, Z.M.; Moukareb, H.A.; Soliman, W.; El Sagheer, G.; Abd El Hamid, S.R.; Abdl Hamid, H. Clinical significances and diagnostic utilities of both mir-215 and squamous cell carcinoma antigen-igm versus alpha-fetoprotein in egyptian patients with hepatitis c virus-induced hepatocellular carcinoma. Clin. Exp. Gastroenterol. 2019, 12, 51–66. [Google Scholar] [CrossRef] [PubMed]
- Ezzat, W.M.; Amr, K.S.; Elhosary, Y.A.; Hegazy, A.E.; Fahim, H.H.; Eltaweel, N.H.; Kamel, R.R. Detection of DNA methylated micrornas in hepatocellular carcinoma. Gene 2019, 702, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Hetta, H.F.; Zahran, A.M.; Shafik, E.A.; El-Mahdy, R.I.; Mohamed, N.A.; Nabil, E.E.; Esmaeel, H.M.; Alkady, O.A.; Elkady, A.; Mohareb, D.A.; et al. Circulating mirna-21 and mirna-23a expression signature as potential biomarkers for early detection of non-small-cell lung cancer. Microrna 2019, 8, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, E.H.; Fawzy, A.; Elshimy, R.A.A. Serum microrna-21 negatively relates to expression of programmed cell death-4 in patients with epithelial ovarian cancer. Asian Pac. J. Cancer Prev. 2018, 19, 33–38. [Google Scholar] [PubMed]
- Sabry, D.; El-Deek, S.E.M.; Maher, M.; El-Baz, M.A.H.; El-Bader, H.M.; Amer, E.; Hassan, E.A.; Fathy, W.; El-Deek, H.E.M. Role of mirna-210, mirna-21 and mirna-126 as diagnostic biomarkers in colorectal carcinoma: Impact of hif-1alpha-vegf signaling pathway. Mol. Cell. Biochem. 2019, 454, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Swellam, M.; Mahmoud, M.S.; Hashim, M.; Hassan, N.M.; Sobeih, M.E.; Nageeb, A.M. Clinical aspects of circulating mirna-335 in breast cancer patients: A prospective study. J. Cell. Biochem. 2019, 120, 8975–8982. [Google Scholar] [CrossRef] [PubMed]
- Swellam, M.; El Magdoub, H.M.; Hassan, N.M.; Hefny, M.M.; Sobeih, M.E. Potential diagnostic role of circulating mirnas in breast cancer: Implications on clinicopathological characters. Clin. Biochem. 2018, 56, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Toraih, E.A.; Alghamdi, S.A.; El-Wazir, A.; Hosny, M.M.; Hussein, M.H.; Khashana, M.S.; Fawzy, M.S. Dual biomarkers long non-coding rna gas5 and microrna-34a co-expression signature in common solid tumors. PLoS ONE 2018, 13, e0198231. [Google Scholar] [CrossRef] [PubMed]
- Zekri, A.N.; El-Sisi, E.R.; Youssef, A.S.E.; Kamel, M.M.; Nassar, A.; Ahmed, O.S.; El Kassas, M.; Barakat, A.B.; Abd El-Motaleb, A.I.; Bahnassy, A.A. Microrna signatures for circulating cd133-positive cells in hepatocellular carcinoma with hcv infection. PLoS ONE 2018, 13, e0193709. [Google Scholar] [CrossRef] [PubMed]
- Demerdash, H.M.; Hussien, H.M.; Hassouna, E.; Arida, E.A. Detection of microrna in hepatic cirrhosis and hepatocellular carcinoma in hepatitis c genotype-4 in egyptian patients. Biomed. Res. Int. 2017, 2017, 1806069. [Google Scholar] [CrossRef] [PubMed]
- Elemeery, M.N.; Badr, A.N.; Mohamed, M.A.; Ghareeb, D.A. Validation of a serum microrna panel as biomarkers for early diagnosis of hepatocellular carcinoma post-hepatitis c infection in egyptian patients. World J. Gastroenterol. 2017, 23, 3864. [Google Scholar] [CrossRef] [PubMed]
- Elhamamsy, A.R.; El Sharkawy, M.S.; Zanaty, A.F.; Mahrous, M.A.; Mohamed, A.E.; Abushaaban, E.A. Circulating mir-92a, mir-143 and mir-342 in plasma are novel potential biomarkers for acute myeloid leukemia. Int. J. Mol. Cell. Med. 2017, 6, 77–86. [Google Scholar]
- Elshafei, A.; Shaker, O.; Abd El-Motaal, O.; Salman, T. The expression profiling of serum mir-92a, mir-375, and mir-760 in colorectal cancer: An egyptian study. Tumour Biol. 2017, 39. [Google Scholar] [CrossRef] [PubMed]
- Khairy, A.; Hamza, I.; Shaker, O.; Yosry, A. Serum mirna panel in egyptian patients with chronic hepatitis c related hepatocellular carcinoma. Asian Pac. J. Cancer Prev. 2016, 17, 2699–2703. [Google Scholar]
- Motawi, T.K.; Rizk, S.M.; Ibrahim, T.M.; Ibrahim, I.A. Circulating micrornas, mir-92a, mir-100 and mir-143, as non-invasive biomarkers for bladder cancer diagnosis. Cell Biochem. Funct. 2016, 34, 142–148. [Google Scholar] [CrossRef]
- Motawi, T.M.; Sadik, N.A.; Shaker, O.G.; Ghaleb, M.H. Elevated serum microrna-122/222 levels are potential diagnostic biomarkers in egyptian patients with chronic hepatitis c but not hepatic cancer. Tumour Biol. 2016, 37, 9865–9874. [Google Scholar] [CrossRef]
- Swellam, M.; El-Khazragy, N. Clinical impact of circulating micrornas as blood-based marker in childhood acute lymphoblastic leukemia. Tumour Biol. 2016, 37, 10571–10576. [Google Scholar] [CrossRef]
- Zekri, A.R.; Youssef, A.S.; Lotfy, M.M.; Gabr, R.; Ahmed, O.S.; Nassar, A.; Hussein, N.; Omran, D.; Medhat, E.; Eid, S.; et al. Circulating serum mirnas as diagnostic markers for colorectal cancer. PLoS ONE 2016, 11, e0154130. [Google Scholar] [CrossRef] [PubMed]
- Alnoanmany, W.; Ismail, H.A.; El-Said, H.; Obada, M.; Sakr, M.A.; Elfert, A.Y. Diagnostic potential of circulating microrna-21 in hepatocellular carcinoma. Int. J. Sci. Technol. Res. 2015, 4, 429–433. [Google Scholar]
- Eissa, S.; Matboli, M.; Hegazy, M.G.; Kotb, Y.M.; Essawy, N.O. Evaluation of urinary microrna panel in bladder cancer diagnosis: Relation to bilharziasis. Transl. Res. 2015, 165, 731–739. [Google Scholar] [CrossRef] [PubMed]
- El-Abd, N.E.; Fawzy, N.A.; El-Sheikh, S.M.; Soliman, M.E. Circulating mirna-122, mirna-199a, and mirna-16 as biomarkers for early detection of hepatocellular carcinoma in egyptian patients with chronic hepatitis c virus infection. Mol. Diagn. Ther. 2015, 19, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Hagrass, H.A.; Sharaf, S.; Pasha, H.F.; Tantawy, E.A.; Mohamed, R.H.; Kassem, R. Circulating micrornas—A new horizon in molecular diagnosis of breast cancer. Genes Cancer 2015, 6, 281–287. [Google Scholar] [PubMed]
- Motawi, T.K.; Shaker, O.G.; El-Maraghy, S.A.; Senousy, M.A. Serum micrornas as potential biomarkers for early diagnosis of hepatitis c virus-related hepatocellular carcinoma in egyptian patients. PLoS ONE 2015, 10, e0137706. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, M.A.; Haj-Ahmad, Y. Promising candidate urinary microrna biomarkers for the early detection of hepatocellular carcinoma among high-risk hepatitis c virus egyptian patients. J. Cancer 2012, 3, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Hamdi, K.; Blancato, J.; Goerlitz, D.; Islam, M.; Neili, B.; Abidi, A.; Gat, A.; Ayed, F.B.; Chivi, S.; Loffredo, C.; et al. Circulating cell-free mirna expression and its association with clinicopathologic features in inflammatory and non-inflammatory breast cancer. Asian Pac. J. Cancer Prev. 2016, 17, 1801–1810. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Abdelgawad, I.A.; Mossallam, G.I.; Radwan, N.H.; Elzawahry, H.M.; Elhifnawy, N.M. Can glypican3 be diagnostic for early hepatocellular carcinoma among egyptian patients? Asian Pac. J. Cancer Prev. 2013, 14, 7345–7349. [Google Scholar] [CrossRef]
