HSPA8/HSC70 in Immune Disorders: A Molecular Rheostat that Adjusts Chaperone-Mediated Autophagy Substrates
Abstract
1. Introduction
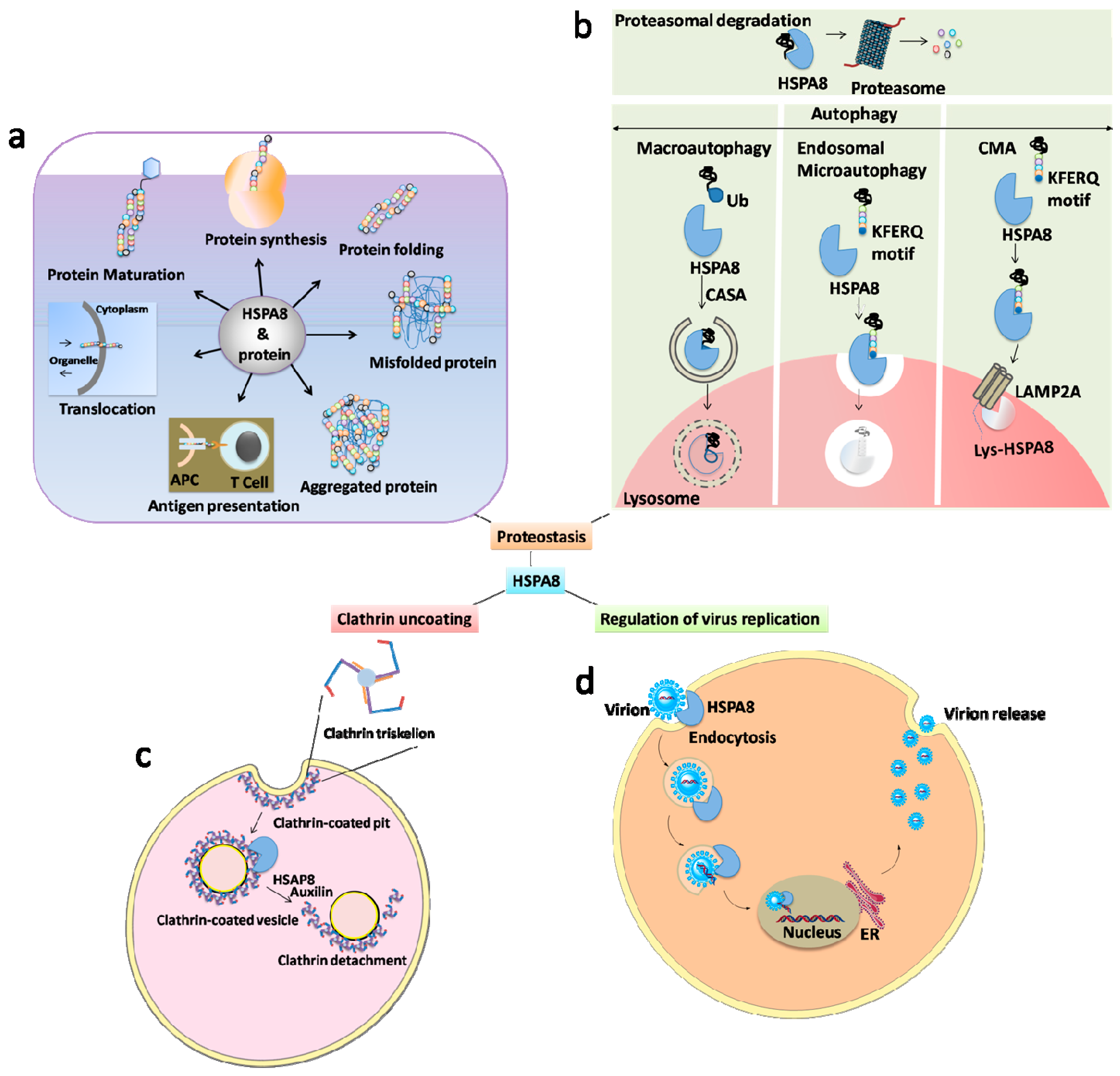
2. HSPA8 and Autophagy
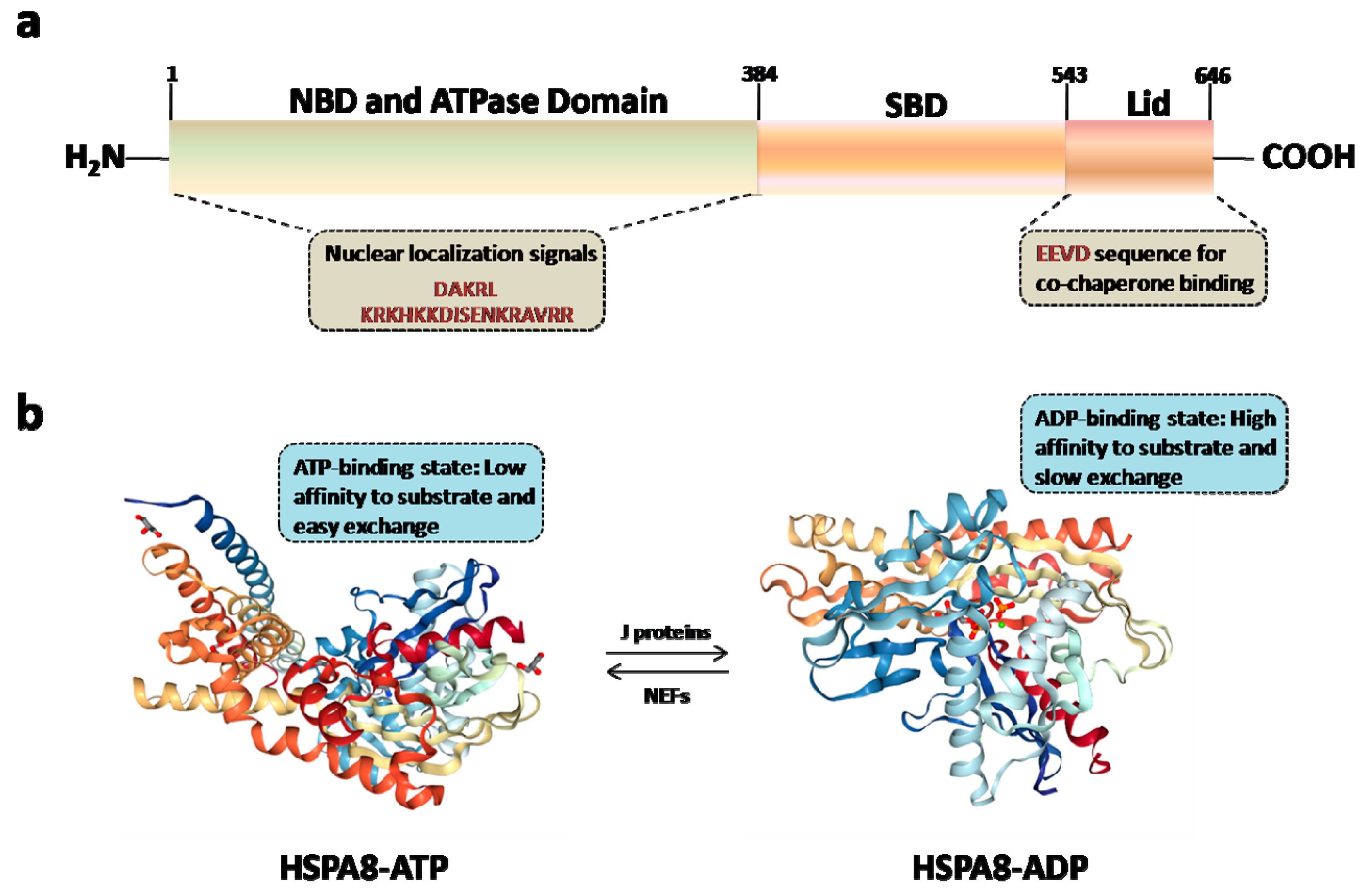
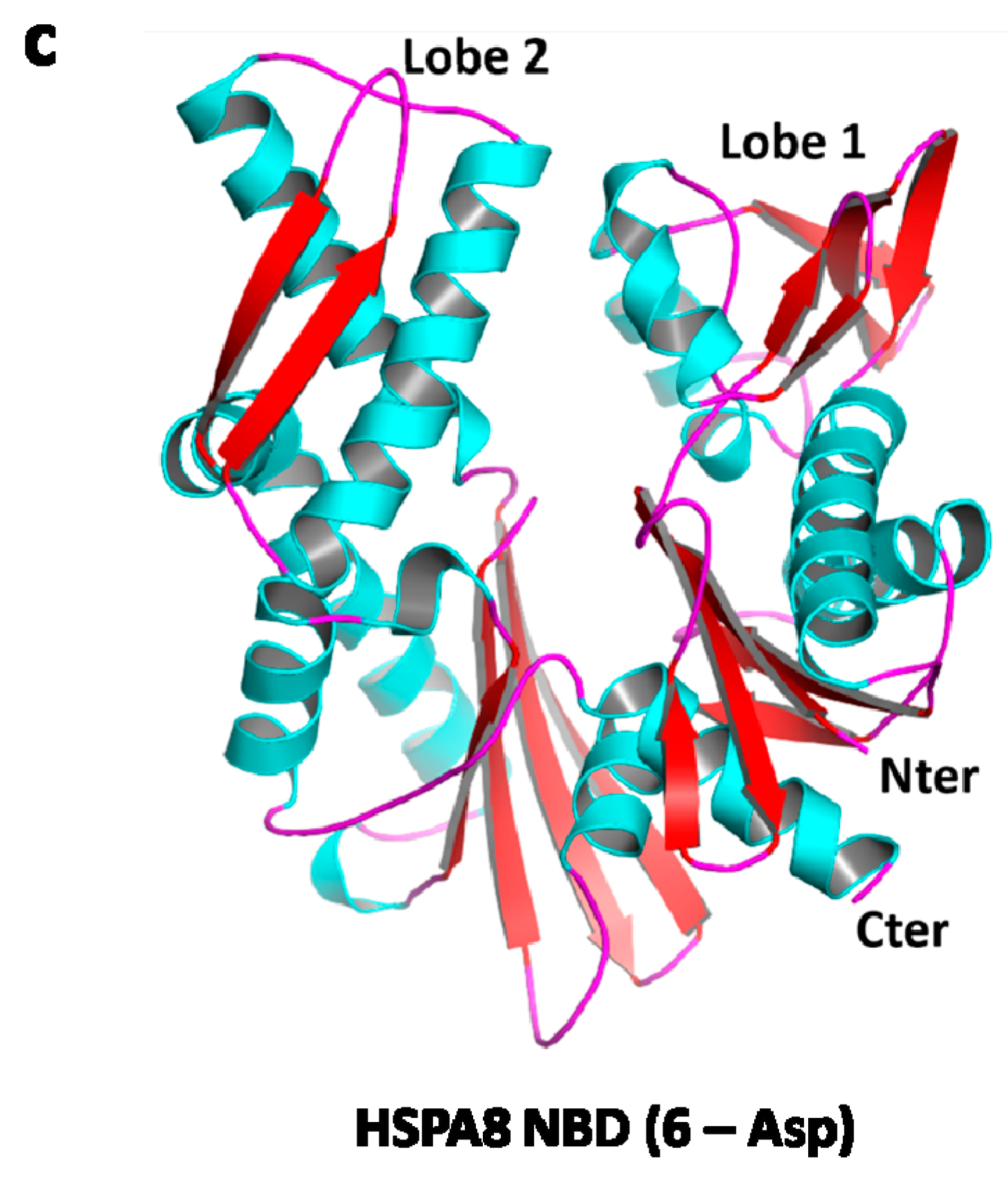
3. HSPA8 Structure and Structure-Function Relationships
4. HSPA8 and Immune Disorders
5. HSPA8 Chemical Activators
6. HSPA8/HSPA8 Chemical Inhibitors
7. HSPA8/HSPA8 as a Therapeutic Target in Clinical Trials
8. Conclusion and Future Prospects
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, T.; Daniels, C.K.; Cao, S. Comprehensive review on the HSC70 functions, interactions with related molecules and involvement in clinical diseases and therapeutic potential. Pharmacol. Ther. 2012, 136, 354–374. [Google Scholar] [CrossRef] [PubMed]
- Stricher, F.; Macri, C.; Ruff, M.; Muller, S. HSPA8/HSC70 chaperone protein: Structure, function, and chemical targeting. Autophagy 2013, 9, 1937–1954. [Google Scholar] [CrossRef] [PubMed]
- Radons, J. The human HSP70 family of chaperones: Where do we stand? Cell Stress Chaperones 2016, 21, 379–404. [Google Scholar] [CrossRef] [PubMed]
- Bukau, B.; Weissman, J.; Horwich, A. Molecular chaperones and protein quality control. Cell 2006, 125, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.P.; Gierasch, L.M. Recent advances in the structural and mechanistic aspects of Hsp70 molecular chaperones. J. Biol. Chem. 2019, 294, 2085–2097. [Google Scholar] [CrossRef] [PubMed]
- Auger, I.; Escola, J.M.; Gorvel, J.P.; Roudier, J. HLA–DR4 and HLA–DR10 motifs that carry susceptibility to rheumatoid arthritis bind 70–KD heat shock proteins. Nat. Med. 1996, 2, 306–310. [Google Scholar] [CrossRef] [PubMed]
- Panjwani, N.; Akbari, O.; Garcia, S.; Brazil, M.; Stockinger, B. The HSC73 molecular chaperone: Involvement in MHC class II antigen presentation. J. Immunol. 1999, 163, 1936–1942. [Google Scholar]
- Chang, H.C.; Newmyer, S.L.; Hull, M.J.; Ebersold, M.; Schmid, S.L.; Mellman, I. Hsc70 is required for endocytosis and clathrin function in Drosophila. J. Cell Biol. 2002, 159, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Böcking, T.; Wolf, M.; Grigorieff, N.; Kirchhausen, T.; Harrison, S.C. Structure of clathrin coat with bound Hsc70 and auxilin: Mechanism of Hsc70-facilitated disassembly. EMBO J. 2010, 29, 655–665. [Google Scholar] [CrossRef] [PubMed]
- Rothnie, A.; Clarke, A.R.; Kuzmic, P.; Cameron, A.; Smith, C.J. A sequential mechanism for clathrin cage disassembly by 70-kDa heat-shock cognate protein (Hsc70) and auxilin. Proc. Natl. Acad. Sci. USA 2011, 108, 6927–6932. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, R.; Nillegoda, N.B.; Mayer, M.P.; Bukau, B. The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 2019, in press. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, S.; Cuervo, A.M. Chaperone-mediated autophagy: A unique way to enter the lysosome world. Trends Cell Biol. 2012, 22, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Penke, B.; Bogár, F.; Crul, T.; Sántha, M.; Tóth, M.; Vígh, L. Heat shock proteins and autophagy pathways in neuroprotection: From molecular bases to pharmacological interventions. Int. J. Mol. Sci. 2018, 19, 325. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, S.; Cuervo, A.M. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 365–381. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, S.; Cuervo, A.M. Chaperones in autophagy. Pharmacol. Res. 2012, 66, 484–493. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.-T.; Simoneschi, D.; Keegan, S.; Melville, D.; Adler, N.S.; Saraf, A.; Florens, L.; Washburn, M.P.; Cavasotto, C.N.; Fenyö, D. The ULK1-FBXW5-SEC23B nexus controls autophagy. eLife 2018, 7, e42253. [Google Scholar] [CrossRef]
- Shima, T.; Kirisako, H.; Nakatogawa, H. COPII vesicles contribute to autophagosomal membranes. J. Cell Biol. 2019, 218, 1503–1510. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Eskelinen, E.-L.; Deretic, V. Autophagosomes, phagosomes, autolysosomes, phagolysosomes, autophagolysosomes… wait, I’m confused. Autophagy 2014, 10, 549–551. [Google Scholar] [CrossRef]
- Tekirdag, K.; Cuervo, A.M. Chaperone-mediated autophagy and endosomal microautophagy: Jointed by a chaperone. J. Biol. Chem. 2018, 293, 5414–5424. [Google Scholar] [CrossRef]
- Fullgrabe, J.; Klionsky, D.J.; Joseph, B. The return of the nucleus: Transcriptional and epigenetic control of autophagy. Nat. Rev. Mol. Cell Biol. 2014, 15, 65–74. [Google Scholar] [CrossRef]
- Feng, Y.; Yao, Z.; Klionsky, D.J. How to control self-digestion: Transcriptional, post-transcriptional, and post-translational regulation of autophagy. Trends Cell Biol. 2015, 25, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.H.; Kim, K.I. Epigenetic Control of Autophagy: Nuclear Events Gain More Attention. Mol. Cell 2017, 65, 781–785. [Google Scholar] [CrossRef] [PubMed]
