Computational Assessment of Bacterial Protein Structures Indicates a Selection Against Aggregation
Abstract
:1. Introduction
2. Material and Methods
2.1. Selection Criteria for the Analysis of Bacterial Proteome
2.2. Structural and Sequential Aggregation Propensity Predictions
2.3. Datasets
2.4. Definition of the Supersaturation Index (SSI)
2.5. Statistical Analysis
3. Results
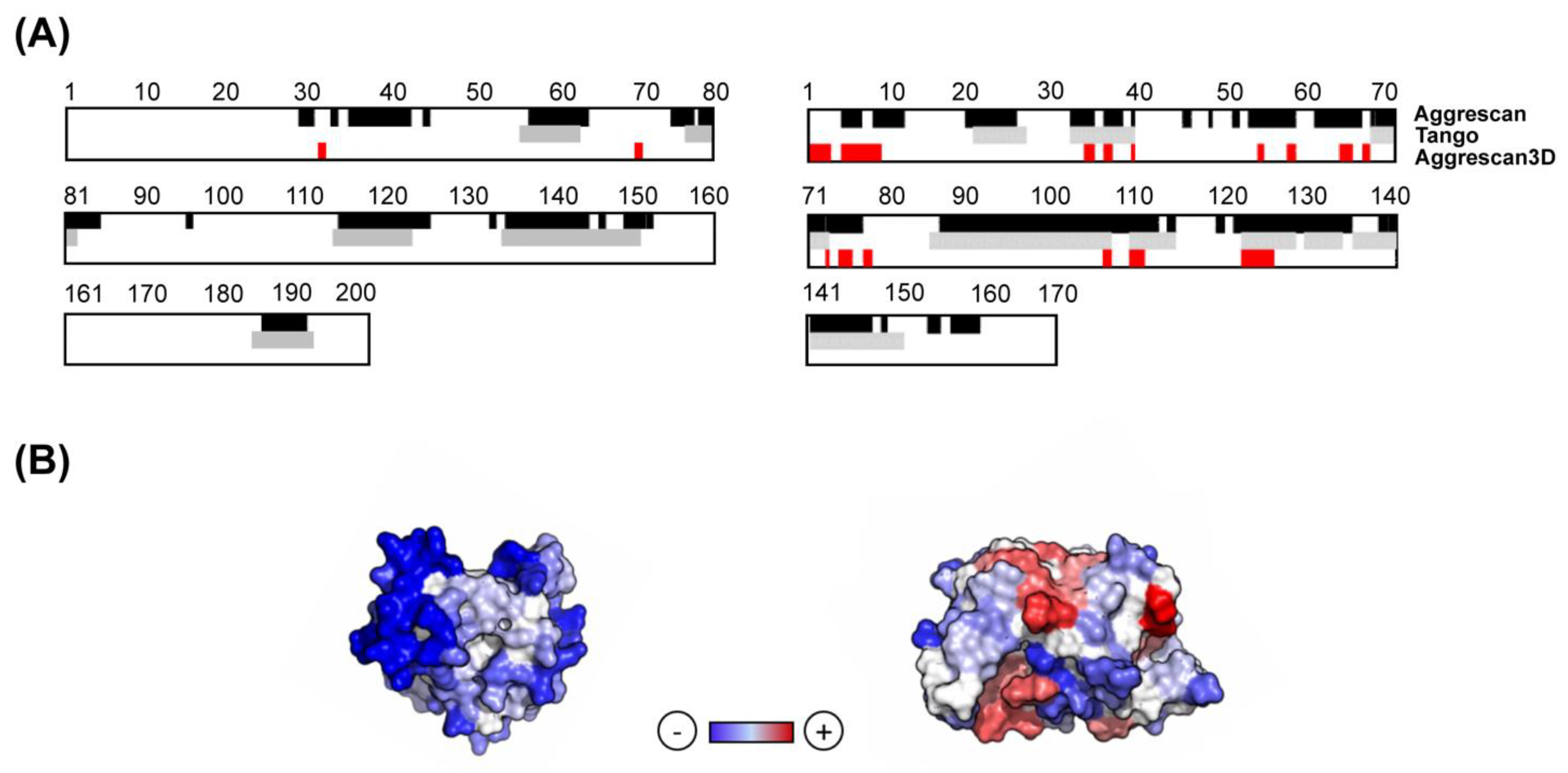
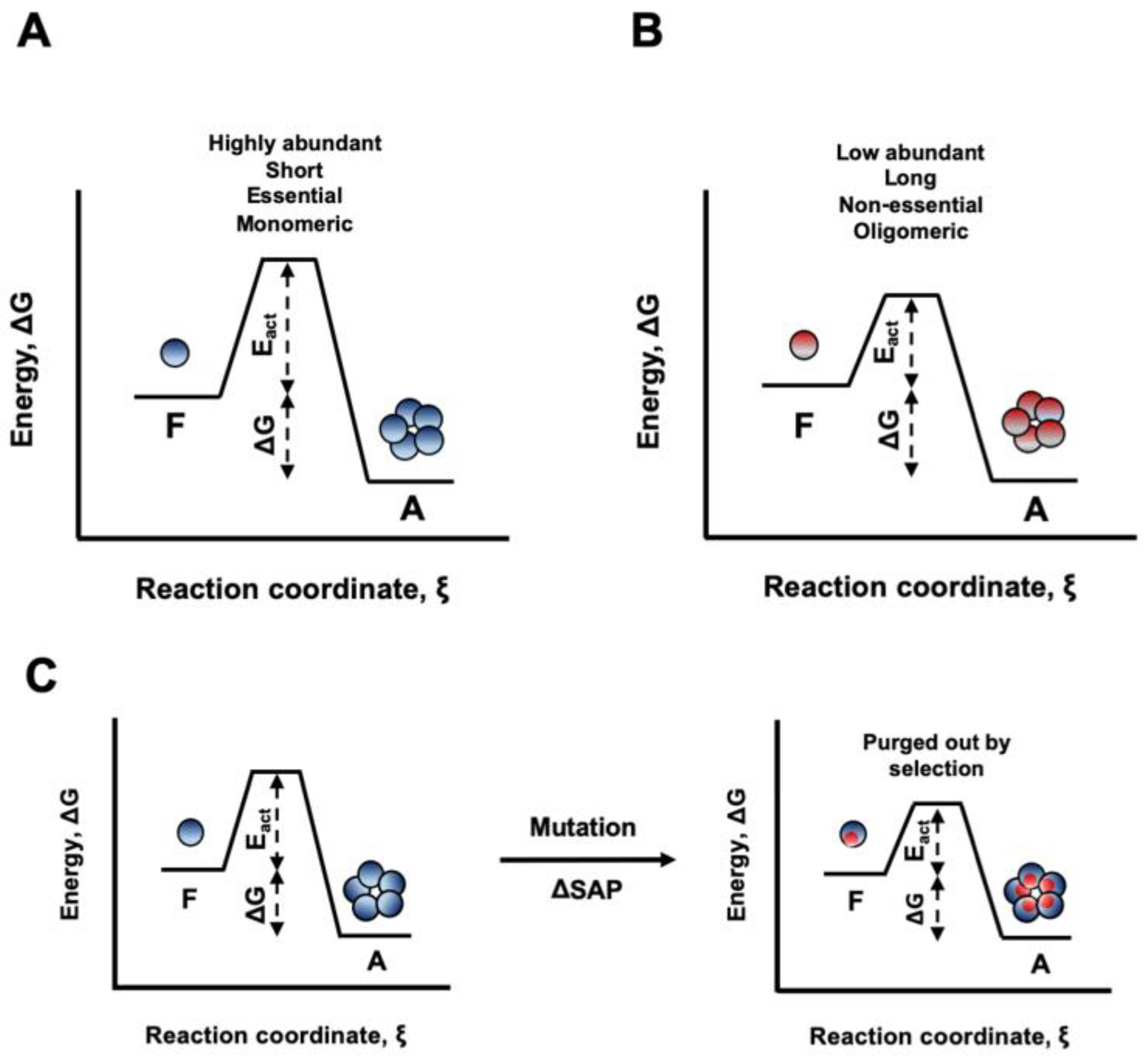
3.1. A3D Analysis Rationale
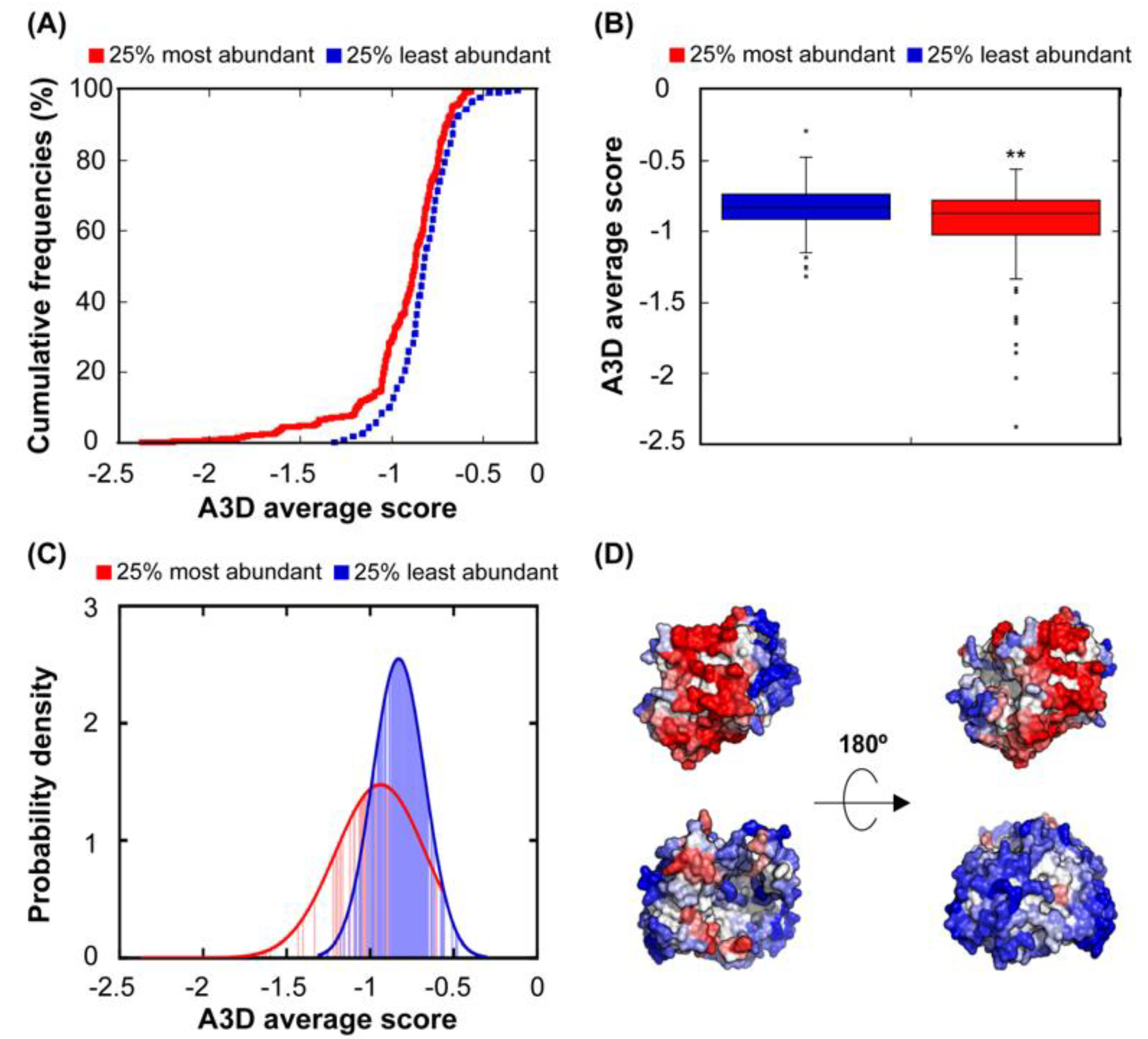
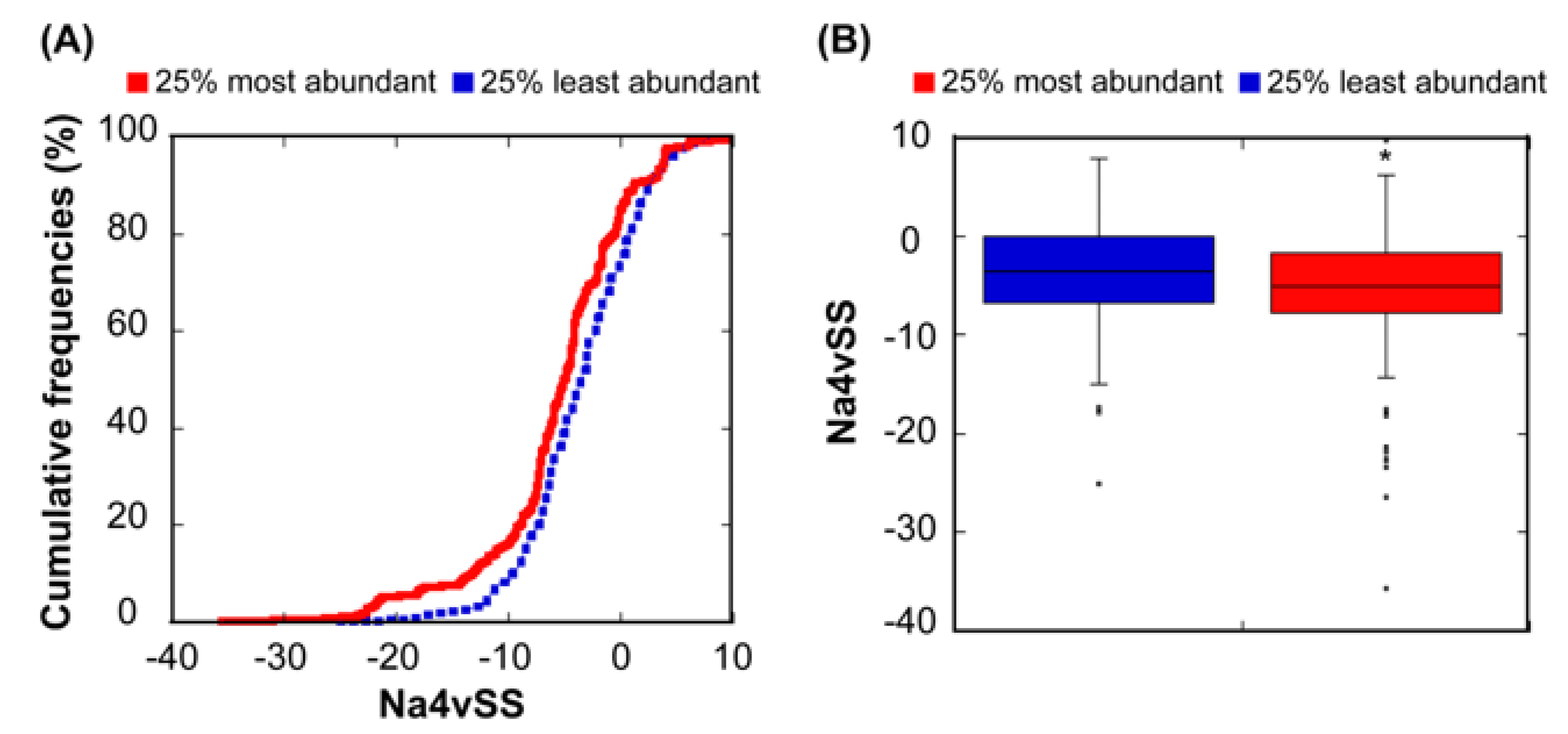
3.2. Relationship Between Protein Abundance and Structural Aggregation Propensity
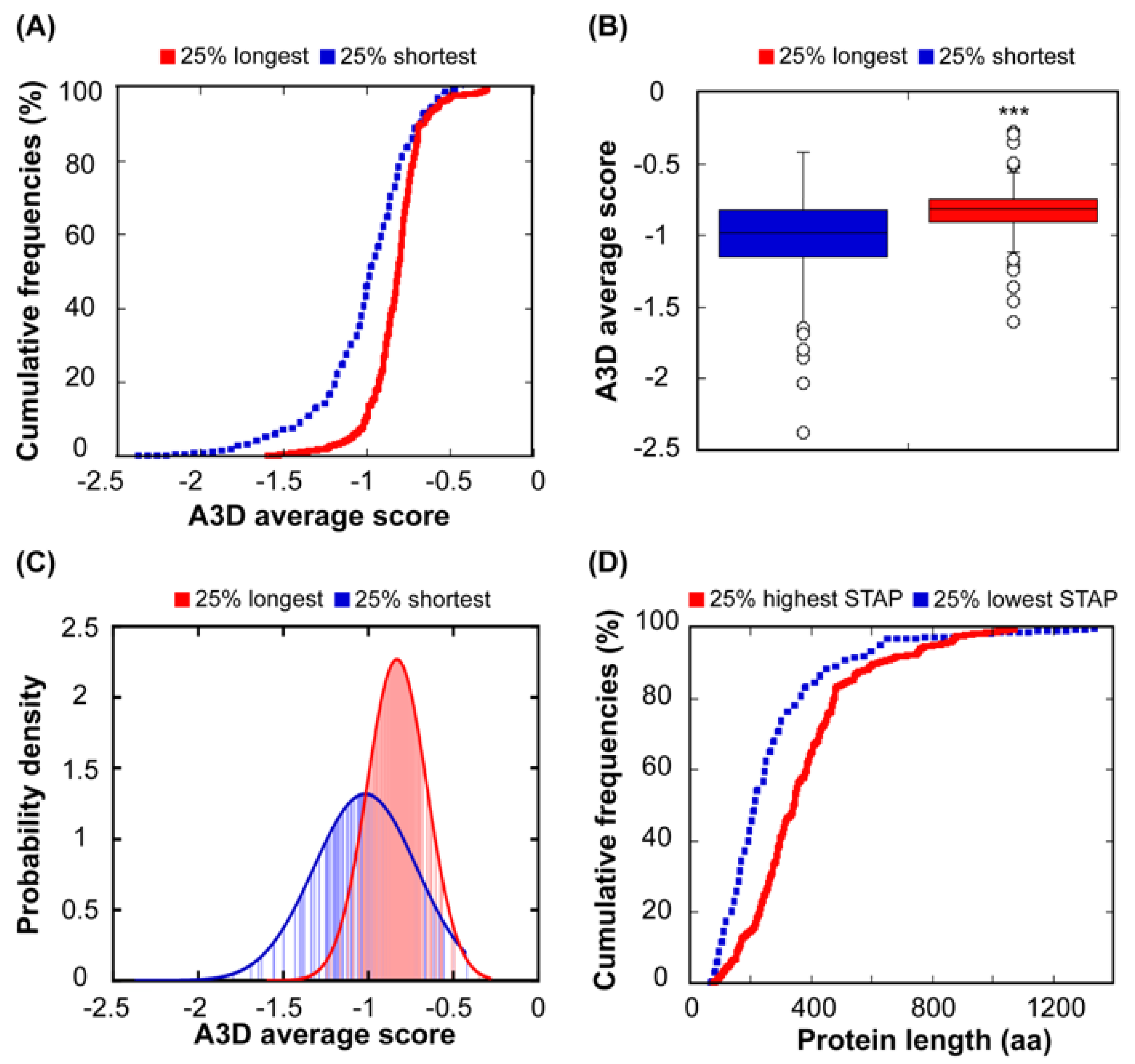
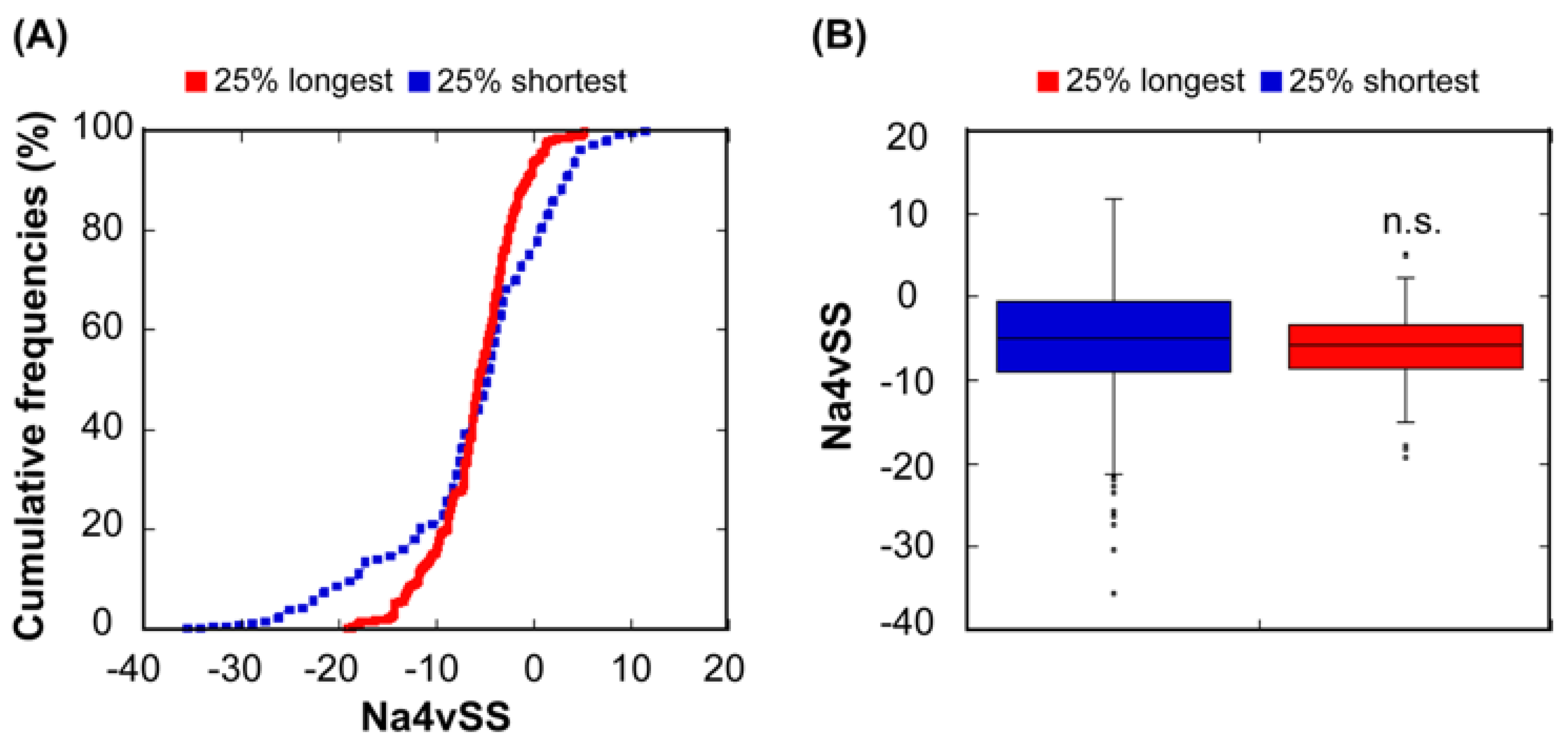
3.3. Relationship Between Protein Length and Structural Aggregation Propensity
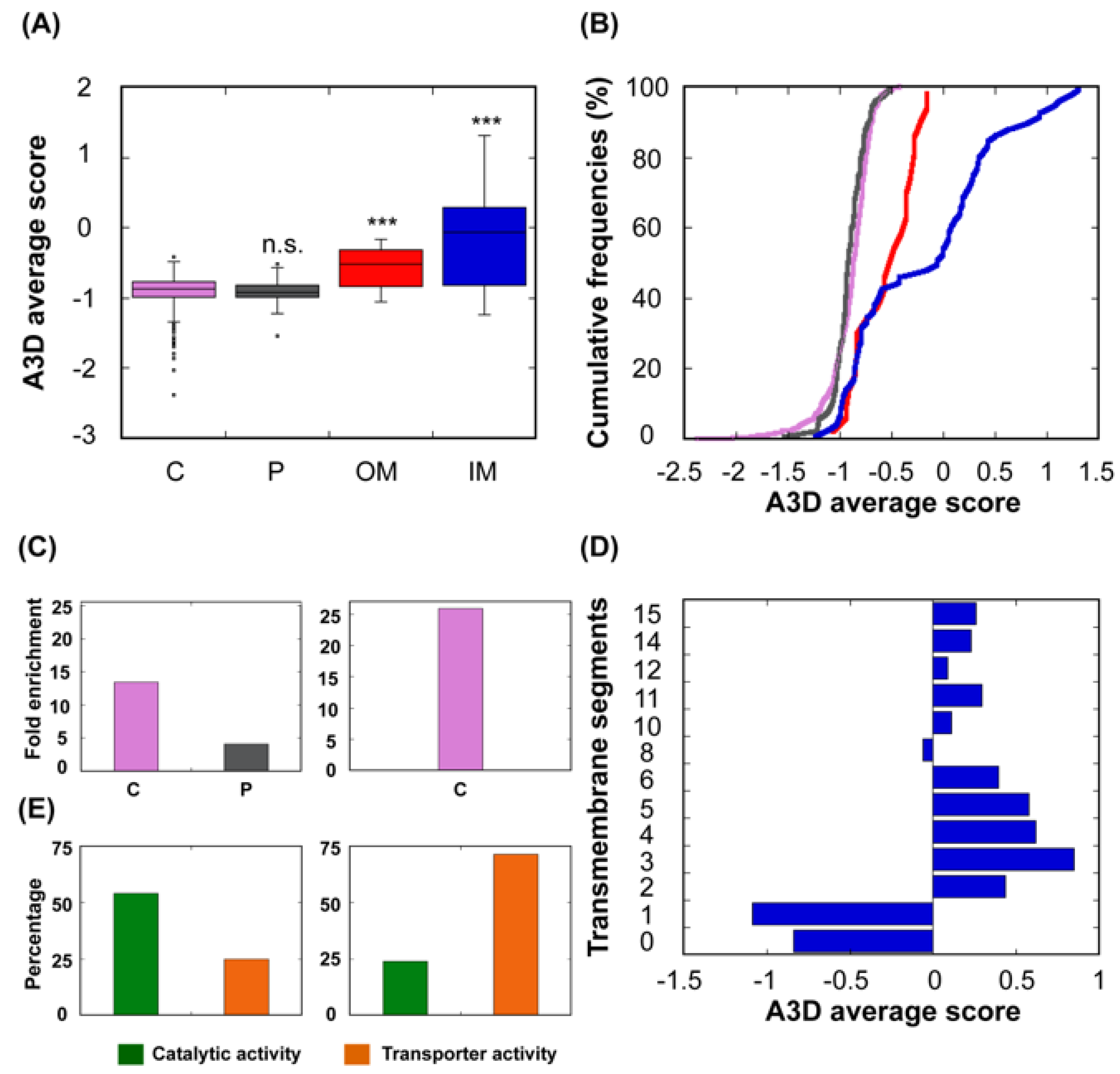
3.4. Relationship Between Structural Aggregation Propensity and Protein Function
3.5. Effect of Subcellular Location on the Structural Aggregation Propensity
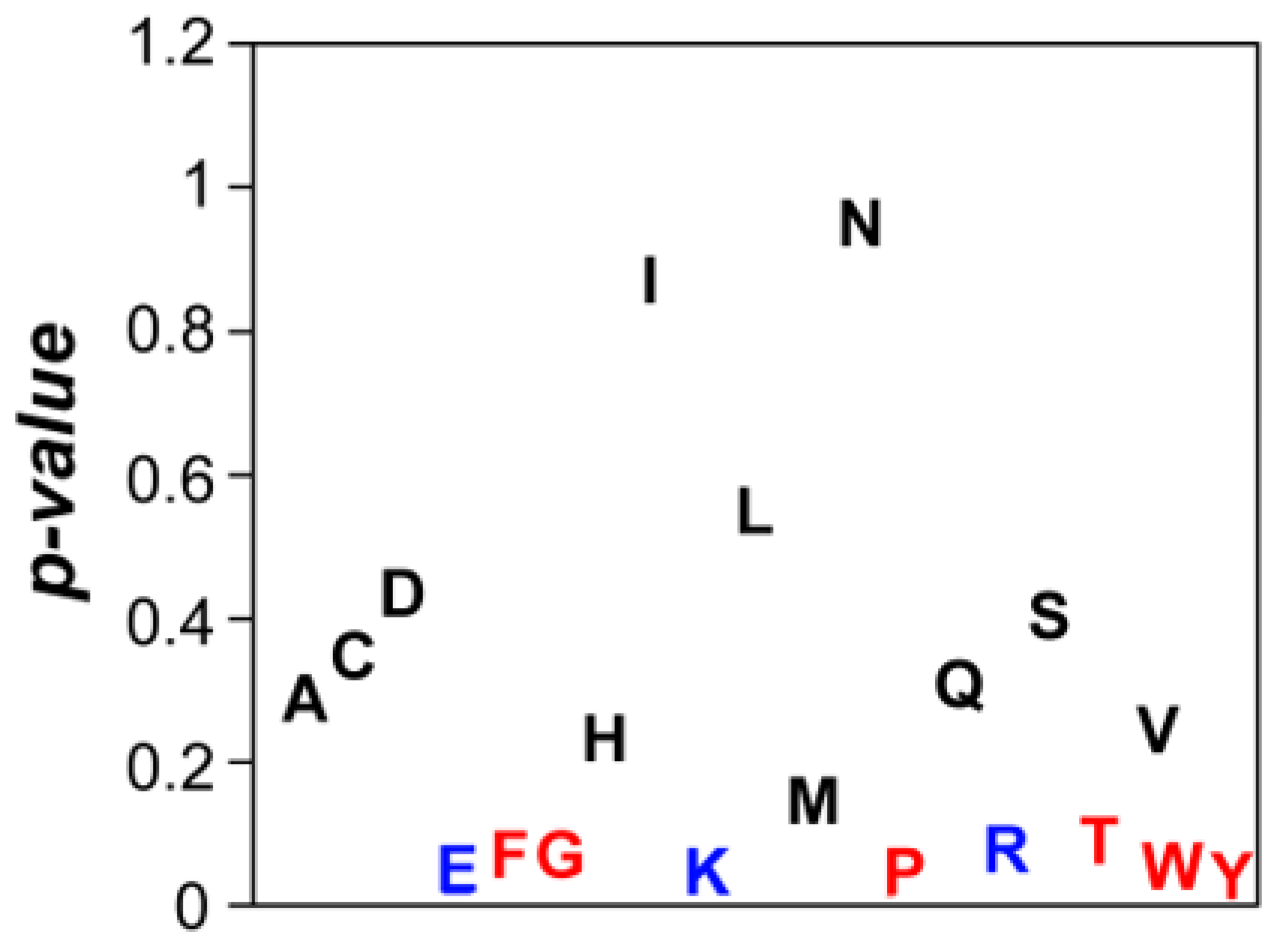
3.6. Compositional Determinants of Protein Structural Aggregation Propensity
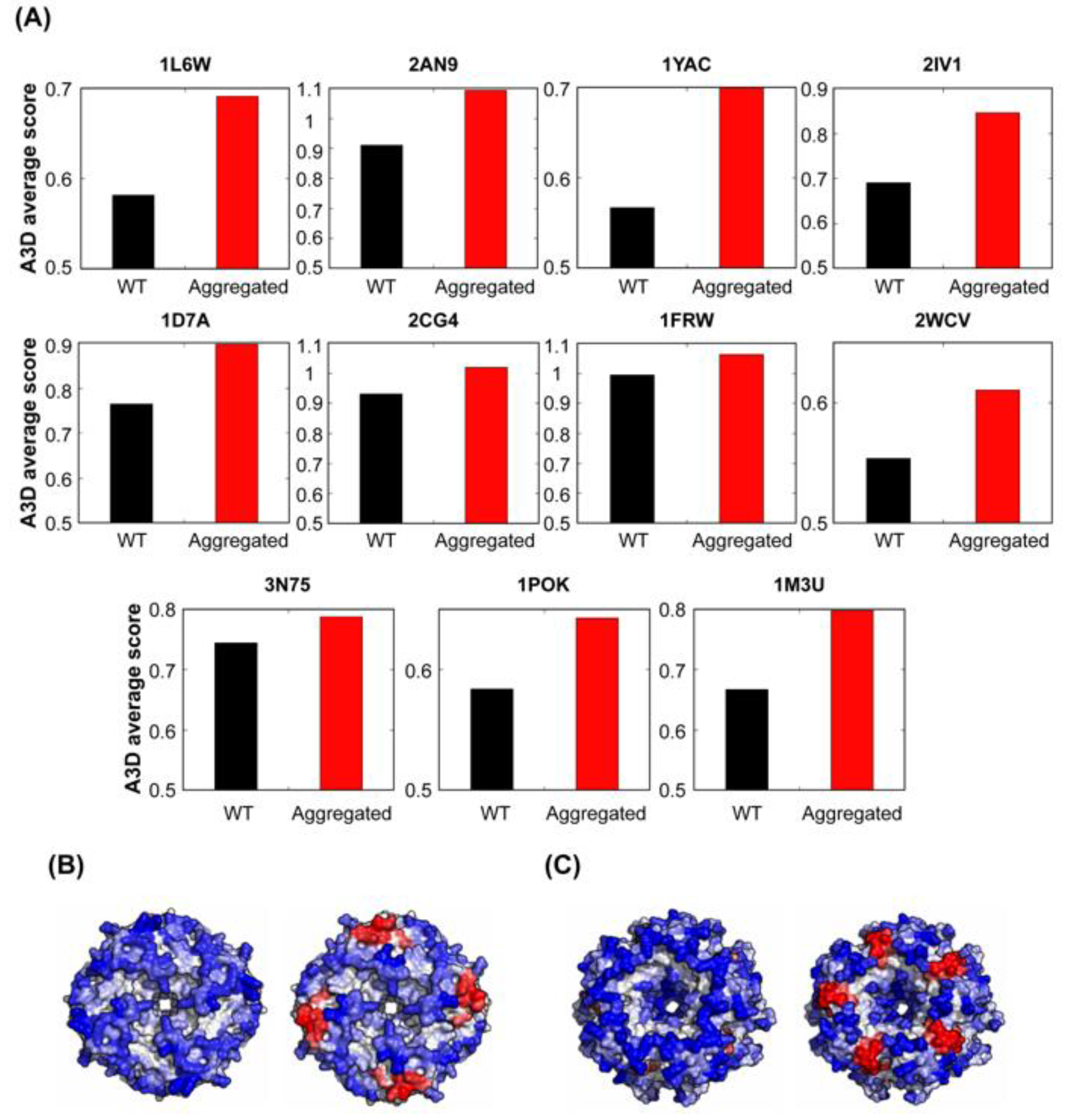
3.7. Relationship Between Functional Protein Assemblies and Protein Structural Aggregation Propensity
3.8. Protein Structural Aggregation Propensity and Bacterial Symmetric Complexes Self-Assembly
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Deeds, E.J.; Ashenberg, O.; Gerardin, J.; Shakhnovich, E.I. Robust protein protein interactions in crowded cellular environments. Proc. Natl. Acad. Sci. USA 2007, 104, 14952–14957. [Google Scholar] [CrossRef] [PubMed]
- Levy, E.D.; De, S.; Teichmann, S.A. Cellular crowding imposes global constraints on the chemistry and evolution of proteomes. Proc. Natl. Acad. Sci. USA 2012, 109, 20461–20466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartl, F.U.; Hayer-Hartl, M. Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 2009, 16, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef] [PubMed]