- Ibrahim, G.H.; Mahmoud, M.A.; Aly, N.M. Evaluation of circulating transforming growth factor-beta1, glypican-3 and golgi protein-73 mrnas expression as predictive markers for hepatocellular carcinoma in egyptian patients. Mol. Biol. Rep. 2013, 40, 7069–7075. [Google Scholar] [CrossRef]
- Zidan, H.E.; Karam, R.A.; El-Seifi, O.S.; Abd Elrahman, T.M. Circulating long non-coding rna malat1 expression as molecular biomarker in egyptian patients with breast cancer. Cancer Genet. 2018, 220, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Shaker, O.G.; Senousy, M.A.; Elbaz, E.M. Association of rs6983267 at 8q24, hulc rs7763881 polymorphisms and serum lncrnas ccat2 and hulc with colorectal cancer in egyptian patients. Sci. Rep. 2017, 7, 16246. [Google Scholar] [CrossRef] [PubMed]
- El-Tawdi, A.H.; Matboli, M.; Shehata, H.H.; Tash, F.; El-Khazragy, N.; Azazy Ael, S.; Abdel-Rahman, O. Evaluation of circulatory rna-based biomarker panel in hepatocellular carcinoma. Mol. Diagn. Ther. 2016, 20, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Hashad, D.; Elbanna, A.; Ibrahim, A.; Khedr, G. Evaluation of the role of circulating long non-coding rna h19 as a promising novel biomarker in plasma of patients with gastric cancer. J. Clin. Lab. Anal. 2016, 30, 1100–1105. [Google Scholar] [CrossRef] [PubMed]
- Abd El Gwad, A.; Matboli, M.; El-Tawdi, A.; Habib, E.K.; Shehata, H.; Ibrahim, D.; Tash, F. Role of exosomal competing endogenous rna in patients with hepatocellular carcinoma. J. Cell. Biochem. 2018, 119, 8600–8610. [Google Scholar] [CrossRef] [PubMed]
- Khalil, F.; Shehata, H.H.; Osman, N.; Abdel-Rahman, O.; Ali, M.; Mohamed, G. Evaluation of the diagnostic role of non-coding rna and exosomal related gene association in lung cancer. Egypt. J. Hosp. Med. 2018, 72, 4148–4153. [Google Scholar]
- Weigelt, B.; Peterse, J.L.; van’t Veer, L.J. Breast cancer metastasis: Markers and models. Nat. Rev. Cancer 2005, 5, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef]
- Cohen, S.J.; Punt, C.J.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C.; et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008, 26, 3213–3221. [Google Scholar] [CrossRef]
- Goldkorn, A.; Ely, B.; Quinn, D.I.; Tangen, C.M.; Fink, L.M.; Xu, T.; Twardowski, P.; Van Veldhuizen, P.J.; Agarwal, N.; Carducci, M.A.; et al. Circulating tumor cell counts are prognostic of overall survival in swog s0421: A phase iii trial of docetaxel with or without atrasentan for metastatic castration-resistant prostate cancer. J. Clin. Oncol. 2014, 32, 1136–1142. [Google Scholar] [CrossRef]
- De Bono, J.S.; Scher, H.I.; Montgomery, R.B.; Parker, C.; Miller, M.C.; Tissing, H.; Doyle, G.V.; Terstappen, L.W.; Pienta, K.J.; Raghavan, D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 2008, 14, 6302–6309. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Hayes, D.F.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Reuben, J.M.; Doyle, G.V.; Matera, J.; Allard, W.J.; Miller, M.C.; et al. Circulating tumor cells: A novel prognostic factor for newly diagnosed metastatic breast cancer. J. Clin. Oncol. 2005, 23, 1420–1430. [Google Scholar] [CrossRef] [PubMed]
- Fiorelli, A.; Accardo, M.; Carelli, E.; Angioletti, D.; Santini, M.; Di Domenico, M. Circulating tumor cells in diagnosing lung cancer: Clinical and morphologic analysis. Ann. Thorac. Surg. 2015, 99, 1899–1905. [Google Scholar] [CrossRef] [PubMed]
- Rhim, A.D.; Mirek, E.T.; Aiello, N.M.; Maitra, A.; Bailey, J.M.; McAllister, F.; Reichert, M.; Beatty, G.L.; Rustgi, A.K.; Vonderheide, R.H.; et al. Emt and dissemination precede pancreatic tumor formation. Cell 2012, 148, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Ilie, M.; Hofman, V.; Long-Mira, E.; Selva, E.; Vignaud, J.M.; Padovani, B.; Mouroux, J.; Marquette, C.H.; Hofman, P. “Sentinel” circulating tumor cells allow early diagnosis of lung cancer in patients with chronic obstructive pulmonary disease. PLoS ONE 2014, 9, e111597. [Google Scholar] [CrossRef]
- Gulbahce, N.; Magbanua, M.J.M.; Chin, R.; Agarwal, M.R.; Luo, X.; Liu, J.; Hayden, D.M.; Mao, Q.; Ciotlos, S.; Li, Z.; et al. Quantitative whole genome sequencing of circulating tumor cells enables personalized combination therapy of metastatic cancer. Cancer Res. 2017, 77, 4530–4541. [Google Scholar] [CrossRef]
- Lin, E.; Cao, T.; Nagrath, S.; King, M.R. Circulating tumor cells: Diagnostic and therapeutic applications. Annu. Rev. Biomed. Eng. 2018, 20, 329–352. [Google Scholar] [CrossRef]
- Beije, N.; Jager, A.; Sleijfer, S. Circulating tumor cell enumeration by the cellsearch system: The clinician’s guide to breast cancer treatment? Cancer Treat. Rev. 2015, 41, 144–150. [Google Scholar] [CrossRef]
- Kraan, J.; Sleijfer, S.; Strijbos, M.H.; Ignatiadis, M.; Peeters, D.; Pierga, J.Y.; Farace, F.; Riethdorf, S.; Fehm, T.; Zorzino, L.; et al. External quality assurance of circulating tumor cell enumeration using the cellsearch® system: A feasibility study. Cytometry B Clin. Cytom. 2011, 80, 112–118. [Google Scholar] [CrossRef]
- Gorges, T.M.; Tinhofer, I.; Drosch, M.; Rose, L.; Zollner, T.M.; Krahn, T.; von Ahsen, O. Circulating tumour cells escape from epcam-based detection due to epithelial-to-mesenchymal transition. BMC Cancer 2012, 12, 178. [Google Scholar] [CrossRef]
- Sieuwerts, A.M.; Kraan, J.; Bolt, J.; van der Spoel, P.; Elstrodt, F.; Schutte, M.; Martens, J.W.; Gratama, J.W.; Sleijfer, S.; Foekens, J.A. Anti-epithelial cell adhesion molecule antibodies and the detection of circulating normal-like breast tumor cells. J. Natl. Cancer Inst. 2009, 101, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, A.J.; Marengo, M.S.; Oltean, S.; Kemeny, G.; Bitting, R.L.; Turnbull, J.D.; Herold, C.I.; Marcom, P.K.; George, D.J.; Garcia-Blanco, M.A. Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Mol. Cancer Res. 2011, 9, 997–1007. [Google Scholar] [CrossRef] [PubMed]
- Pantel, K.; Denève, E.; Nocca, D.; Coffy, A.; Vendrell, J.P.; Maudelonde, T.; Riethdorf, S.; Alix-Panabières, C. Circulating epithelial cells in patients with benign colon diseases. Clin. Chem. 2012, 58, 936–940. [Google Scholar]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.; Uhr, J.W.; Terstappen, L.W. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef] [PubMed]
- Riethdorf, S.; Fritsche, H.; Muller, V.; Rau, T.; Schindlbeck, C.; Rack, B.; Janni, W.; Coith, C.; Beck, K.; Janicke, F.; et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: A validation study of the cellsearch system. Clin. Cancer Res. 2007, 13, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Punnoose, E.A.; Atwal, S.K.; Spoerke, J.M.; Savage, H.; Pandita, A.; Yeh, R.-F.; Pirzkall, A.; Fine, B.M.; Amler, L.C.; Chen, D.S.; et al. Molecular biomarker analyses using circulating tumor cells. PLoS ONE 2010, 5, e12517. [Google Scholar] [CrossRef] [PubMed]