- Hargarten, J.C.; Williamson, P.R. Epigenetic Regulation of Autophagy: A Path to the Control of Autoimmunity. Front. Immunol. 2018, 9, 1864. [Google Scholar] [CrossRef] [PubMed]
- Agarraberes, F.A.; Dice, J.F. A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J. Cell Sci. 2001, 114, 2491–2499. [Google Scholar] [PubMed]
- Dengjel, J.; Schoor, O.; Fischer, R.; Reich, M.; Kraus, M.; Müller, M.; Kreymborg, K.; Altenberend, F.; Brandenburg, J.; Kalbacher, H.; et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 7922–7927. [Google Scholar] [CrossRef] [PubMed]
- Aichinger, M.; Wu, C.; Nedjic, J.; Klein, L. Macroautophagy substrates are loaded onto MHC class II of medullary thymic epithelial cells for central tolerance. J. Exp. Med. 2013, 210, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Blum, J.S.; Wearsch, P.A.; Cresswell, P. Pathways of Antigen Processing. Annu. Rev. Immunol. 2013, 31, 443–473. [Google Scholar] [CrossRef] [PubMed]
- Deffit, S.N.; Blum, J.S. Macronutrient deprivation modulates antigen trafficking and immune recognition through HSC70 accessibility. J. Immunol. 2015, 194, 1446–1453. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Zhao, X.; Song, Y.; Cheng, H.; Zhou, R. Nuclear autophagy: An evolutionarily conserved mechanism of nuclear degradation in the cytoplasm. Autophagy 2016, 12, 1973–1983. [Google Scholar] [CrossRef]
- Anding, A.L.; Baehrecke, E.H. Cleaning House: Selective Autophagy of Organelles. Dev. Cell 2017, 41, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Gatica, D.; Lahiri, V.; Klionsky, D.J. Cargo recognition and degradation by selective autophagy. Nat. Cell Biol. 2018, 20, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J.; Debnath, J. Autophagy at the crossroads of catabolism and anabolism. Nat. Rev. Mol. Cell Biol. 2015, 16, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Janssen, A.F.J.; Katrukha, E.A.; van Straaten, W.; Verlhac, P.; Reggiori, F.; Kapitein, L.C. Probing aggrephagy using chemically-induced protein aggregates. Nat. Commun. 2018, 9, 4245. [Google Scholar] [CrossRef] [PubMed]
- Sahu, R.; Kaushik, S.; Clement, C.C.; Cannizzo, E.S.; Scharf, B.; Follenzi, A.; Potolicchio, I.; Nieves, E.; Cuervo, A.M.; Santambrogio, L. Microautophagy of Cytosolic Proteins by Late Endosomes. Dev. Cell 2011, 20, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xue, Y.; Xu, X.; Wang, G.; Liu, Y.; Wu, H.; Li, W.; Wang, Y.; Chen, Z.; Zhang, W. A mitochondrial FUNDC1/HSC70 interaction organizes the proteostatic stress response at the risk of cell morbidity. EMBO J. 2019, 38, e98786. [Google Scholar] [CrossRef] [PubMed]
- Kettern, N.; Dreiseidler, M.; Tawo, R.; Höhfeld, J. Chaperone-assisted degradation: Multiple paths to destruction. Biol. Chem. 2010, 391, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Arndt, V.; Dick, N.; Tawo, R.; Dreiseidler, M.; Wenzel, D.; Hesse, M.; Fürst, D.O.; Saftig, P.; Saint, R.; Fleischmann, B.K.; et al. Chaperone-Assisted Selective Autophagy Is Essential for Muscle Maintenance. Curr. Biol. 2010, 20, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Bonam, S.R.; Schall, N.; Kuhn, L.; Hammann, P.; Chaloin, O.; Madinier, J.-B.; Briand, J.-P.; Page, N.; Muller, S. Blocking nuclear export of HSPA8 after heat shock stress severely alters cell survival. Sci. Rep. 2018, 8, 16820. [Google Scholar] [CrossRef] [PubMed]
- Aprile, F.A.; Dhulesia, A.; Stengel, F.; Roodveldt, C.; Benesch, J.L.; Tortora, P.; Robinson, C.V.; Salvatella, X.; Dobson, C.M.; Cremades, N. Hsp70 oligomerization is mediated by an interaction between the interdomain linker and the substrate-binding domain. PLoS ONE 2013, 8, e67961. [Google Scholar] [CrossRef]
- Akhter, S.; Chakraborty, S.; Moutinho, D.; Álvarez-Coiradas, E.; Rosa, I.; Viñuela, J.; Domínguez, E.; García, A.; Requena, J.R. The human VGF-derived bioactive peptide TLQP-21 binds heat shock 71 kDa protein 8 (HSPA8) on the surface of SH-SY5Y cells. PLoS ONE 2017, 12, e0185176. [Google Scholar] [CrossRef]
- Zhang, Z.; Cellitti, J.; Teriete, P.; Pellecchia, M.; Stec, B. New crystal structures of HSC-70 ATP binding domain confirm the role of individual binding pockets and suggest a new method of inhibition. Biochimie 2015, 108, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.M.; Westwood, I.M.; Osborne, J.D.; Matthews, T.P.; Cheeseman, M.D.; Rowlands, M.G.; Jeganathan, F.; Burke, R.; Lee, D.; Kadi, N. A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms. Sci. Rep. 2016, 6, 34701. [Google Scholar] [CrossRef] [PubMed]
- Williamson, D.S.; Borgognoni, J.; Clay, A.; Daniels, Z.; Dokurno, P.; Drysdale, M.J.; Foloppe, N.; Francis, G.L.; Graham, C.J.; Howes, R. Novel adenosine-derived inhibitors of 70 kDa heat shock protein, discovered through structure-based design. J. Med. Chem. 2009, 52, 1510–1513. [Google Scholar] [CrossRef] [PubMed]
- Rauch, J.N.; Gestwicki, J.E. Binding of human nucleotide exchange factors to heat shock protein 70 (Hsp70) generates functionally distinct complexes in vitro. J. Biol. Chem. 2014, 289, 1402–1414. [Google Scholar] [CrossRef] [PubMed]