- Chapman, E.; Farr, G.W.; Usaite, R.; Furtak, K.; Fenton, W.A.; Chaudhuri, T.K.; Hondorp, E.R.; Matthews, R.G.; Wolf, S.G.; Yates, J.R.; et al. Global aggregation of newly translated proteins in an Escherichia coli strain deficient of the chaperonin GroEL. Proc. Natl. Acad. Sci. USA 2006, 103, 15800–15805. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, J.D.; Tsechansky, M.; Royall, A.; Boutz, D.R.; Ellington, A.D.; Marcotte, E.M. A proteomic survey of widespread protein aggregation in yeast. Mol. Biosyst. 2014, 10, 851–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- David, D.C.; Ollikainen, N.; Trinidad, J.C.; Cary, M.P.; Burlingame, A.L.; Kenyon, C. Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol. 2010, 8, 47–48. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Stefani, M.; Taddei, N.; Ramponi, G.; Dobson, C.M. Rationalization of the effects of mutations on peptide andprotein aggregation rates. Nature 2003, 424, 805–808. [Google Scholar] [CrossRef]
- Ventura, S. Sequence determinants of protein aggregation: Tools to increase protein solubility. Microb. Cell Fact. 2005, 4, 11. [Google Scholar] [CrossRef]
- Reumers, J.; Maurer-Stroh, S.; Schymkowitz, J.; Rousseau, F. Protein sequences encode safeguards against aggregation. Hum. Mutat. 2009, 30, 431–437. [Google Scholar] [CrossRef]
- Trainor, K.; Broom, A.; Meiering, E.M. Exploring the relationships between protein sequence, structure and solubility. Curr. Opin. Struct. Biol. 2017, 42, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Tartaglia, G.G.; Pechmann, S.; Dobson, C.M.; Vendruscolo, M. Life on the edge: A link between gene expression levels and aggregation rates of human proteins. Trends Biochem. Sci. 2007, 32, 204–206. [Google Scholar] [CrossRef] [PubMed]