- Budd, G.T.; Cristofanilli, M.; Ellis, M.J.; Stopeck, A.; Borden, E.; Miller, M.C.; Matera, J.; Repollet, M.; Doyle, G.V.; Terstappen, L.W. Circulating tumor cells versus imaging—Predicting overall survival in metastatic breast cancer. Clin. Cancer Res. 2006, 12, 6403–6409. [Google Scholar] [CrossRef]
- Heller, G.; McCormack, R.; Kheoh, T.; Molina, A.; Smith, M.R.; Dreicer, R.; Saad, F.; de Wit, R.; Aftab, D.T.; Hirmand, M.; et al. Circulating tumor cell number as a response measure of prolonged survival for metastatic castration-resistant prostate cancer: A comparison with prostate-specific antigen across five randomized phase iii clinical trials. J. Clin. Oncol. 2018, 36, 572–580. [Google Scholar] [CrossRef]
- Yan, W.T.; Cui, X.; Chen, Q.; Li, Y.F.; Cui, Y.H.; Wang, Y.; Jiang, J. Circulating tumor cell status monitors the treatment responses in breast cancer patients: A meta-analysis. Sci. Rep. 2017, 7, 43464. [Google Scholar] [CrossRef]
- Krebs, M.G.; Renehan, A.G.; Backen, A.; Gollins, S.; Chau, I.; Hasan, J.; Valle, J.W.; Morris, K.; Beech, J.; Ashcroft, L.; et al. Circulating tumor cell enumeration in a phase ii trial of a four-drug regimen in advanced colorectal cancer. Clin. Colorectal Cancer 2015, 14, 115–122. [Google Scholar] [CrossRef]
- Molloy, T.J.; Bosma, A.J.; Baumbusch, L.O.; Synnestvedt, M.; Borgen, E.; Russnes, H.G.; Schlichting, E.; van’t Veer, L.J.; Naume, B. The prognostic significance of tumour cell detection in the peripheral blood versus the bone marrow in 733 early-stage breast cancer patients. Breast Cancer Res. 2011, 13, R61. [Google Scholar] [CrossRef] [PubMed]
- Daskalaki, A.; Agelaki, S.; Perraki, M.; Apostolaki, S.; Xenidis, N.; Stathopoulos, E.; Kontopodis, E.; Hatzidaki, D.; Mavroudis, D.; Georgoulias, V. Detection of cytokeratin-19 mrna-positive cells in the peripheral blood and bone marrow of patients with operable breast cancer. Br. J. Cancer 2009, 101, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Skillrud, D.M.; Offord, K.P.; Miller, R.D. Higher risk of lung cancer in chronic obstructive pulmonary disease. A prospective, matched, controlled study. Ann. Intern. Med. 1986, 105, 503–507. [Google Scholar] [CrossRef] [PubMed]
- Bardelli, A.; Pantel, K. Liquid biopsies, what we do not know (yet). Cancer Cell 2017, 31, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Li, C.; Lu, S.; Zhou, W.; Tang, F.; Xie, X.S.; Huang, Y. Uniform and accurate single-cell sequencing based on emulsion whole-genome amplification. Proc. Natl. Acad. Sci. USA 2015, 112, 11923–11928. [Google Scholar] [CrossRef] [PubMed]
- Lohr, J.G.; Adalsteinsson, V.A.; Cibulskis, K.; Choudhury, A.D.; Rosenberg, M.; Cruz-Gordillo, P.; Francis, J.M.; Zhang, C.Z.; Shalek, A.K.; Satija, R.; et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat. Biotechnol. 2014, 32, 479–484. [Google Scholar] [CrossRef] [PubMed]
- D’Avola, D.; Villacorta-Martin, C.; Martins-Filho, S.N.; Craig, A.; Labgaa, I.; von Felden, J.; Kimaada, A.; Bonaccorso, A.; Tabrizian, P.; Hartmann, B.M.; et al. High-density single cell mrna sequencing to characterize circulating tumor cells in hepatocellular carcinoma. Sci. Rep. 2018, 8, 11570. [Google Scholar] [CrossRef] [PubMed]
- Kong, S.L.; Liu, X.; Suhaimi, N.M.; Koh, K.J.H.; Hu, M.; Lee, D.Y.S.; Cima, I.; Phyo, W.M.; Lee, E.X.W.; Tai, J.A.; et al. Molecular characterization of circulating colorectal tumor cells defines genetic signatures for individualized cancer care. Oncotarget 2017, 8, 68026–68037. [Google Scholar] [CrossRef] [PubMed]
- Aceto, N.; Bardia, A.; Wittner, B.S.; Donaldson, M.C.; O’Keefe, R.; Engstrom, A.; Bersani, F.; Zheng, Y.; Comaills, V.; Niederhoffer, K.; et al. Ar expression in breast cancer ctcs associates with bone metastases. Mol. Cancer Res. 2018, 16, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Shiratsuchi, H.; Palanisamy, N.; Nagrath, S.; Ramnath, N. Expanded circulating tumor cells from a patient with alk-positive lung cancer present with eml4-alk rearrangement along with resistance mutation and enable drug sensitivity testing: A case study. J. Thorac. Oncol. 2017, 12, 397–402. [Google Scholar] [CrossRef]
- Mandel, P. Les acides nucleiques du plasma sanguin chez 1 homme. C.R. Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar]
- Leon, S.A.; Shapiro, B.; Sklaroff, D.M.; Yaros, M.J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar] [PubMed]
- Agassi, R.; Czeiger, D.; Shaked, G.; Avriel, A.; Sheynin, J.; Lavrenkov, K.; Ariad, S.; Douvdevani, A. Measurement of circulating cell-free DNA levels by a simple fluorescent test in patients with breast cancer. Am. J. Clin. Pathol. 2015, 143, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Boddy, J.L.; Gal, S.; Malone, P.R.; Harris, A.L.; Wainscoat, J.S. Prospective study of quantitation of plasma DNA levels in the diagnosis of malignant versus benign prostate disease. Clin. Cancer Res. 2005, 11, 1394–1399. [Google Scholar] [CrossRef] [PubMed]
- Gordian, E.; Ramachandran, K.; Reis, I.M.; Manoharan, M.; Soloway, M.S.; Singal, R. Serum free circulating DNA is a useful biomarker to distinguish benign versus malignant prostate disease. Cancer Epidem. Biomar. 2010, 19, 1984–1991. [Google Scholar] [CrossRef] [PubMed]
- Fleischhacker, M.; Schmidt, B. Circulating nucleic acids (cnas) and cancer—A survey. BBA-Rev. Cancer 2007, 1775, 181–232. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Chan, C.W.; Chan, K.C.; Cheng, S.H.; Wong, J.; Wong, V.W.; Wong, G.L.; Chan, S.L.; Mok, T.S.; Chan, H.L.; et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc. Natl. Acad. Sci. USA 2015, 112, E1317–E1325. [Google Scholar] [CrossRef]
- Snyder, M.W.; Kircher, M.; Hill, A.J.; Daza, R.M.; Shendure, J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 2016, 164, 57–68. [Google Scholar] [CrossRef]
- Stroun, M.; Lyautey, J.; Lederrey, C.; Olson-Sand, A.; Anker, P. About the possible origin and mechanism of circulating DNA—Apoptosis and active DNA release. Clin. Chim. Acta 2001, 313, 139–142. [Google Scholar] [CrossRef]
- Umetani, N.; Giuliano, A.E.; Hiramatsu, S.H.; Amersi, F.; Nakagawa, T.; Martino, S.; Hoon, D.S. Prediction of breast tumor progression by integrity of free circulating DNA in serum. J. Clin. Oncol. 2006, 24, 4270–4276. [Google Scholar] [CrossRef]
- Giacona, M.B.; Ruben, G.C.; Iczkowski, K.A.; Roos, T.B.; Porter, D.M.; Sorenson, G.D. Cell-free DNA in human blood plasma: Length measurements in patients with pancreatic cancer and healthy controls. Pancreas 1998, 17, 89–97. [Google Scholar] [CrossRef]
- Wang, B.G.; Huang, H.Y.; Chen, Y.C.; Bristow, R.E.; Kassauei, K.; Cheng, C.C.; Roden, R.; Sokoll, L.J.; Chan, D.W.; Shih Ie, M. Increased plasma DNA integrity in cancer patients. Cancer Res. 2003, 63, 3966–3968. [Google Scholar] [PubMed]