- Rauch, J.N.; Zuiderweg, E.R.; Gestwicki, J.E. Non-canonical interactions between heat shock cognate protein 70 (Hsc70) and Bcl2-associated anthanogene (BAG) co-chaperones are important for client release. J. Biol. Chem. 2016, 291, 19848–19857. [Google Scholar] [CrossRef] [PubMed]
- Bański, P.; Mahboubi, H.; Kodiha, M.; Shrivastava, S.; Kanagaratham, C.; Stochaj, U. Nucleolar targeting of the chaperone hsc70 is regulated by stress, cell signaling, and a composite targeting signal which is controlled by autoinhibition. J. Biol. Chem. 2010, 285, 21858–21867. [Google Scholar] [CrossRef] [PubMed]
- Page, N.; Schall, N.; Strub, J.-M.; Quinternet, M.; Chaloin, O.; Décossas, M.; Cung, M.T.; Van Dorsselaer, A.; Briand, J.-P.; Muller, S. The spliceosomal phosphopeptide P140 controls the lupus disease by interacting with the HSC70 protein and via a mechanism mediated by γδ T cells. PLoS ONE 2009, 4, e5273. [Google Scholar] [CrossRef] [PubMed]
- Macri, C.; Wang, F.; Tasset, I.; Schall, N.; Page, N.; Briand, J.-P.; Cuervo, A.M.; Muller, S. Modulation of deregulated chaperone-mediated autophagy by a phosphopeptide. Autophagy 2015, 11, 472–486. [Google Scholar] [CrossRef]
- Florin, L.; Becker, K.A.; Sapp, C.; Lambert, C.; Sirma, H.; Müller, M.; Streeck, R.E.; Sapp, M. Nuclear translocation of papillomavirus minor capsid protein L2 requires Hsc70. J. Virol. 2004, 78, 5546–5553. [Google Scholar] [CrossRef]
- Rohde, M.; Daugaard, M.; Jensen, M.H.; Helin, K.; Nylandsted, J.; Jäättelä, M. Members of the heat-shock protein 70 family promote cancer cell growth by distinct mechanisms. Genes Dev. 2005, 19, 570–582. [Google Scholar] [CrossRef]
- Bauer, P.O.; Goswami, A.; Wong, H.K.; Okuno, M.; Kurosawa, M.; Yamada, M.; Miyazaki, H.; Matsumoto, G.; Kino, Y.; Nagai, Y.; et al. Harnessing chaperone-mediated autophagy for the selective degradation of mutant huntingtin protein. Nat. Biotechnol. 2010, 28, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Qu, B.; Jia, Y.; Liu, Y.; Wang, H.; Ren, G.; Wang, H. The detection and role of heat shock protein 70 in various nondisease conditions and disease conditions: A literature review. Cell Stress Chaperones 2015, 20, 885–892. [Google Scholar] [CrossRef] [PubMed]
- Milani, A.; Basirnejad, M.; Bolhassani, A. Heat-shock proteins in diagnosis and treatment: An overview of different biochemical and immunological functions. Immunotherapy 2019, 11, 215–239. [Google Scholar] [CrossRef]
- Crotzer, V.L.; Blum, J.S. Autophagy and intracellular surveillance: Modulating MHC class II antigen presentation with stress. Proc. Natl. Acad. Sci. USA 2005, 102, 7779–7780. [Google Scholar] [CrossRef] [PubMed]
- Kettern, N.; Rogon, C.; Limmer, A.; Schild, H.; Höhfeld, J. The Hsc/Hsp70 co-chaperone network controls antigen aggregation and presentation during maturation of professional antigen presenting cells. PLoS ONE 2011, 6, e16398. [Google Scholar] [CrossRef] [PubMed]
- Deffit, S.N.; Blum, J.S. A central role for HSC70 in regulating antigen trafficking and MHC class II presentation. Mol. Immunol. 2015, 68, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Page, N.; Gros, F.; Schall, N.; Décossas, M.; Bagnard, D.; Briand, J.-P.; Muller, S. HSC70 blockade by the therapeutic peptide P140 affects autophagic processes and endogenous MHCII presentation in murine lupus. Ann. Rheum. Dis. 2011, 70, 837–843. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Muller, S. Manipulating autophagic processes in autoimmune diseases: A special focus on modulating chaperone-mediated autophagy, an emerging therapeutic target. Front. Immunol. 2015, 6, 252. [Google Scholar] [CrossRef] [PubMed]
- Raghawan, A.K.; Ramaswami, R.; Radha, V.; Swarup, G. HSC70 regulates cold-induced caspase-1 hyperactivation by an autoinflammation-causing mutant of cytoplasmic immune receptor NLRC4. BioRxiv 2019, 578138. [Google Scholar]
- Brosnan, C.F.; Raine, C.S. Mechanisms of immune injury in multiple sclerosis. Brain Pathol. 1996, 6, 243–257. [Google Scholar] [CrossRef]
- Turturici, G.; Tinnirello, R.; Sconzo, G.; Asea, A.; Savettieri, G.; Ragonese, P.; Geraci, F. Positive or negative involvement of heat shock proteins in multiple sclerosis pathogenesis: An overview. J. Neuropathol. Exp. Neurol. 2014, 73, 1092–1106. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Coyne, A.N.; Lorenzini, I.; Chou, C.-C.; Torvund, M.; Rogers, R.S.; Starr, A.; Zaepfel, B.L.; Levy, J.; Johannesmeyer, J.; Schwartz, J.C. Post-transcriptional inhibition of Hsc70-4/HSPA8 expression leads to synaptic vesicle cycling defects in multiple models of ALS. Cell Rep. 2017, 21, 110–125. [Google Scholar] [CrossRef] [PubMed]