- Gsponer, J.; Babu, M.M. Cellular Strategies for Regulating Functional and Nonfunctional Protein Aggregation. Cell Rep. 2012, 2, 1425–1437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rousseau, F.; Serrano, L.; Schymkowitz, J.W.H. How evolutionary pressure against protein aggregation shaped chaperone specificity. J. Mol. Biol. 2006, 355, 1037–1047. [Google Scholar] [CrossRef] [PubMed]
- Espargaró, A.; Castillo, V.; de Groot, N.S.; Ventura, S. The in vivo and in vitro aggregation properties of globular proteins correlate with their conformational stability: The SH3 case. J. Mol. Biol. 2008, 378, 1116–1131. [Google Scholar] [CrossRef]
- Drummond, D.A.; Wilke, C.O. Mistranslation-Induced Protein Misfolding as a Dominant Constraint on Coding-Sequence Evolution. Cell 2008, 134, 341–352. [Google Scholar] [CrossRef] [Green Version]
- Drummond, D.A.; Wilke, C.O. The evolutionary consequences of erroneous protein synthesis. Nat. Rev. Genet. 2009, 10, 715–724. [Google Scholar] [CrossRef] [Green Version]
- Falsone, A.; Falsone, S.F. Legal but lethal: Functional protein aggregation at the verge of toxicity. Front. Cell. Neurosci. 2015, 9, 1–16. [Google Scholar] [CrossRef]
- Bruce, J.B.; West, S.A.; Griffin, A.S. Functional amyloids promote retention of public goods in bacteria. Proc. R. Soc. B 2019, 286. [Google Scholar] [CrossRef]
- Marcon, G.; Plakoutsi, G.; Canale, C.; Relini, A.; Taddei, N.; Dobson, C.M.; Ramponi, G.; Chiti, F. Amyloid formation from HypF-N under conditions in which the protein is initially in its native state. J. Mol. Biol. 2005, 347, 323–335. [Google Scholar] [CrossRef]
- Garcia-Pardo, J.; Graña-Montes, R.; Fernandez-Mendez, M.; Ruyra, A.; Roher, N.; Avilés, F.X.; Lorenzo, J.; Ventura, S. Amyloid formation by human carboxypeptidase D transthyretin-like domain under physiological conditions. J. Biol. Chem. 2014, 289, 33783–33796. [Google Scholar] [CrossRef] [PubMed]
- McGuffee, S.R.; Elcock, A.H. Diffusion, crowding & protein stability in a dynamic molecular model of the bacterial cytoplasm. PLoS Comput. Biol. 2010, 6, e1000694. [Google Scholar]