- Schwarzenbach, H.; Hoon, D.S.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Mouliere, F.; Rosenfeld, N. Circulating tumor-derived DNA is shorter than somatic DNA in plasma. Proc. Natl. Acad. Sci. USA 2015, 112, 3178–3179. [Google Scholar] [CrossRef] [PubMed]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Su, L.; Qian, C. Circulating tumor DNA: A promising biomarker in the liquid biopsy of cancer. Oncotarget 2016, 7, 48832–48841. [Google Scholar] [CrossRef]
- Husain, H.; Nykin, D.; Bui, N.; Quan, D.; Gomez, G.; Woodward, B.; Venkatapathy, S.; Duttagupta, R.; Fung, E.; Lippman, S.M.; et al. Cell-free DNA from ascites and pleural effusions: Molecular insights into genomic aberrations and disease biology. Mol. Cancer Ther. 2017, 16, 948–955. [Google Scholar] [CrossRef]
- Fujiwara, K.; Fujimoto, N.; Tabata, M.; Nishii, K.; Matsuo, K.; Hotta, K.; Kozuki, T.; Aoe, M.; Kiura, K.; Ueoka, H.; et al. Identification of epigenetic aberrant promoter methylation in serum DNA is useful for early detection of lung cancer. Clin. Cancer Res. 2005, 11, 1219–1225. [Google Scholar]
- Stroun, M.; Anker, P.; Maurice, P.; Lyautey, J.; Lederrey, C.; Beljanski, M. Neoplastic characteristics of the DNA found in the plasma of cancer-patients. Oncology 1989, 46, 318–322. [Google Scholar] [CrossRef]
- Wang, J.Y.; Hsieh, J.S.; Chang, M.Y.; Huang, T.J.; Chen, F.M.; Cheng, T.L.; Alexandersen, K.; Huang, Y.S.; Tzou, W.S.; Lin, S.R. Molecular detection of apc, k-ras, and p53 mutations in the serum of colorectal cancer patients as circulating biomarkers. World J. Surg. 2004, 28, 721–726. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, R.; Yan, C.; Liu, L.; Tong, Z.; Jiang, W.; Yao, M.; Fang, W.; Chen, Z. Advantage of next-generation sequencing in dynamic monitoring of circulating tumor DNA over droplet digital pcr in cetuximab treated colorectal cancer patients. Transl. Oncol. 2019, 12, 426–431. [Google Scholar] [CrossRef]
- Wyatt, A.W.; Annala, M.; Aggarwal, R.; Beja, K.; Feng, F.; Youngren, J.; Foye, A.; Lloyd, P.; Nykter, M.; Beer, T.M.; et al. Concordance of circulating tumor DNA and matched metastatic tissue biopsy in prostate cancer. J. Natl. Cancer Inst. 2017, 109, djx118. [Google Scholar] [CrossRef] [PubMed]
- Kidess-Sigal, E.; Liu, H.E.; Triboulet, M.M.; Che, J.; Ramani, V.C.; Visser, B.C.; Poultsides, G.A.; Longacre, T.A.; Marziali, A.; Vysotskaia, V.; et al. Enumeration and targeted analysis of kras, braf and pik3ca mutations in ctcs captured by a label-free platform: Comparison to ctdna and tissue in metastatic colorectal cancer. Oncotarget 2016, 7, 85349–85364. [Google Scholar] [CrossRef] [PubMed]
- Malapelle, U.; Sirera, R.; Jantus-Lewintre, E.; Reclusa, P.; Calabuig-Fariñas, S.; Blasco, A.; Pisapia, P.; Rolfo, C.; Camps, C. Profile of the roche cobas® egfr mutation test v2 for non-small cell lung cancer. Expert Rev. Mol. Diagn. 2017, 17, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-L.; Lee, V.; Liam, C.-K.; Lu, S.; Park, K.; Srimuninnimit, V.; Wang, J.; Zhou, C.; Appius, A.; Button, P.; et al. Clinical utility of a blood-based egfr mutation test in patients receiving first-line erlotinib therapy in the ensure, fastact-2, and aspiration studies. Lung Cancer 2018, 126, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Potter, N.T.; Hurban, P.; White, M.N.; Whitlock, K.D.; Lofton-Day, C.E.; Tetzner, R.; Koenig, T.; Quigley, N.B.; Weiss, G. Validation of a real-time pcr–based qualitative assay for the detection of methylated sept9 DNA in human plasma. Clin. Chem. 2014, 60, 1183–1191. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Jia, J.; Peng, X.; Xiao, W.; Li, Y. The performance of the sept9 gene methylation assay and a comparison with other crc screening tests: A meta-analysis. Sci. Rep. 2017, 7, 3032. [Google Scholar] [CrossRef]
- Ou, S.-H.I.; Nagasaka, M.; Zhu, V.W. Liquid biopsy to identify actionable genomic alterations. ASCO Educ. Book 2018, 38, 978–997. [Google Scholar] [CrossRef] [PubMed]
- Stroun, M.; Anker, P.; Beljanski, M.; Henri, J.; Lederrey, C.; Ojha, M.; Maurice, P.A. Presence of rna in the nucleoprotein complex spontaneously released by human lymphocytes and frog auricles in culture. Cancer Res. 1978, 38, 3546–3554. [Google Scholar]
- Colombo, M.; Raposo, G.; Thery, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell. Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Tsui, N.B.; Ng, E.K.; Lo, Y.M. Stability of endogenous and added rna in blood specimens, serum, and plasma. Clin. Chem. 2002, 48, 1647–1653. [Google Scholar]
- Suraj, S.; Dhar, C.; Srivastava, S. Circulating nucleic acids: An analysis of their occurrence in malignancies. Biomed. Rep. 2017, 6, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Umu, S.U.; Langseth, H.; Bucher-Johannessen, C.; Fromm, B.; Keller, A.; Meese, E.; Lauritzen, M.; Leithaug, M.; Lyle, R.; Rounge, T.B. A comprehensive profile of circulating rnas in human serum. RNA Biol. 2018, 15, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Rapisuwon, S.; Vietsch, E.E.; Wellstein, A. Circulating biomarkers to monitor cancer progression and treatment. Comput. Struct. Biotechnol. J. 2016, 14, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Garcia, V.; Garcia, J.M.; Pena, C.; Silva, J.; Dominguez, G.; Lorenzo, Y.; Diaz, R.; Espinosa, P.; de Sola, J.G.; Cantos, B.; et al. Free circulating mrna in plasma from breast cancer patients and clinical outcome. Cancer Lett. 2008, 263, 312–320. [Google Scholar] [CrossRef] [PubMed]
- March-Villalba, J.A.; Martinez-Jabaloyas, J.M.; Herrero, M.J.; Santamaria, J.; Alino, S.F.; Dasi, F. Cell-free circulating plasma htert mrna is a useful marker for prostate cancer diagnosis and is associated with poor prognosis tumor characteristics. PLoS ONE 2012, 7, e43470. [Google Scholar] [CrossRef] [PubMed]
- Sahengbieke, S.; Wang, J.; Li, X.; Wang, Y.; Lai, M.; Wu, J. Circulating cell-free high mobility group at-hook 2 mrna as a detection marker in the serum of colorectal cancer patients. J. Clin. Lab. Anal. 2018, 32, e22332. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Kong, W.; Wu, Y.; Ren, H.; Wei, J.; Yang, Y.; Yang, Y.; Yu, L.; Guan, W.; Liu, B. Plasma mrna as liquid biopsy predicts chemo-sensitivity in advanced gastric cancer patients. J. Cancer 2017, 8, 434–442. [Google Scholar] [CrossRef] [PubMed]
- De Souza, M.F.; Kuasne, H.; de Camargo Barros-Filho, M.; Cilião, H.L.; Marchi, F.A.; Fuganti, P.E.; Paschoal, A.R.; Rogatto, S.R.; de Syllos Cólus, I.M. Circulating mrnas and mirnas as candidate markers for the diagnosis and prognosis of prostate cancer. PloS ONE 2017, 12, e0184094. [Google Scholar] [CrossRef] [PubMed]