- Tsukimi, Y.; Okabe, S. Recent advances in gastrointestinal pathophysiology: Role of heat shock proteins in mucosal defense and ulcer healing. Biol. Pharm. Bull. 2001, 24, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hirota, K.; Nakamura, H.; Masutani, H.; Yodoi, J. Thioredoxin superfamily and thioredoxin-inducing agents. Ann. N. Y. Acad. Sci. 2002, 957, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Lennikov, A.; Kitaichi, N.; Kase, S.; Noda, K.; Horie, Y.; Nakai, A.; Ohno, S.; Ishida, S. Induction of heat shock protein 70 ameliorates ultraviolet-induced photokeratitis in mice. Int. J. Mol. Sci. 2013, 14, 2175–2189. [Google Scholar] [CrossRef] [PubMed]
- Jacquier-Sarlin, M.R.; Jornot, L.; Polla, B.S. Differential expression and regulation of hsp70 and hsp90 by phorbol esters and heat shock. J. Biol. Chem. 1995, 270, 14094–14099. [Google Scholar] [CrossRef]
- Evans, C.; Wisén, S.; Gestwicki, J. Heat shock proteins 70 and 90 inhibit early stages of amyloid beta-(1-42) aggregation in vitro. J. Biol. Chem. 2006, 281, 33182–33191. [Google Scholar] [CrossRef] [PubMed]
- Jinwal, U.K.; Miyata, Y.; Koren, J.; Jones, J.R.; Trotter, J.H.; Chang, L.; O’Leary, J.; Morgan, D.; Lee, D.C.; Shults, C.L. Chemical manipulation of hsp70 ATPase activity regulates tau stability. J. Neurosci. 2009, 29, 12079–12088. [Google Scholar] [CrossRef]
- Brochu, C.; Haimeur, A.; Ouellette, M. The heat shock protein HSP70 and heat shock cognate protein HSC70 contribute to antimony tolerance in the protozoan parasite Leishmania. Cell Stress Chaperones 2004, 9, 294. [Google Scholar] [CrossRef]
- Shao, H.; Li, X.; Moses, M.A.; Gilbert, L.A.; Kalyanaraman, C.; Young, Z.T.; Chernova, M.; Journey, S.N.; Weissman, J.S.; Hann, B. Exploration of Benzothiazole Rhodacyanines as Allosteric Inhibitors of Protein–Protein Interactions with Heat Shock Protein 70 (Hsp70). J. Med. Chem. 2018, 61, 6163–6177. [Google Scholar] [CrossRef]
- Yang, X.; Tohda, C. Diosgenin restores Aβ-induced axonal degeneration by reducing the expression of heat shock cognate 70 (HSC70). Sci. Rep. 2018, 8, 11707. [Google Scholar] [CrossRef]
- Baek, K.-H.; Zhang, H.; Lee, B.R.; Kwon, Y.-G.; Ha, S.-J.; Shin, I. A small molecule inhibitor for ATPase activity of Hsp70 and Hsc70 enhances the immune response to protein antigens. Sci. Rep. 2015, 5, 17642. [Google Scholar] [CrossRef]
- Ko, S.-K.; Kim, J.; Na, D.C.; Park, S.; Park, S.-H.; Hyun, J.Y.; Baek, K.-H.; Kim, N.D.; Kim, N.-K.; Park, Y.N. A small molecule inhibitor of ATPase activity of HSP70 induces apoptosis and has antitumor activities. Chem. Biol. 2015, 22, 391–403. [Google Scholar] [CrossRef]
- Cho, H.J.; Kim, G.-H.; Park, S.-H.; Hyun, J.Y.; Kim, N.-K.; Shin, I. Probing the effect of an inhibitor of an ATPase domain of Hsc70 on clathrin-mediated endocytosis. Mol. Biosyst. 2015, 11, 2763–2769. [Google Scholar] [CrossRef]
- Ohlman, S.; Gannedahl, G.; Tyden, G.; Tufveson, G.; Groth, C. Treatment of renal transplant rejection with 15-deoxyspergualin—A dose-finding study in man. Transplant. Proc. 1992, 24, 318–320. [Google Scholar]
- Hoeger, P.H.; Tepper, M.A.; Faith, A.; Higgins, J.A.; Lamb, J.R.; Geha, R.S. Immunosuppressant deoxyspergualin inhibits antigen processing in monocytes. J. Immunol. 1994, 153, 3908–3916. [Google Scholar]
- Nadeau, K.; Nadler, S.G.; Saulnier, M.; Walsh, C.T.; Tepper, M.A. Quantitation of the interaction of the immunosuppressant deoxyspergualin and analogs with Hsc70 and Hsp90. Biochemistry 1994, 33, 2561–2567. [Google Scholar] [CrossRef]
- Nadler, S.G.; Dischino, D.D.; Malacko, A.R.; Cleaveland, J.S.; Fujihara, S.M.; Marquardt, H. Identification of a binding site on Hsc70 for the immunosuppressant 15-deoxyspergualin. Biochem. Biophys. Res. Commun. 1998, 253, 176–180. [Google Scholar] [CrossRef]
- Holcombe, H.; Mellman, I.; Janeway, C.A.; Bottomly, K.; Dittel, B.N. The immunosuppressive agent 15-deoxyspergualin functions by inhibiting cell cycle progression and cytokine production following naive T cell activation. J. Immunol. 2002, 169, 4982–4989. [Google Scholar] [CrossRef]
- Morikawa, A.; Kato, Y.; Sugiyama, T.; Koide, N.; Kawai, M.; Fukada, M.; Yoshida, T.; Yokochi, T. Altered expression of constitutive type and inducible type heat shock proteins in response of D-galactosamine-sensitized mice to lipopolysaccharide as an experimental endotoxic shock model. FEMS Immunol. Med. Microbiol. 1998, 21, 37–45. [Google Scholar]
- Gao, L.-M.; Han, Y.-X.; Wang, Y.-P.; Li, Y.-H.; Shan, Y.-Q.; Li, X.; Peng, Z.-G.; Bi, C.-W.; Zhang, T.; Du, N.-N. Design and synthesis of oxymatrine analogues overcoming drug resistance in hepatitis B virus through targeting host heat stress cognate 70. J. Med. Chem. 2011, 54, 869–876. [Google Scholar] [CrossRef]
- Peng, Z.G.; Fan, B.; Du, N.N.; Wang, Y.P.; Gao, L.M.; Li, Y.H.; Li, Y.H.; Liu, F.; You, X.F.; Han, Y.X. Small molecular compounds that inhibit hepatitis C virus replication through destabilizing heat shock cognate 70 messenger RNA. Hepatology 2010, 52, 845–853. [Google Scholar] [CrossRef]