- Gazit, E. The “correctly folded” state of proteins: Is it a metastable state? Angew. Chem. Int. Ed. 2002, 41, 257–259. [Google Scholar] [CrossRef]
- Ciryam, P.; Kundra, R.; Morimoto, R.I.; Dobson, C.M.; Vendruscolo, M. Supersaturation is a major driving force for protein aggregation in neurodegenerative diseases. Trends Pharmacol. Sci. 2015, 36, 72–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buell, A.K.; Dhulesia, A.; White, D.A.; Knowles, T.P.J.; Dobson, C.M.; Welland, M.E. Detailed Analysis of the Energy Barriers for Amyloid Fibril Growth. Angew. Chem. Int. Ed. 2012, 51, 5247–5251. [Google Scholar] [CrossRef] [PubMed]
- Zambrano, R.; Jamroz, M.; Szczasiuk, A.; Pujols, J.; Kmiecik, S.; Ventura, S. AGGRESCAN3D (A3D): Server for prediction of aggregation properties of protein structures. Nucleic Acids Res. 2015, 43, W306–W313. [Google Scholar] [CrossRef] [PubMed]
- Pujols, J.; Peña-Díaz, S.; Ventura, S. AGGRESCAN3D: Toward the Prediction of the Aggregation Propensities of Protein Structures. In Computational Drug Discovery and Design; Gore, M., Jagtap, U.B., Eds.; Springer: New York, NY, USA, 2018; pp. 427–443. ISBN 978-1-4939-7756-7. [Google Scholar]
- Kuriata, A.; Iglesias, V.; Pujols, J.; Kurcinski, M.; Kmiecik, S.; Ventura, S. Aggrescan3D (A3D) 2.0: Prediction and engineering of protein solubility. Nucleic Acids Res. 2019, 47, 300–307. [Google Scholar] [CrossRef]
- Kuriata, A.; Iglesias, V.; Kurcinski, M.; Ventura, S.; Kmiecik, S. Structural bioinformatics Aggrescan3D standalone package for structure-based prediction of protein aggregation properties. Bioinformatics 2019, btz143. [Google Scholar]
- Conchillo-Solé, O.; de Groot, N.S.; Avilés, F.X.; Vendrell, J.; Daura, X.; Ventura, S. AGGRESCAN: A server for the prediction and evaluation of “hot spots” of aggregation in polypeptides. BMC Bioinform. 2007, 8, 65. [Google Scholar] [CrossRef]
- de Groot, N.S.; Pallarés, I.; Avilés, F.X.; Vendrell, J.; Ventura, S. Prediction of “hot spots” of aggregation in disease-linked polypeptides. BMC Struct. Biol. 2005, 5, 18. [Google Scholar] [CrossRef]