- Tani, N.; Ichikawa, D.; Ikoma, D.; Tomita, A.; Sai, S.; Ikoma, H.; Okamoto, K.; Ochiai, T.; Ueda, Y.; Otsuji, E.; et al. Circulating cell-free mrna in plasma as a tumor marker for patients with primary and recurrent gastric cancer. Anticancer Res. 2007, 27, 1207–1212. [Google Scholar] [PubMed]
- Fok, E.T.; Scholefield, J.; Fanucchi, S.; Mhlanga, M.M. The emerging molecular biology toolbox for the study of long noncoding rna biology. Epigenomics 2017, 9, 1317–1327. [Google Scholar] [CrossRef] [PubMed]
- Bhat, S.A.; Ahmad, S.M.; Mumtaz, P.T.; Malik, A.A.; Dar, M.A.; Urwat, U.; Shah, R.A.; Ganai, N.A. Long non-coding rnas: Mechanism of action and functional utility. Non-coding RNA Res. 2016, 1, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Wang, S.; Li, Q.; Ji, Q.; Guo, P.; Liu, X. Malat1: A long non-coding rna highly associated with human cancers. Oncol. Lett. 2018, 16, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Feng, Y.; Wang, X. Lncrna zeb1-as1 expression in cancer prognosis: Review and meta-analysis. Clin. Chim. Acta 2018, 484, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Zhang, Y.; Du, L.; Jiang, X.; Yan, S.; Duan, W.; Li, J.; Zhan, Y.; Wang, L.; Zhang, S. Circulating long noncoding rna act as potential novel biomarkers for diagnosis and prognosis of non-small cell lung cancer. Mol. Oncol. 2018, 12, 648–658. [Google Scholar] [CrossRef] [PubMed]
- Lv, R.; Zhang, J.; Zhang, W.; Huang, Y.; Wang, N.; Zhang, Q.; Qu, S. Circulating hotair expression predicts the clinical response to neoadjuvant chemotherapy in patients with breast cancer. Cancer Biomark. 2018, 22, 249–256. [Google Scholar]
- Aubin, S.M.J.; Reid, J.; Sarno, M.J.; Blase, A.; Aussie, J.; Rittenhouse, H.; Rittmaster, R.; Andriole, G.L.; Groskopf, J. Pca3 molecular urine test for predicting repeat prostate biopsy outcome in populations at risk: Validation in the placebo arm of the dutasteride reduce trial. J. Urology 2010, 184, 1947–1952. [Google Scholar] [CrossRef] [PubMed]
- De La Taille, A. Progensa™ pca3 test for prostate cancer detection. Expert Rev. Mol. Diagn. 2007, 7, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Sohel, M.H. Extracellular/circulating micrornas: Release mechanisms, functions and challenges. Achiev. Life Sci. 2016, 10, 175–186. [Google Scholar] [CrossRef]
- Catalanotto, C.; Cogoni, C.; Zardo, G. Microrna in control of gene expression: An overview of nuclear functions. Int. J. Mol. Sci. 2016, 17, 1712. [Google Scholar] [CrossRef]
- Bortoluzzi, S.; Lovisa, F.; Gaffo, E.; Mussolin, L. Small rnas in circulating exosomes of cancer patients: A minireview. High-Throughput 2017, 6, 13. [Google Scholar] [CrossRef]
- Iftikhar, H.; Carney, G.E. Evidence and potential in vivo functions for biofluid mirnas: From expression profiling to functional testing: Potential roles of extracellular mirnas as indicators of physiological change and as agents of intercellular information exchange. BioEssays 2016, 38, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Iguchi, H.; Ochiya, T. Circulating microrna in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010, 101, 2087–2092. [Google Scholar] [CrossRef] [PubMed]
- Zaborowski, M.P.; Balaj, L.; Breakefield, X.O.; Lai, C.P. Extracellular vesicles: Composition, biological relevance, and methods of study. Bioscience 2015, 65, 783–797. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating micrornas as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Roth, C.; Rack, B.; Muller, V.; Janni, W.; Pantel, K.; Schwarzenbach, H. Circulating micrornas as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010, 12, R90. [Google Scholar] [CrossRef] [PubMed]
- Cuk, K.; Zucknick, M.; Heil, J.; Madhavan, D.; Schott, S.; Turchinovich, A.; Arlt, D.; Rath, M.; Sohn, C.; Benner, A.; et al. Circulating micrornas in plasma as early detection markers for breast cancer. Int. J. Cancer 2013, 132, 1602–1612. [Google Scholar] [CrossRef]
- Song, C.J.; Chen, H.; Chen, L.Z.; Ru, G.M.; Guo, J.J.; Ding, Q.N. The potential of micrornas as human prostate cancer biomarkers: A meta-analysis of related studies. J. Cell. Biochem. 2018, 119, 2763–2786. [Google Scholar] [CrossRef]
- Wang, Y.N.; Chen, Z.H.; Chen, W.C. Novel circulating micrornas expression profile in colon cancer: A pilot study. Eur. J. Med. Res. 2017, 22, 51. [Google Scholar] [CrossRef]
- Motawi, T.M.; Sadik, N.A.; Shaker, O.G.; El Masry, M.R.; Mohareb, F. Study of micrornas-21/221 as potential breast cancer biomarkers in egyptian women. Gene 2016, 590, 210–219. [Google Scholar] [CrossRef]
- Fattah, H.I.A.; Mahmoud, N.H.; Elzoghby, D.M.; Matar, M.M.; El-Shaer, I.M.M. Clinical utility of circulating microrna-21 in breast cancer. Egypt. J. Hosp. Med. 2018, 71, 2950–2955. [Google Scholar]
- Fernando, M.R.; Jiang, C.; Krzyzanowski, G.D.; Ryan, W.L. New evidence that a large proportion of human blood plasma cell-free DNA is localized in exosomes. PLoS ONE 2017, 12, e0183915. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Harding, C.V.; Heuser, J.E.; Stahl, P.D. Exosomes: Looking back three decades and into the future. J. Cell Biol. 2013, 200, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Rai, A.; Chen, M.; Suwakulsiri, W.; Greening, D.W.; Simpson, R.J. Extracellular vesicles in cancer—Implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 2018, 15, 617–638. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, C. Exosome cancer diagnostic reaches market. Nat. Biotechnol. 2016, 34, 359–360. [Google Scholar] [CrossRef]
- Vanni, I.; Alama, A.; Grossi, F.; Dal Bello, M.G.; Coco, S. Exosomes: A new horizon in lung cancer. Drug Discov. Today 2017, 22, 927–936. [Google Scholar] [CrossRef]
- Witwer, K.W.; Buzas, E.I.; Bemis, L.T.; Bora, A.; Lasser, C.; Lotvall, J.; Nolte-’t Hoen, E.N.; Piper, M.G.; Sivaraman, S.; Skog, J.; et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles 2013, 2, 20360. [Google Scholar] [CrossRef]
- Xu, R.; Greening, D.W.; Zhu, H.J.; Takahashi, N.; Simpson, R.J. Extracellular vesicle isolation and characterization: Toward clinical application. J. Clin. Investig. 2016, 126, 1152–1162. [Google Scholar] [CrossRef]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell. Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef]
- Skog, J.; Wurdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport rna and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell. Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef]
- Di Meo, A.; Pasic, M.D.; Yousef, G.M. Proteomics and peptidomics: Moving toward precision medicine in urological malignancies. Oncotarget 2016, 7, 52460–52474. [Google Scholar] [CrossRef] [PubMed]
- Pasic, M.D.; Yousef, G.M.; Diamandis, E.P. The proteomic revolution in laboratory medicine. Clin. Biochem. 2013, 46, 397–398. [Google Scholar] [CrossRef] [PubMed]