- Du, N.-N.; Li, X.; Wang, Y.-P.; Liu, F.; Liu, Y.-X.; Li, C.-X.; Peng, Z.-G.; Gao, L.-M.; Jiang, J.-D.; Song, D.-Q. Synthesis, structure—Activity relationship and biological evaluation of novel N-substituted matrinic acid derivatives as host heat-stress cognate 70 (Hsc70) down-regulators. Bioorg. Med. Chem. Lett. 2011, 21, 4732–4735. [Google Scholar] [CrossRef]
- Young, Z.T.; Rauch, J.N.; Assimon, V.A.; Jinwal, U.K.; Ahn, M.; Li, X.; Dunyak, B.M.; Ahmad, A.; Carlson, G.A.; Srinivasan, S.R. Stabilizing the Hsp70-Tau complex promotes turnover in models of tauopathy. Cell Chem. Biol. 2016, 23, 992–1001. [Google Scholar] [CrossRef]
- Chen, D.-Z.; Jiang, J.-D.; Zhang, K.-Q.; He, H.-P.; Di, Y.-T.; Zhang, Y.; Cai, J.-Y.; Wang, L.; Li, S.-L.; Yi, P. Evaluation of anti-HCV activity and SAR study of (+)-lycoricidine through targeting of host heat-stress cognate 70 (Hsc70). Bioorg. Med. Chem. Lett. 2013, 23, 2679–2682. [Google Scholar] [CrossRef]
- Chen, D.; Cai, J.; Yin, J.; Jiang, J.; Jing, C.; Zhu, Y.; Cheng, J.; Di, Y.; Zhang, Y.; Cao, M. Lycorine-derived phenanthridine downregulators of host Hsc70 as potential hepatitis C virus inhibitors. Future Med. Chem. 2015, 7, 561–570. [Google Scholar] [CrossRef]
- Adam, C.; Baeurle, A.; Brodsky, J.L.; Wipf, P.; Schrama, D.; Becker, J.C.; Houben, R. The HSP70 modulator MAL3-101 inhibits Merkel cell carcinoma. PLoS ONE 2014, 9, e92041. [Google Scholar] [CrossRef]
- Shrestha, L.; Patel, H.J.; Chiosis, G. Chemical tools to investigate mechanisms associated with HSP90 and HSP70 in disease. Cell Chem. Biol. 2016, 23, 158–172. [Google Scholar] [CrossRef]
- Tian, Y.; Xu, H.; Farooq, A.A.; Nie, B.; Chen, X.; Su, S.; Yuan, R.; Qiao, G.; Li, C.; Li, X. Maslinic acid induces autophagy by down-regulating HSPA8 in pancreatic cancer cells. Phytother. Res. 2018, 32, 1320–1331. [Google Scholar] [CrossRef]
- Wang, Y.-P.; Liu, F.; He, H.-W.; Han, Y.-X.; Peng, Z.-G.; Li, B.-W.; You, X.-F.; Song, D.-Q.; Li, Z.-R.; Yu, L.-Y. Heat stress cognate 70 host protein as a potential drug target against drug resistance in hepatitis B virus. Antimicrob. Agents Chemother. 2010, 54, 2070–2077. [Google Scholar] [CrossRef]
- Tang, S.; Peng, Z.-G.; Zhang, X.; Cheng, X.-Y.; Li, W.-J.; Jiang, J.-D.; Li, Y.-H.; Song, D.-Q. Synthesis and biological evaluation of 12-benzyl matrinic amide derivatives as a novel family of anti-HCV agents. Chin. Chem. Lett. 2016, 27, 1052–1057. [Google Scholar] [CrossRef]
- Li, Y.H.; Tang, S.; Li, Y.H.; Cheng, X.Y.; Zhang, X.; Wang, Y.X.; Su, F.; Song, D.Q. Novel 12N-substituted matrinanes as potential anti-coxsackievirus agents. Bioorg. Med. Chem. Lett. 2017, 27, 829–833. [Google Scholar] [CrossRef]
- Li, Y.-H.; Wu, Z.-Y.; Tang, S.; Zhang, X.; Wang, Y.-X.; Jiang, J.-D.; Peng, Z.-G.; Song, D.-Q. Evolution of matrinic ethanol derivatives as anti-HCV agents from matrine skeleton. Bioorg. Med. Chem. Lett. 2017, 27, 1962–1966. [Google Scholar] [CrossRef]
- Tang, S.; Peng, Z.-G.; Li, Y.-H.; Zhang, X.; Fan, T.-Y.; Jiang, J.-D.; Wang, Y.-X.; Song, D.-Q. Synthesis and biological evaluation of tricyclic matrinic derivatives as a class of novel anti-HCV agents. Chem. Cent. J. 2017, 11, 94. [Google Scholar] [CrossRef]
- Du, N.-N.; Peng, Z.-G.; Bi, C.-W.; Tang, S.; Li, Y.-H.; Li, J.-R.; Zhu, Y.-P.; Zhang, J.-P.; Wang, Y.-X.; Jiang, J.-D. N-substituted benzyl matrinic acid derivatives inhibit hepatitis C virus (HCV) replication through down-regulating host heat-stress cognate 70 (Hsc70) expression. PLoS ONE 2013, 8, e58675. [Google Scholar] [CrossRef]
- Stigliano, A.; Cerquetti, L.; Borro, M.; Gentile, G.; Bucci, B.; Misiti, S.; Piergrossi, P.; Brunetti, E.; Simmaco, M.; Toscano, V. Modulation of proteomic profile in H295R adrenocortical cell line induced by mitotane. Endocr. Relat. Cancer 2008, 15, 1–10. [Google Scholar] [CrossRef]
- Rousaki, A.; Miyata, Y.; Jinwal, U.K.; Dickey, C.A.; Gestwicki, J.E.; Zuiderweg, E.R. Allosteric drugs: The interaction of antitumor compound MKT-077 with human Hsp70 chaperones. J. Mol. Biol. 2011, 411, 614–632. [Google Scholar] [CrossRef]
- Fewell, S.W.; Day, B.W.; Brodsky, J.L. Identification of an inhibitor of hsc70-mediated protein translocation and ATP hydrolysis. J. Biol. Chem. 2001, 276, 910–914. [Google Scholar] [CrossRef]
- Kreiling, J.A.; Creton, R.; Reinisch, C. Early embryonic exposure to polychlorinated biphenyls disrupts heat-shock protein 70 cognate expression in zebrafish. J. Toxicol. Environ. Health Part A 2007, 70, 1005–1013. [Google Scholar] [CrossRef]
- Na, Y.-R.; Seok, S.-H.; Baek, M.-W.; Lee, H.-Y.; Kim, D.-J.; Park, S.-H.; Lee, H.-K.; Park, J.-H. Protective effects of vitamin E against 3, 3′, 4, 4′, 5-pentachlorobiphenyl (PCB126) induced toxicity in zebrafish embryos. Ecotoxicol. Environ. Saf. 2009, 72, 714–719. [Google Scholar] [CrossRef]
- Leu, J.-J.; Pimkina, J.; Frank, A.; Murphy, M.E.; George, D.L. A small molecule inhibitor of inducible heat shock protein 70. Mol. Cell 2009, 36, 15–27. [Google Scholar] [CrossRef]