- Belli, M.; Ramazzotti, M.; Chiti, F. Prediction of amyloid aggregation in vivo. EMBO Rep. 2011, 12, 657–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishihama, Y.; Schmidt, T.; Rappsilber, J.; Mann, M.; Hartl, F.U.; Kerner, M.J.; Frishman, D. Protein abundance profiling of the Escherichia coli cytosol. BMC Genom. 2008, 9, 102. [Google Scholar] [CrossRef] [PubMed]
- De Groot, N.S.; Ventura, S. Protein aggregation profile of the bacterial cytosol. PLoS ONE 2010, 5, e9383. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.; Bourne, P. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Weiss, M.; Simonovic, M.; Haertinger, G.; Schrimpf, S.P.; Hengartner, M.O.; von Mering, C. PaxDb, a Database of Protein Abundance Averages Across All Three Domains of Life. Mol. Cell. Proteom. 2012, 11, 492–500. [Google Scholar] [CrossRef] [Green Version]
- Boeckmann, B.; Bairoch, A.; Apweiler, R.; Blatter, M.C.; Estreicher, A.; Gasteiger, E.; Martin, M.J.; Michoud, K.; O’Donovan, C.; Phan, I.; et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003, 31, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Dennis, G.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, R60. [Google Scholar] [CrossRef]
- Gerdes, S.; Scholle, M.D.; Campbell, J.W.; Balázsi, G.; Ravasz, E.; Daugherty, M.D.; Somera, A.L.; Kyrpides, N.C.; Anderson, I.; Gelfand, M.S.; et al. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 2003, 185, 5673–5684. [Google Scholar] [CrossRef]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2. [Google Scholar] [CrossRef]
- Zhou, J.; Rudd, K.E. EcoGene 3.0. Nucleic Acids Res. 2013, 41, D613–D624. [Google Scholar] [CrossRef]
- Huerta, A.M.; Salgado, H.; Thieffry, D.; Collado-Vides, J. RegulonDB: A database on transcriptional regulation in Escherichia coli. Nucleic Acids Res. 1998, 26, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX web server: An online force field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef] [PubMed]