- Abdel Wahab, A.H.A.; El-Halawany, M.S.; Emam, A.A.; Elfiky, A.; Abd Elmageed, Z.Y. Identification of circulating protein biomarkers in patients with hepatocellular carcinoma concomitantly infected with chronic hepatitis c virus. Biomarkers 2017, 22, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Alkady, M.M.; Abdel-Messeih, P.L.; Nosseir, N.M. Assessment of serum levels of the adipocytokine chemerin in colorectal cancer patients. J. Med. Biochem. 2018, 37, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Aref, S.; Osman, E.; Mansy, S.; Omer, N.; Azmy, E.; Goda, T.; El-Sherbiny, M. Prognostic relevance of circulating matrix metalloproteinase-2 in acute myeloid leukaemia patients. Hematol. Oncol. 2007, 25, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Ben Anes, A.; ben Nasr, H.; Hammann, P.; Kuhn, L.; Trimeche, M.; Hamrita, B.; Bougmiza, I.; Chaieb, A.; Khairi, H.; Chahed, K. Assessment of the clinical significance of antigenic and functional levels of α1-proteinase inhibitor (α1-pi) in infiltrating ductal breast carcinomas. Clin. Biochem. 2012, 45, 1421–1431. [Google Scholar] [CrossRef] [PubMed]
- El-Bassiouni, N.E.; Nosseir, M.M.; Madkour, M.E.; Zoheiry, M.M.; Bekheit, I.W.; Ibrahim, R.A.; Ibrahim, I.M.; El Bassiouny, A.E. Role of fibrogenic markers in chronic hepatitis c and associated hepatocellular carcinoma. Mol. Biol. Rep. 2012, 39, 6843–6850. [Google Scholar] [CrossRef] [PubMed]
- El-Mesallamy, H.O.; Hamdy, N.M.; Zaghloul, A.S.; Sallam, A.M. Clinical value of circulating lipocalins and insulin-like growth factor axis in pancreatic cancer diagnosis. Pancreas 2013, 42, 149–154. [Google Scholar] [CrossRef]
- El-Shal, A.S.; Zidan, H.E.; Rashad, N.M.; Wadea, F.M. Angiopoietin-like protein 3 and 4 expression 4 and their serum levels in hepatocellular carcinoma. Cytokine 2017, 96, 75–86. [Google Scholar] [CrossRef]
- Eltaher, S.M.; El-Gil, R.; Fouad, N.; Mitwali, R.; El-Kholy, H. Evaluation of serum levels and significance of soluble cd40 ligand in screening patients with hepatitis c virus-related hepatocellular carcinoma. East. Mediterr. Health J. 2016, 22, 603–610. [Google Scholar] [CrossRef]
- Hamdy, K.; Al Swaff, R.; Hussein, H.A.; Gamal, M. Assessment of serum adiponectin in egyptian patients with hcv-related cirrhosis and hepatocellular carcinoma. J. Endocrinol. Investig. 2015, 38, 1225–1231. [Google Scholar] [CrossRef] [PubMed]
- Hamrita, B.; Nasr, H.B.; Hammann, P.; Kuhn, L.; Guillier, C.L.; Chaieb, A.; Khairi, H.; Chahed, K. An elongation factor-like protein (ef-tu) elicits a humoral response in infiltrating ductal breast carcinomas: An immunoproteomics investigation. Clin. Biochem. 2011, 44, 1097–1104. [Google Scholar] [CrossRef] [PubMed]
- M’Hamdi, H.; Baizig, N.M.; OE, E.L.; M’Hamdi, N.; Attia, Z.; Gritli, S.; Gamoudi, A.; El May, M.V.; May, A.E. Usefulness of igf-1 serum levels as diagnostic marker of nasopharyngeal carcinoma. Immunobiology 2016, 221, 1304–1308. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, A.A.; Soliman, H.; Ismail, M.; Ziada, D.; Farid, T.M.; Aref, A.M.; Al Daly, M.E.; Abd Elmageed, Z.Y. Evaluation of circulating adh and mic-1 as diagnostic markers in egyptian patients with pancreatic cancer. Pancreatology 2015, 15, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Shaarawy, M.; El-Sharkawy, S.A. Biomarkers of intrinsic angiogenic and anti-angiogenic activity in patients with endometrial hyperplasia and endometrial cancer. Acta Oncol. 2001, 40, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Shaker, O.G.; Ay El-Deen, M.A.; Abd El-Rahim, M.T.; Talaat, R.M. Gene expression of e-selectin in tissue and its protein level in serum of breast cancer patients. Tumori 2006, 92, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Talaat, R.M.; Salem, T.A.; El-Masry, S.; Imbarek, A.; Mokhles, M.; Abdel-Aziz, A. Circulating pro-and anti-angiogenic mediators in patients infected with hepatitis c at different stages of hepatocellular carcinoma. J. Med. Virol. 2014, 86, 1120–1129. [Google Scholar] [CrossRef] [PubMed]
- Zekri, A.; Bakr, Y.M.; Ezzat, M.M.; Zakaria, M.; Elbaz, T.M. Circulating levels of adipocytokines as potential biomarkers for early detection of colorectal carcinoma in egyptian patients. Asian Pac. J. Cancer Prev. 2015, 16, 6923–6928. [Google Scholar] [CrossRef] [PubMed]
- Zohny, S.F.; Fayed, S.T. Clinical utility of circulating matrix metalloproteinase-7 (mmp-7), cc chemokine ligand 18 (ccl18) and cc chemokine ligand 11 (ccl11) as markers for diagnosis of epithelial ovarian cancer. Med. Oncol. 2010, 27, 1246–1253. [Google Scholar] [CrossRef]
- Adeola, H.A.; Blackburn, J.M.; Rebbeck, T.R.; Zerbini, L.F. Emerging proteomics biomarkers and prostate cancer burden in africa. Oncotarget 2017, 8, 37991–38007. [Google Scholar] [CrossRef]
- Kinyua, P. Kenya-third African country to use blood-based tests to detect cancer. Jamhuri News, 21 May 2018. [Google Scholar]
- Adeola, H.A.; Adefuye, A.O.; Jimoh, S.A. Potential latitudinal variation in orodigestive tract cancers in Africa. S. Afr. Med. J. 2018, 108, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Neumann, M.H.D.; Bender, S.; Krahn, T.; Schlange, T. Ctdna and ctcs in liquid biopsy—Current status and where we need to progress. Comput. Struct. Biotechnol. J. 2018, 16, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Perakis, S.; Speicher, M.R. Emerging concepts in liquid biopsies. BMC Med. 2017, 15, 75. [Google Scholar] [CrossRef] [PubMed]
- Arneth, B. Update on the types and usage of liquid biopsies in the clinical setting: A systematic review. BMC Cancer 2018, 18, 527. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Wang, J.; Sun, Y. Circulating tumor DNA as biomarkers for cancer detection. Egypt. J. Med. Hum. Genet. 2017, 15, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Babayan, A.; Pantel, K. Advances in liquid biopsy approaches for early detection and monitoring of cancer. Genome Med. 2018, 10, 21. [Google Scholar] [CrossRef] [PubMed]
- Kapeleris, J.; Kulasinghe, A.; Warkiani, M.E.; Vela, I.; Kenny, L.; O’Byrne, K.; Punyadeera, C. The prognostic role of circulating tumor cells (ctcs) in lung cancer. Front. Oncol. 2018, 8, 311. [Google Scholar] [CrossRef] [PubMed]
- Tanos, R.; Thierry, A.R. Clinical relevance of liquid biopsy for cancer screening. Transl. Cancer Res. 2018, 7, S105–S129. [Google Scholar] [CrossRef]
- Alix-Panabieres, C.; Pantel, K. Real-time liquid biopsy: Circulating tumor cells versus circulating tumor DNA. Ann. Transl. Med. 2013, 1, 18. [Google Scholar] [PubMed]
- Gao, Y.; Zhu, Y.; Yuan, Z. Circulating tumor cells and circulating tumor DNA provide new insights into pancreatic cancer. Int. J. Med. Sci. 2016, 13, 902–913. [Google Scholar] [CrossRef]
- Heitzer, E.; Perakis, S.; Geigl, J.B.; Speicher, M.R. The potential of liquid biopsies for the early detection of cancer. NPJ Precis. Oncol. 2017, 1, 36. [Google Scholar] [CrossRef] [PubMed]