- Leu, J.-J.; Pimkina, J.; Pandey, P.; Murphy, M.E.; George, D.L. HSP70 inhibition by the small-molecule 2-phenylethynesulfonamide impairs protein clearance pathways in tumor cells. Mol. Cancer Res. 2011, 9, 936–947. [Google Scholar] [CrossRef]
- Schlecht, R.; Scholz, S.R.; Dahmen, H.; Wegener, A.; Sirrenberg, C.; Musil, D.; Bomke, J.; Eggenweiler, H.-M.; Mayer, M.P.; Bukau, B. Functional analysis of Hsp70 inhibitors. PLoS ONE 2013, 8, e78443. [Google Scholar] [CrossRef]
- Zeng, F.; Tee, C.; Liu, M.; Sherry, J.P.; Dixon, B.; Duncker, B.P.; Bols, N.C. The p53/HSP70 inhibitor, 2-phenylethynesulfonamide, causes oxidative stress, unfolded protein response and apoptosis in rainbow trout cells. Aquat. Toxicol. 2014, 146, 45–51. [Google Scholar] [CrossRef]
- Ishaq, M.; Ojha, R.; Sharma, K.; Sharma, G.; Singh, S.K.; Majumdar, S. Functional inhibition of Hsp70 by Pifithrin-μ switches gambogic acid induced caspase dependent cell death to caspase independent cell death in human bladder cancer cells. Biochim. Biophys. Acta Mol. Cell Res. 2016, 1863, 2560–2573. [Google Scholar] [CrossRef]
- Aquino, D.A.; Peng, D.; Lopez, C.; Farooq, M. The constitutive heat shock protein-70 is required for optimal expression of myelin basic protein during differentiation of oligodendrocytes. Neurochem. Res. 1998, 23, 413–420. [Google Scholar] [CrossRef]
- Tatsuta, T.; Hosono, M.; Ogawa, Y.; Inage, K.; Sugawara, S.; Nitta, K. Downregulation of Hsp70 inhibits apoptosis induced by sialic acid-binding lectin (leczyme). Oncol. Rep. 2014, 31, 13–18. [Google Scholar] [CrossRef]
- Sala, G.; Marinig, D.; Riva, C.; Arosio, A.; Stefanoni, G.; Brighina, L.; Formenti, M.; Alberghina, L.; Colangelo, A.M.; Ferrarese, C. Rotenone down-regulates HSPA8/hsc70 chaperone protein in vitro: A new possible toxic mechanism contributing to Parkinson’s disease. Neurotoxicology 2016, 54, 161–169. [Google Scholar] [CrossRef]
- Rubenstein, R.C.; Zeitlin, P.L. Sodium 4-phenylbutyrate downregulates Hsc70: Implications for intracellular trafficking of ΔF508-CFTR. Am. J. Physiol. Cell Physiol. 2000, 278, C259–C267. [Google Scholar] [CrossRef]
- Powers, M.V.; Clarke, P.A.; Workman, P. Dual targeting of HSC70 and HSP72 inhibits HSP90 function and induces tumor-specific apoptosis. Cancer Cell 2008, 14, 250–262. [Google Scholar] [CrossRef]
- Yang, X.; Tohda, C. Heat shock cognate 70 inhibitor, VER-155008, reduces memory deficits and axonal degeneration in a mouse model of Alzheimer’s disease. Front. Pharmacol. 2018, 9, 48. [Google Scholar] [CrossRef]
- Li, X.; Srinivasan, S.R.; Connarn, J.; Ahmad, A.; Young, Z.T.; Kabza, A.M.; Zuiderweg, E.R.; Sun, D.; Gestwicki, J.E. Analogues of the allosteric heat shock protein 70 (Hsp70) inhibitor, MKT-077, as anti-cancer agents. ACS Med. Chem. Lett. 2013, 4, 1042–1047. [Google Scholar] [CrossRef]
- Maeda, K.; Umeda, Y.; Saino, T. Synthesis and background chemistry of 15-deoxyspergualin. Ann. N. Y. Acad. Sci. 1993, 685, 123–135. [Google Scholar] [CrossRef]
- Perenyei, M.; Jayne, D.R.; Floßmann, O. Gusperimus: Immunological mechanism and clinical applications. Rheumatology 2014, 53, 1732–1741. [Google Scholar] [CrossRef]
- Kawada, M.; Masuda, T.; Ishizuka, M.; Takeuchi, T. 15-Deoxyspergualin inhibits Akt kinase activation and phosphatidylcholine synthesis. J. Biol. Chem. 2002, 277, 27765–27771. [Google Scholar] [CrossRef]
- Lorenz, H.; Grunke, M.; Wendler, J.; Heinzel, P.; Kalden, J. Safety of 15-deoxyspergualin in the treatment of glomerulonephritis associated with active systemic lupus erythematosus. Ann. Rheum. Dis. 2005, 64, 1517–1519. [Google Scholar] [CrossRef]
- Lorenz, H.-M.; Schmitt, W.H.; Tesar, V.; Müller-Ladner, U.; Tarner, I.; Hauser, I.A.; Hiepe, F.; Alexander, T.; Woehling, H.; Nemoto, K. Treatment of active lupus nephritis with the novel immunosuppressant 15-deoxyspergualin: An open-label dose escalation study. Arthritis Res. Ther. 2011, 13, R36. [Google Scholar] [CrossRef]
- Imai, H.; Hotta, O.; Yoshimura, M.; Konta, T.; Tsubakihara, Y.; Miyazaki, M.; Tomida, C.; Kobayashi, M.; Suzuki, S.; Shiiki, H. Deoxyspergualin, an immunosuppressant, in patients suffering from nephropathies with crescent formation: An open-label trial to evaluate safety and efficacy. Clin. Exp. Nephrol. 2006, 10, 40–54. [Google Scholar] [CrossRef]
- Ohlman, S.; Zilg, H.; Schindel, F.; Lindholm, A. Pharmacokinetics of 15-deoxyspergualin studied in renal transplant patients receiving the drug during graft rejection. Transpl. Int. 1994, 7, 5–10. [Google Scholar] [CrossRef]
- Floßmann, O.; Jayne, D.R. Long-term treatment of relapsing Wegener’s granulomatosis with 15-deoxyspergualin. Rheumatology 2009, 49, 556–562. [Google Scholar] [CrossRef]
- Monneaux, F.; Lozano, J.M.; Patarroyo, M.E.; Briand, J.P.; Muller, S. T cell recognition and therapeutic effect of a phosphorylated synthetic peptide of the 70K snRNP protein administered in MRL/lpr mice. Eur. J. Immunol. 2003, 33, 287–296. [Google Scholar] [CrossRef]