- Meisl, G.; Kirkegaard, J.B.; Arosio, P.; Michaels, T.C.T.; Vendruscolo, M.; Dobson, C.M.; Linse, S.; Knowles, T.P.J. Molecular mechanisms of protein aggregation from global fitting of kinetic models. Nat. Protoc. 2016, 11, 252–272. [Google Scholar] [CrossRef] [PubMed]
- Selinger, D.W.; Cheung, K.J.; Mei, R.; Johansson, E.M.; Richmond, C.S.; Blattner, F.R.; Lockhart, D.J.; Church, G.M. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat. Biotechnol. 2000, 18, 1262–1268. [Google Scholar] [CrossRef] [PubMed]
- Monsellier, E.; Ramazzotti, M.; Taddei, N.; Chiti, F. Aggregation propensity of the human proteome. PLoS Comput. Biol. 2008, 4, e1000199. [Google Scholar] [CrossRef] [PubMed]
- Tartaglia, G.G.; Vendruscolo, M. Correlation between mRNA expression levels and protein aggregation propensities in subcellular localisations. Mol. Biosyst. 2009, 5, 1873–1876. [Google Scholar] [CrossRef]
- Olzscha, H.; Schermann, S.M.; Woerner, A.C.; Pinkert, S.; Hecht, M.H.; Tartaglia, G.G.; Vendruscolo, M.; Hayer-Hartl, M.; Hartl, F.U.; Vabulas, R.M. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 2011, 144, 67–78. [Google Scholar] [CrossRef]
- Albu, R.F.; Chan, G.T.; Zhu, M.; Wong, E.T.C.; Taghizadeh, F.; Hu, X.; Mehran, A.E.; Johnson, J.D.; Gsponer, J.; Mayor, T. A feature analysis of lower solubility proteins in three eukaryotic systems. J. Proteom. 2015, 118, 21–38. [Google Scholar] [CrossRef]
- Chen, Y.; Dokholyan, N. V Natural selection against protein aggregation on self-interacting and essential proteins in yeast, fly, and worm. Mol. Biol. Evolut. 2008, 25, 1530–1533. [Google Scholar] [CrossRef] [PubMed]
- Linding, R.; Schymkowitz, J.; Rousseau, F.; Diella, F.; Serrano, L. A comparative study of the relationship between protein structure and β-aggregation in globular and intrinsically disordered proteins. J. Mol. Biol. 2004, 342, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Dougan, D.A.; Mogk, A.; Bukau, B. Protein folding and degradation in bacteria: To degrade or not to degrade? That is the question. Cell. Mol. Life Sci. 2002, 59, 1607–1616. [Google Scholar] [CrossRef] [PubMed]