- Azubuike, S.O.; Muirhead, C.; Hayes, L.; McNally, R. Rising global burden of breast cancer: The case of sub-saharan africa (with emphasis on nigeria) and implications for regional development: A. review. World J. Surg. Oncol. 2018, 16, 63. [Google Scholar] [CrossRef] [PubMed]
- Chirenje, Z.M.; Rusakaniko, S.; Akino, V.; Mlingo, M. A review of cervical cancer patients presenting in harare and parirenyatwa hospitals in 1998. Cent. Afr. J. Med. 2000, 46, 264–267. [Google Scholar] [CrossRef] [PubMed]
- Rambau, P.F.; Chalya, P.L.; Manyama, M.M.; Jackson, K.J. Pathological features of breast cancer seen in northwestern tanzania: A nine years retrospective study. BMC Res. Notes 2011, 4, 214. [Google Scholar] [CrossRef] [PubMed]
- Amosu, A.M.; Degun, A.M.; Thomas, A.M.; Babalola, A.O. Assessment of awareness, perception, specific knowledge, and screening behaviour regarding breast cancer among rural women in ipokia local government area, ogun state, nigeria. Arch. Appl. Sci. Res. 2011, 3, 253–265. [Google Scholar]
- Clegg-Lamptey, J.; Dakubo, J.; Attobra, Y.N. Why do breast cancer patients report late or abscond during treatment in ghana? A pilot study. Ghana Med. J. 2009, 43, 127–131. [Google Scholar] [PubMed]
- Mbuka-Ongona, D.; Tumbo, J.M. Knowledge about breast cancer and reasons for late presentation by cancer patients seen at princess marina hospital, gaborone, botswana. Afr. J. Prim. Health Care Fam. Med. 2013, 5, 465. [Google Scholar] [CrossRef][Green Version]
| Samples | Study Design | Cancer Type | Downstream Analysis | Country | References |
|---|---|---|---|---|---|
| CTC | |||||
| Blood | 75 BC patients 20 healthy controls | Breast cancer | Circulating endothelial progenitor cells count CD14, CD133 and VEGFR2 expression levels (flow cytometry) | Egypt | Montaser, et al. [16] |
| Blood | 50 BC patients 14 healthy controls | Breast cancer | mRNA expression levels (qPCR) | Egypt | Elnagdy, et al. [17] |
| Blood | 51 BC patients | Breast cancer | CTC and CSC count (flow cytometry) | Egypt | Sayed, et al. [18] |
| Peripheral blood | 70 HCC patients 30 CHC patients 33 healthy controls | Liver cancer | CTC and CSC countProtein expression levels of CK19, CD133, CD90 (flow cytometry) | Egypt | Bahnassy, et al. [19] |
| Blood | 50 HCC patients 20 healthy controls | Liver cancer | CTC count (flow cytometry) | Egypt | Mansour, et al. [20] |
| Blood | 40 BC patients | Breast cancer | CTC and CSC count (flow cytometry) | Egypt | Zedan, et al. [21] |
| Blood | 50 BC patients 30 healthy controls | Breast cancer | mRNA expression levels (qPCR) | Egypt | Ebeed, et al. [22] |
| Blood | 36 CRC patients 18 healthy controls | Colorectal cancer | mRNA expression levels (qPCR) | Egypt | Teama and Agwa [23] |
| Blood | 147 BC patients 94 healthy controls (41 U.S. healthy volunteers) | Breast cancer | mRNA expression levels (qPCR) | Senegal | Zehentner, et al. [24] |
| Peripheral blood | 143 primary melanoma patients | Melanoma | The use of qPCR to determine the presence of tyrosinase mRNA in peripheral blood | South Africa | Hanekom, et al. [25] |
| cfDNA | |||||
| Plasma | 195 HCC patients 263 CLD control patients 49 healthy controls | Liver cancer | cfDNA mutational analysis using droplet digital PCR | Cameroon, Central African Republic | Marchio, et al. [26] |
| Plasma | 40 BC patients 10 healthy controls | Breast cancer | cfDNA quantification and Integrity index using qPCR | Egypt | Hussein, et al. [27] |
| Serum | 60 LC patients 40 COPD patients 40 healthy controls | Lung cancer | cfDNA quantification and Integrity index using qPCR | Egypt | Soliman, et al. [28] |
| Plasma | 50 PCa patients 25 BPH patients 30 healthy controls | Prostate cancer | cfDNA quantification and Integrity index using qPCR | Egypt | Fawzy, et al. [29] |
| Serum | 40 BC patients 40 healthy controls | Breast cancer | cfDNA quantification using qPCR | Egypt | Ibrahim, et al. [30] |
| Serum | 50 CRC patients 10 colonic polyps’ patients 20 healthy controls | Colorectal cancer | cfDNA quantification and Integrity index using qPCR | Egypt | El-Gayar, et al. [31] |
| Plasma | 50 BC patients 30 benign breast lesions 20 healthy controls | Breast cancer | Quantification of cfDNA andmtDNA using multiplex qPCR | Egypt | Mahmoud, et al. [32] |
| Plasma | 120 cancer patients 120 patients with benign diseases 120 healthy controls | Breast, Lung, Colon and Liver cancers | cfDNA quantification and Integrity index using qPCR | Egypt | Zaher, et al. [33] |
| Plasma | 42 BC patients 30 benign lesion patients 27 healthy controls | Breast cancer | cfDNA quantification and Integrity index using qPCR | Egypt | Hashad, et al. [34] |
| Serum | 25 HCV-related HCC patients 25 chronic HCV patients 15 healthy controls | Liver cancer | cfDNA quantification and Integrity index using qPCR | Egypt | El-Shazly, et al. [35] |
| Plasma | 28 HCC patients | Liver cancer | Methylation profile determined for five genes using qPCR | Egypt | Iyer, et al. [36] |
| Serum | 20 NHL patients 20 healthy controls | non-Hodgkin’s lymphoma | cfDNA quantification using Fluorometric assay | Egypt | Hosny, et al. [37] |
| Serum | 76 HCC patients 110 CLD patients 69 healthy controls | Liver cancer | cfDNA quantification and sequencing of the positive RFLP fragments using nested PCR | Egypt | Hosny, et al. [38] |
| Plasma | 216 HCC patients 121 liver cirrhosis patients 408 healthy controls | Liver cancer | cfDNA quantification and sequencing using nested PCR | Gambia | Kirk, et al. [39] |
| Plasma | 29 HCC patients | Liver cancer | cfDNA quantification and sequencing using nested PCR | Gambia | Szymanska, et al. [40] |
| Plasma | 12 PCa patients 10 healthy controls | Prostate cancer | cfDNA quantification and parallel tagged sequencing | South Africa | van der Vaart, et al. [41] |
| Plasma | 1 BC patient 1 healthy control | Breast cancer | Cloning and sequencing of cfDNA | South Africa | van der Vaart and Pretorius [42] |
| miRNA | |||||
| Serum | 65 LC patients 29 pulmonary tuberculosis patients 29 pneumonia 37 healthy controls | Lung cancer | Expression levels of miR-21, miR-155, miR-182, and miR-197 assessed using qPCR | Egypt | Abd-El-Fattah, et al. [43] |
| Serum | 60 HCV-related HCC patients 60 HCV-related liver cirrhosis patients 60 healthy controls | Liver cancer | Expression levels of miRNAs determined using qPCR | Egypt | Ali, et al. [44] |
| Serum | 60 HCC patients 30 healthy controls | Liver cancer | Expression levels of microRNAs 191, 203 and 335 determined using qPCR | Egypt | Ezzat, et al. [45] |
| Plasma | 45 LC patients 40 healthy controls | Lung cancer | The expression level of miR-21 and miR-23a was detected by qPCR | Egypt | Hetta, et al. [46] |
| Serum | 60 ovarian cancer patients 30 healthy controls | Ovarian cancer | Serum miR-21 levels were measured by TaqMan-qPCR | Egypt | Mahmoud, et al. [47] |