- Schall, N.; Muller, S. Resetting the autoreactive immune system with a therapeutic peptide in lupus. Lupus 2015, 24, 412–418. [Google Scholar] [CrossRef]
- Monneaux, F.; Parietti, V.; Briand, J.-P.; Muller, S. Importance of spliceosomal RNP1 motif for intermolecular TB cell spreading and tolerance restoration in lupus. Arthritis Res. Ther. 2007, 9, R111. [Google Scholar] [CrossRef]
- Schall, N.; Page, N.; Macri, C.; Chaloin, O.; Briand, J.-P.; Muller, S. Peptide-based approaches to treat lupus and other autoimmune diseases. J. Autoimmun. 2012, 39, 143–153. [Google Scholar] [CrossRef]
- Gros, F.; Muller, S. Pharmacological regulators of autophagy and their link with modulators of lupus disease. Br. J. Pharmacol. 2014, 171, 4337–4359. [Google Scholar] [CrossRef]
- Bonam, S.R.; Wang, F.; Muller, S. Autophagy: A new concept in autoimmunity regulation and a novel therapeutic option. J. Autoimmun. 2018, 94, 16–32. [Google Scholar] [CrossRef]
- Retnakumar, S.V.; Muller, S. Pharmacological Autophagy Regulators as Therapeutic Agents for Inflammatory Bowel Diseases. Trends Mol. Med. 2019, 25, 516–537. [Google Scholar] [CrossRef]
- Bendorius, M.; Neeli, I.; Wang, F.; Bonam, S.R.; Dombi, E.; Buron, N.; Borgne-Sanchez, A.; Poulton, J.; Radic, M.; Muller, S. The mitochondrion-lysosome axis in adaptive and innate immunity: Effect of lupus regulator peptide P140 on mitochondria autophagy and NETosis. Front. Immunol. 2018, 9, 2158. [Google Scholar] [CrossRef]
- Li, B.; Wang, F.; Schall, N.; Muller, S. Rescue of autophagy and lysosome defects in salivary glands of MRL/lpr mice by a therapeutic phosphopeptide. J. Autoimmun. 2018, 90, 132–145. [Google Scholar] [CrossRef]
- Wilhelm, M.; Wang, F.; Schall, N.; Kleinmann, J.-F.; Faludi, M.; Nashi, E.P.; Sibilia, J.; Martin, T.; Schaeffer, E.; Muller, S. Lupus regulator peptide P140 represses B cell differentiation by reducing HLA class II molecule overexpression. Arthritis Rheumatol. 2018, 70, 1077–1088. [Google Scholar] [CrossRef]
- Zimmer, R.; Scherbarth, H.; Rillo, O.; Gomez-Reino, J.; Muller, S. Lupuzor/P140 peptide in patients with systemic lupus erythematosus: A randomised, double-blind, placebo-controlled phase IIb clinical trial. Ann. Rheum. Dis. 2013, 72, 1830–1835. [Google Scholar] [CrossRef]
- Muller, S.; Monneaux, F.; Schall, N.; Rashkov, R.K.; Oparanov, B.A.; Wiesel, P.; Geiger, J.M.; Zimmer, R. Spliceosomal peptide P140 for immunotherapy of systemic lupus erythematosus: Results of an early phase II clinical trial. Arthritis Rheum. 2008, 58, 3873–3883. [Google Scholar] [CrossRef]
- Muller, S.; Brun, S.; René, F.; De Sèze, J.; Loeffler, J.-P.; Jeltsch-David, H. Autophagy in neuroinflammatory diseases. Autoimmun. Rev. 2017, 16, 856–874. [Google Scholar] [CrossRef]
- Bendorius, M.; Po, C.; Muller, S.; Jeltsch-David, H. From systemic inflammation to neuroinflammation: The case of neurolupus. Int. J. Mol. Sci. 2018, 19, 3588. [Google Scholar] [CrossRef]
- Brun, S.; Schall, N.; Bonam, S.R.; Bigaut, K.; Mensah-Nyagan, A.G.; De Seze, J.; Muller, S. An autophagy-targeting peptide to treat chronic inflammatory demyelinating polyneuropathies. J. Autoimmun. 2018, 92, 114–125. [Google Scholar] [CrossRef]
- Zhao, Z.; Faden, A.I.; Loane, D.J.; Lipinski, M.M.; Sabirzhanov, B.; Stoica, B.A. Neuroprotective effects of geranylgeranylacetone in experimental traumatic brain injury. J. Cereb. Blood Flow Metab. 2013, 33, 1897–1908. [Google Scholar] [CrossRef]
- Kim, J.Y.; Han, Y.; Lee, J.E.; Yenari, M.A. The 70-kDa heat shock protein (Hsp70) as a therapeutic target for stroke. Expert Opin. Ther. Targets 2018, 22, 191–199. [Google Scholar] [CrossRef]
- Hageman, J.; Kampinga, H.H. Computational analysis of the human HSPH/HSPA/DNAJ family and cloning of a human HSPH/HSPA/DNAJ expression library. Cell Stress Chaperones 2009, 14, 1–21. [Google Scholar] [CrossRef]
- Wang, C.; Wang, H.; Zhang, D.; Luo, W.; Liu, R.; Xu, D.; Diao, L.; Liao, L.; Liu, Z. Phosphorylation of ULK1 affects autophagosome fusion and links chaperone-mediated autophagy to macroautophagy. Nat. Commun. 2018, 9, 3492. [Google Scholar] [CrossRef]
- Jung, C.H.; Jun, C.B.; Ro, S.-H.; Kim, Y.-M.; Otto, N.M.; Cao, J.; Kundu, M.; Kim, D.-H. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 2009, 20, 1992–2003. [Google Scholar] [CrossRef]
- Russell, R.C.; Tian, Y.; Yuan, H.; Park, H.W.; Chang, Y.-Y.; Kim, J.; Kim, H.; Neufeld, T.P.; Dillin, A.; Guan, K.-L. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 2013, 15, 741. [Google Scholar] [CrossRef]
- De Maio, A. Extracellular Hsp70: Export and function. Curr. Protein Pept. Sci. 2014, 15, 225–231. [Google Scholar] [CrossRef]
- Gehrmann, M.; Cervello, M.; Montalto, G.; Cappello, F.; Gulino, A.; Knape, C.; Specht, H.M.; Multhoff, G. Heat shock protein 70 serum levels differ significantly in patients with chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. Front. Immunol. 2014, 5, 307. [Google Scholar] [CrossRef]