- Santoni, V.; Molloy, M.; Rabilloud, T. Membrane proteins and proteomics: Un amour impossible? Electrophoresis 2000, 21, 1054–1070. [Google Scholar] [CrossRef]
- Cowan, S.W.; Schirmer, T.; Rummel, G.; Steiert, M.; Ghosh, R.; Pauptit, R.A.; Jansonius, J.N.; Rosenbusch, J.P. Crystal structures explain functional properties of two E. coli porins. Nature 1992, 358, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.S.; Phillips, K.J.; Liu, D.R. Supercharging proteins can impart unusual resilience. J. Am. Chem. Soc. 2007, 129, 10110–10112. [Google Scholar] [CrossRef] [PubMed]
- Miklos, A.E.; Kluwe, C.; Der, B.S.; Pai, S.; Sircar, A.; Hughes, R.A.; Berrondo, M.; Xu, J.; Codrea, V.; Buckley, P.E.; et al. Structure-based design of supercharged, highly thermoresistant antibodies. Chem. Biol. 2012, 19, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Jeppesen, M.D.; Westh, P.; Otzen, D.E. The role of protonation in protein fibrillation. FEBS Lett. 2010, 584, 780–784. [Google Scholar] [CrossRef] [PubMed]
- Morshedi, D.; Ebrahim-Habibi, A.; Moosavi-Movahedi, A.A.; Nemat-Gorgani, M. Chemical modification of lysine residues in lysozyme may dramatically influence its amyloid fibrillation. Biochim. Biophys. Acta-Proteins Proteom. 2010, 1804, 714–722. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, R.; Degac, J.; Helms, V. Composition of overlapping protein-protein and protein-ligand interfaces. PLoS ONE 2015, 10, e0140965. [Google Scholar] [CrossRef] [PubMed]
- Villar-Piqué, A.; Ventura, S. Modeling amyloids in bacteria. Microb. Cell Fact. 2012, 11, 166. [Google Scholar] [CrossRef] [PubMed]