| Serum | 35 CRC patients 51 patients with benign lesions 101 healthy controls | Colorectal cancer | The expression of miR-210, miR-21 and miR-126 was performed using qPCR | Egypt | Sabry, et al. [48] |
| Serum | 106 BC patients 49 benign breast lesion patients 40 healthy controls | Breast Cancer | The expression level of miR-335 was detected by qPCR | Egypt | Swellam, et al. [49] |
| Serum | 137 BC patients 60 benign breast lesion patients 38 healthy controls | Breast cancer | miRNAs expression levels were determined using reaction qPCR | Egypt | Swellam, et al. [50] |
| Serum | 30 HCC patients 20 healthy controls | Liver cancer | lncRNA GAS5 and miR-34a expression level measured using qPCR | Egypt | Toraih, et al. [51] |
| Blood | 9 CHC patients 6 liver cirrhosis patients 9 HCC patients 8 healthy controls | Liver cancer | miRNAs expression levels were determined using reaction qPCR | Egypt | Zekri, et al. [52] |
| Plasma | 60 HCC patients 60 CHC patients 60 healthy controls | Liver cancer | miRNA expression levels assessed using qPCR | Egypt | Demerdash, et al. [53] |
| Serum | 224 HCC patients 250 CHC patients 84 healthy controls | Liver cancer | miRNAs (hsa-miR-1269, hsa-miR-125b, hsa-miR-138, hsa-miR-214-5p, hsa-miR-494, hsa-miR-375 and hsa-miR-145) were assessed using qPCR | Egypt | Elemeery, et al. [54] |
| Plasma | 65 AML patients 50 healthy controls | Acute myeloid leukemia | Expression of miR-92a, miR-143 and miR-342 was measured using qPCR | Egypt | Elhamamsy, et al. [55] |
| Serum | 64 CRC patients 27 healthy controls | Colorectal cancer | Expression levels of miR-92a, miR-375, and miR-760 assessed using qPCR | Egypt | Elshafei, et al. [56] |
| Serum | 23 HCC patients 25 post-HCV liver cirrhosis patients 30 HCV patients 10 healthy controls | Liver cancer | miRNA expression levels using qPCR | Egypt | Khairy, et al. [57] |
| Plasma | 70 bladder cancer patients 62 healthy controls | Bladder cancer | Expression levels of miR-92a, miR-100 and miR-143 measured using qPCR | Egypt | Motawi, et al. [58] |
| Serum | 60 HCC patients 40 CHC patients 30 healthy controls | Liver cancer | Expression levels of miRNA-122 and miRNA-222 assessed using qPCR | Egypt | Motawi, et al. [59] |
| Peripheral blood mononuclear cells | 85 ALL patients 25 healthy controls | Acute lymphoblastic leukemia | Expression levels of miR-92a, miR-100 and miR-143 were measured using qPCR | Egypt | Swellam and El-Khazragy [60] |
| Serum | 30 CRC patients 18 IBD patients 18 colonic polyps’ patients 24 colonic symptoms patients 100 CRC patients (validation) | Colorectal cancer | miRNAs expression levels were determined using reaction qPCR | Egypt | Zekri, et al. [61] |
| Blood | 30 HCC patients 20 HCV patients 20 healthy controls | Liver cancer | miRNA expression levels assessed using qPCR | Egypt | Alnoanmany, et al. [62] |
| Urine | 188 Bladder cancer patients 88 Benign bladder lesions 92 healthy controls | Bladder cancer | miR-210, miR-10b, miR-29c, miR-221, and miR-23a expression levels assessed using qPCR | Egypt | Eissa, et al. [63] |
| Serum | 40 HCC patients 40 HCV patients 20 Healthy controls | Liver cancer | miRNA expression levels using qPCR | Egypt | El-Abd, et al. [64] |
| Serum | 120 BC patients 50 healthy controls | Breast cancer | Expression levels of miRNAs (miR10b, miR34a, miR155, miR195 and miR16) determined using qPCR | Egypt | Hagrass, et al. [65] |
| Serum | 112 HCV-related HCC patients 125 HCV-related CLD patients 42 healthy controls | Liver cancer | Expression miRNA was measured using qPCR | Egypt | Motawi, et al. [66] |
| Urine | 32 HCC patients with post-HCV infection 74 chronic HCV patients 12 healthy controls | Liver cancer | miRNA whole-genome expression profiling and relative expression profiling for candidate miRNAs using qPCR | Egypt | Abdalla and Haj-Ahmad [67] |
| Serum | 20 Inflammatory BC patients 20 non-inflammatory BC patients 20 healthy controls | Breast cancer | TaqMan qPCR was performed to detect the circulating expression of miRNAs | Tunisia | Hamdi, et al. [68] |
| mRNA | |||||
| Serum | 40 HCC patients 10 healthy controls | Liver cancer | mRNA expression levels using qPCR | Egypt | Abdelgawad, et al. [69] |
| Serum | 25 HCC patients 15 healthy controls | Liver cancer | mRNA expression levels using qPCR | Egypt | Ibrahim, et al. [70] |
| lncRNAs | |||||
| Serum | 80 BC patients 80 healthy controls | Breast cancer | mRNA expression levels using qPCR | Egypt | Zidan, et al. [71] |
| Serum | 120 CRC patients 30 adenomatous polyps’ patients 96 healthy controls | Colorectal cancer | Serum expression levels of lncRNAs and miRNA using qPCR | Egypt | Shaker, et al. [72] |
| Serum | 78 HCC patients 36 CHC patients 44 healthy controls | Liver cancer | mRNA expression levels using qPCR | Egypt | El-Tawdi, et al. [73] |
| Plasma | 32 gastric cancer patients 30 healthy controls | Gastric cancer | mRNA expression levels using qPCR | Egypt | Hashad, et al. [74] |
| Exosomes | |||||
| Serum | 60 HCC patients 42 CHC patients 18 healthy controls | Liver cancer | Expression of exosomal RNA using qPCR | Egypt | Abd El Gwad, et al. [75] |
| Serum | 20 LC patients | Lung cancer | Expression of exosomal RNA using qPCR | Egypt | Khalil, et al. [76] |
| Analysis Capability | CTC | cfDNA | Circulating Tumor RNA | Exosomes |
|---|---|---|---|---|
| Genomic mutations | Yes | Yes | Yes | Yes |
| RNA profiling | Yes | No | Yes | Yes |
| Phenotypic studies of tumor cell | Yes | No | No | No |
| Proteomic analysis | Yes | No | No | Yes |
| Clinical Applications | ||||
| Clinical trials | Phase IV | Phase IV | Phase IV | Phase II |
| Clinical approved techniques | CellSearch |
| Progensa™ PCA3 | No |
| Cost of clinical use (outside Africa) | $350 | $170–470 | $220 | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Temilola, D.O.; Wium, M.; Coulidiati, T.H.; Adeola, H.A.; Carbone, G.M.; Catapano, C.V.; Zerbini, L.F. The Prospect and Challenges to the Flow of Liquid Biopsy in Africa. Cells 2019, 8, 862. https://doi.org/10.3390/cells8080862
Temilola DO, Wium M, Coulidiati TH, Adeola HA, Carbone GM, Catapano CV, Zerbini LF. The Prospect and Challenges to the Flow of Liquid Biopsy in Africa. Cells. 2019; 8(8):862. https://doi.org/10.3390/cells8080862
Chicago/Turabian StyleTemilola, Dada Oluwaseyi, Martha Wium, Tangbadioa Herve Coulidiati, Henry Ademola Adeola, Giuseppina Maria Carbone, Carlo Vittorio Catapano, and Luiz Fernando Zerbini. 2019. "The Prospect and Challenges to the Flow of Liquid Biopsy in Africa" Cells 8, no. 8: 862. https://doi.org/10.3390/cells8080862
APA StyleTemilola, D. O., Wium, M., Coulidiati, T. H., Adeola, H. A., Carbone, G. M., Catapano, C. V., & Zerbini, L. F. (2019). The Prospect and Challenges to the Flow of Liquid Biopsy in Africa. Cells, 8(8), 862. https://doi.org/10.3390/cells8080862