- Ao, L.; Zou, N.; Cleveland, J.C., Jr.; Fullerton, D.A.; Meng, X. Myocardial TLR4 is a determinant of neutrophil infiltration after global myocardial ischemia: Mediating KC and MCP-1 expression induced by extracellular HSC70. Am. J. Physiol. Heart Circ. Physiol. 2009, 297, H21–H28. [Google Scholar] [CrossRef]
- Rubinsztein, D.C.; Codogno, P.; Levine, B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discov. 2012, 11, 709–730. [Google Scholar] [CrossRef]
- Brun, S.; Schall, N.; Jeltsch-David, H.; Seze, J.D.; Muller, S. Assessing autophagy in sciatic nerves of a rat model that develops inflammatory autoimmune peripheral neuropathies. Cells 2017, 6, 30. [Google Scholar] [CrossRef]
- Byun, S.; Lee, E.; Lee, K.W. Therapeutic implications of autophagy inducers in immunological disorders, infection, and cancer. Int. J. Mol. Sci. 2017, 18, 1959. [Google Scholar] [CrossRef]
- Jang, Y.J.; Kim, J.H.; Byun, S. Modulation of Autophagy for Controlling Immunity. Cells 2019, 8, 138. [Google Scholar] [CrossRef]
- Panda, P.K.; Fahrner, A.; Vats, S.; Seranova, E.; Sharma, V.; Chipara, M.; Desai, P.; Torresi, J.; Rosenstock, T.; Kumar, D.; et al. Chemical Screening Approaches Enabling Drug Discovery of Autophagy Modulators for Biomedical Applications in Human Diseases. Front. Cell Dev. Biol. 2019, 7, 38. [Google Scholar] [CrossRef]
- Bonam, S.R.; Wang, F.; Muller, S. Lysosomes as a therapeutic target. Nat. Rev. Drug Discov. 2019, 18, in press. [Google Scholar]

| Characteristics | HSPA1A | HSPA8 |
|---|---|---|
| Molar mass | 70 kDa | 72 kDa |
| Length (amino acid residues) | 641 | 646 |
| Variable region | C-terminal domain | C-terminal domain |
| C-terminal EEVD motif | Yes | Yes |
| Binding of co-chaperones and other HSPs | Yes | Yes |
| Hydrophobic linker DL/VLLLD connecting the nucleotide and substrate binding domains | Yes | Yes |
| Expression | Abundant in stress conditions | Constitutively expressed and relatively less strongly expressed during stress |
| Association with immunogenic peptides | Yes | Moderate |
| Trafficking of ion channels (sodium channels) | Increased | Decreased |
| Interactions with lipid bilayers | Yes | Yes |
| Liposomal aggregation | Moderate | Yes |
| Involvement in drug resistance of cancer cells (e.g., leukemia cells) | Yes | No |
| Involvement in tumor growth | Yes | Not yet explored in depth |
| ATP/ADP-dependent | Yes | Yes |
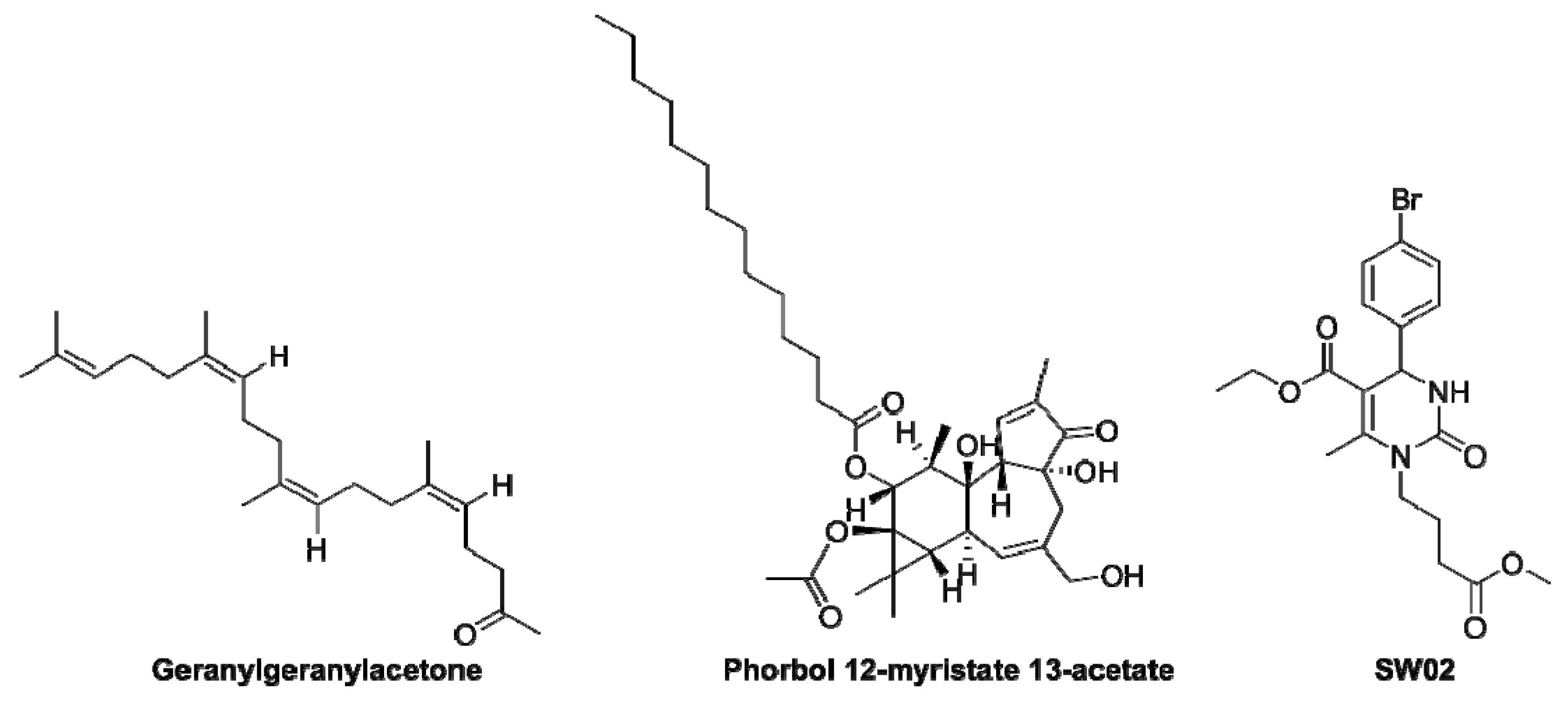
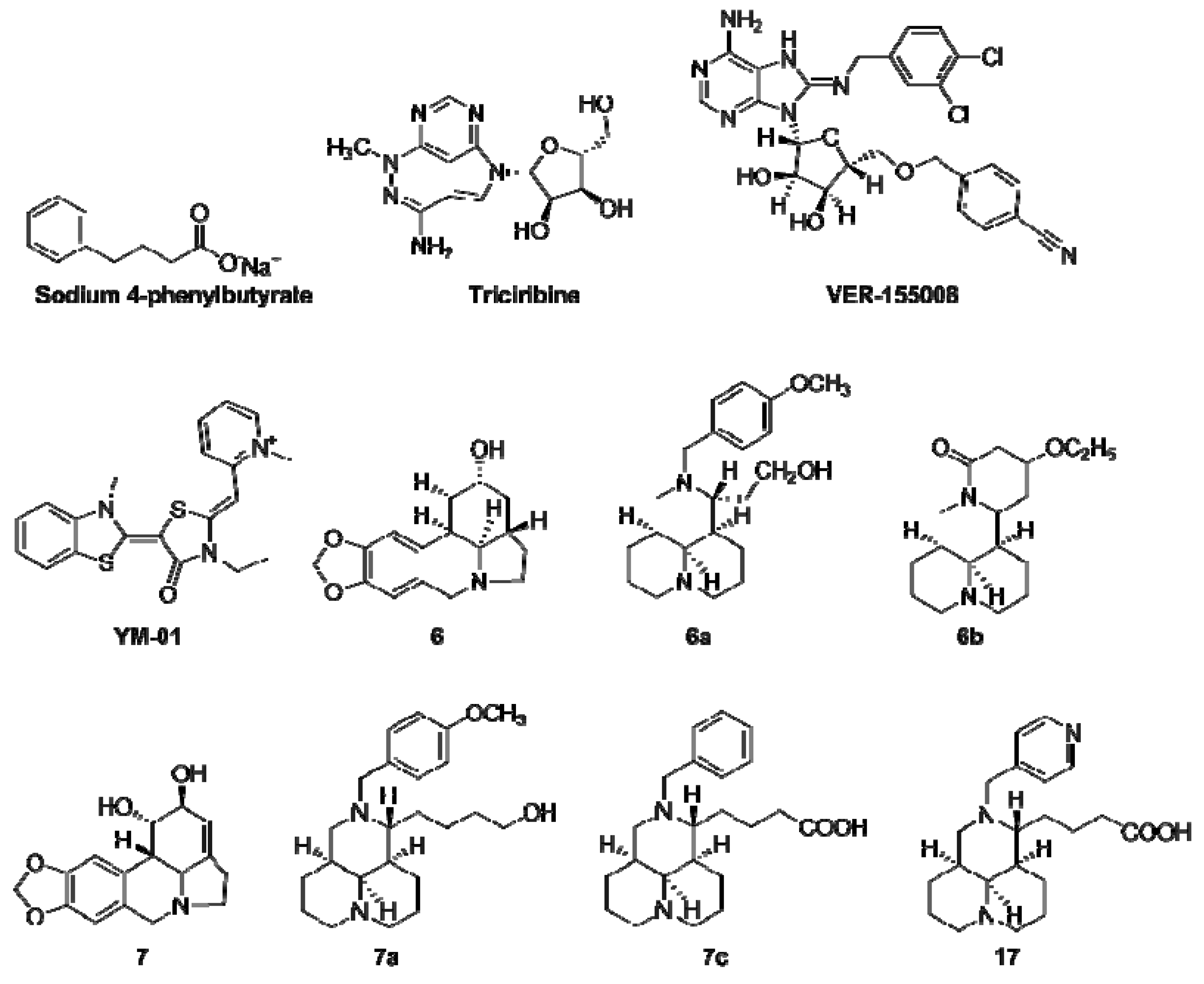
| HSPA8 Activator | Type of Molecule and Effects | Ref. |
|---|---|---|
| Geranylgeranylacetone |
| [62,63,64,65] |
| Phorbol 12-myristate 13-acetate |
| [66] |
| SW02 |
| [67,68] |
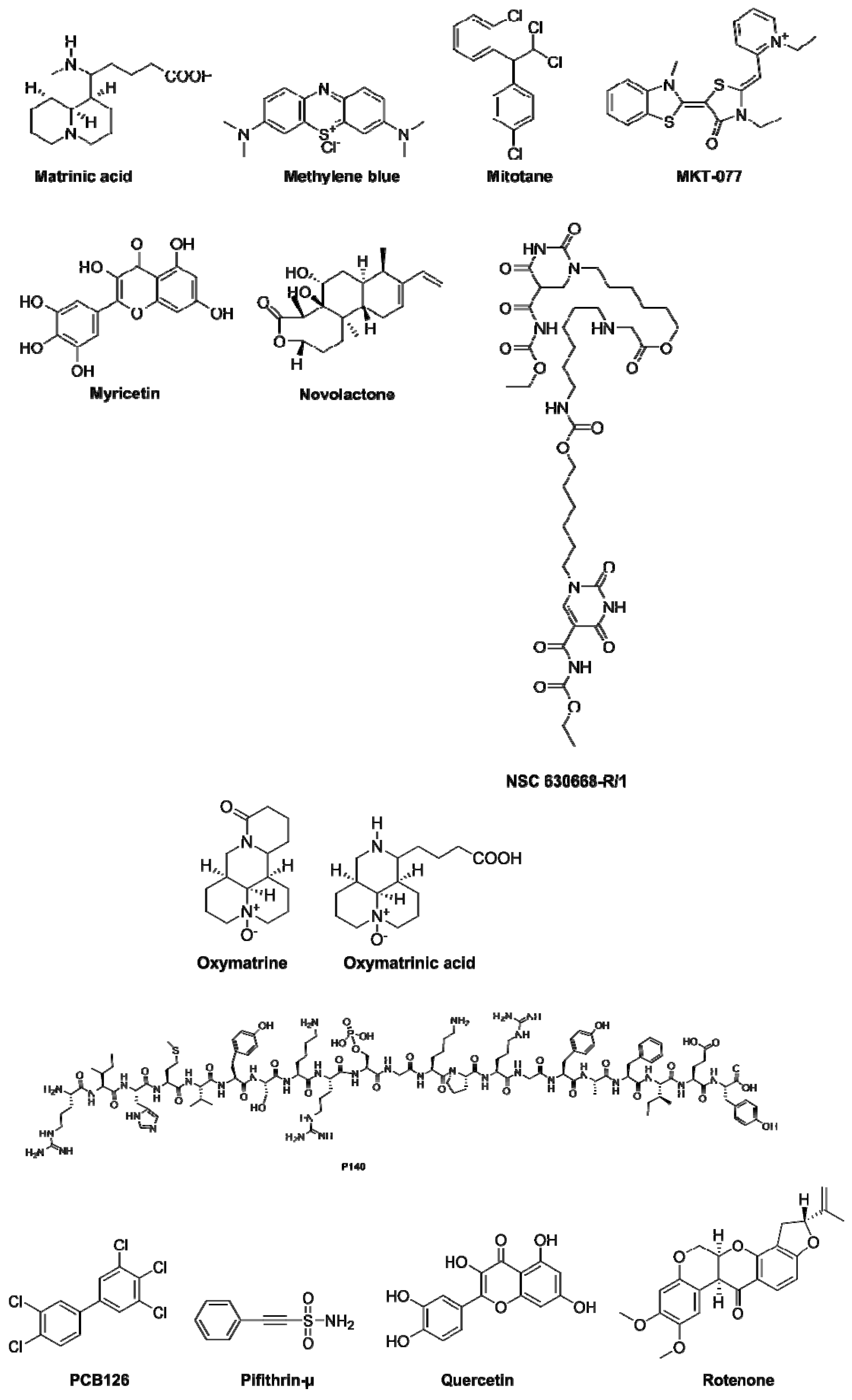
| Compounds | Therapeutic Field * | Observation # | References |
|---|---|---|---|
| Apoptozole | • Vaccine efficacy and cancer |
| [72,73,74] |
| 15-Deoxyspergualin (15-DSG) |
|
| [2,75,76,77,78,79] |
| D-Galactosamine | • Septic shock |
| [80] |
| Diosgenin | • Neurological diseases |
| [71] |
| 13-Ethoxymatrine (6b) | • Infectious diseases |
| [81] |
| Glycerol | • In silico studies |
| [41] |
| IMB-DM122 | • Infectious diseases |
| [82,83] |
| JG-48 | • Stability on model proteins |
| [84] |
| (+)-Lycoricidine | • Infectious diseases |
| [85,86] |
| MAL3-101 | • Cancer |
| [87,88] |
| Maslinic acid | • Cancer |
| [89] |
| Matrine |
|
| [82,90,91,92,93,94] |
| 12-N-4-methylbenzyl matrinic acid | • Infectious diseases |
| [95] |
| Mitotane (Lysodren) | • Cancer |
| [96] |
| MKT-077 | • Cancer |
| [97] |
| NSC 630668-R/1 | • Cancer |
| [98] |
| Oxymatrine | • Infectious diseases |
| [83] |
| P140 (Lupuzor) | • Autoimmunity |
| [38,47] |
| 3,3′,4,4′,5-Pentachlorinated biphenyl 126 (PCB126) | • Zebrafish model |
| [99,100] |
| 2-phenylethynesulfonamide or PES (pifithrin-μ) | • Cancer |
| [101,102,103,104,105] |
| Quercetin | • Neurological diseases |
| [106,107] |
| Rotenone | • Neurodegenerative diseases |
| [108] |
| Sodium 4-phenylbutyrate | • Cystic fibrosis model |
| [109] |
| VER155008 | • Cancer and neurodegenerative diseases |
| [43,103,110,111] |
| YM-01 | • Cancer |
| [112] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bonam, S.R.; Ruff, M.; Muller, S. HSPA8/HSC70 in Immune Disorders: A Molecular Rheostat that Adjusts Chaperone-Mediated Autophagy Substrates. Cells 2019, 8, 849. https://doi.org/10.3390/cells8080849
Bonam SR, Ruff M, Muller S. HSPA8/HSC70 in Immune Disorders: A Molecular Rheostat that Adjusts Chaperone-Mediated Autophagy Substrates. Cells. 2019; 8(8):849. https://doi.org/10.3390/cells8080849
Chicago/Turabian StyleBonam, Srinivasa Reddy, Marc Ruff, and Sylviane Muller. 2019. "HSPA8/HSC70 in Immune Disorders: A Molecular Rheostat that Adjusts Chaperone-Mediated Autophagy Substrates" Cells 8, no. 8: 849. https://doi.org/10.3390/cells8080849
APA StyleBonam, S. R., Ruff, M., & Muller, S. (2019). HSPA8/HSC70 in Immune Disorders: A Molecular Rheostat that Adjusts Chaperone-Mediated Autophagy Substrates. Cells, 8(8), 849. https://doi.org/10.3390/cells8080849