- Ciryam, P.; Tartaglia, G.G.; Morimoto, R.I.; Dobson, C.M.; Vendruscolo, M. Widespread Aggregation and Neurodegenerative Diseases Are Associated with Supersaturated Proteins. Cell Rep. 2013, 5, 781–790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hörnberg, A.; Eneqvist, T.; Olofsson, A.; Lundgren, E.; Sauer-Eriksson, A.E. A comparative analysis of 23 structures of the amyloidogenic protein transthyretin. J. Mol. Biol. 2000, 302, 649–669. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, M.; Durazo, A.; Sohn, S.H.; Strong, C.D.; Gralla, E.B.; Whitelegge, J.P.; Valentine, J.S. Initiation and elongation in fibrillation of ALS-linked superoxide dismutase. Proc. Natl. Acad. Sci. USA 2008, 105, 18663–18668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Seisdedos, H.; Empereur-Mot, C.; Elad, N.; Levy, E.D. Proteins evolve on the edge of supramolecular self-assembly. Nature 2017, 548, 244–247. [Google Scholar] [CrossRef] [PubMed]
- Gil-Garcia, M.; Bañó-Polo, M.; Varejão, N.; Jamroz, M.; Kuriata, A.; Díaz-Caballero, M.; Lascorz, J.; Morel, B.; Navarro, S.; Reverter, D.; et al. Combining Structural Aggregation Propensity and Stability Predictions To Redesign Protein Solubility. Mol. Pharm. 2018, 15, 3846–3859. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carija, A.; Pinheiro, F.; Iglesias, V.; Ventura, S. Computational Assessment of Bacterial Protein Structures Indicates a Selection Against Aggregation. Cells 2019, 8, 856. https://doi.org/10.3390/cells8080856
Carija A, Pinheiro F, Iglesias V, Ventura S. Computational Assessment of Bacterial Protein Structures Indicates a Selection Against Aggregation. Cells. 2019; 8(8):856. https://doi.org/10.3390/cells8080856
Chicago/Turabian StyleCarija, Anita, Francisca Pinheiro, Valentin Iglesias, and Salvador Ventura. 2019. "Computational Assessment of Bacterial Protein Structures Indicates a Selection Against Aggregation" Cells 8, no. 8: 856. https://doi.org/10.3390/cells8080856
APA StyleCarija, A., Pinheiro, F., Iglesias, V., & Ventura, S. (2019). Computational Assessment of Bacterial Protein Structures Indicates a Selection Against Aggregation. Cells, 8(8), 856. https://doi.org/10.3390/cells8080856





