Berberine Promotes Beige Adipogenic Signatures of 3T3-L1 Cells by Regulating Post-transcriptional Events
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture, In Vitro Differentiation, and Chemicals
2.2. miRNA-seq Analyses
2.3. Plasmid Construction
2.4. Poly(A) Tailing of Small RNA
2.5. Transient Transfection, Reverse-Transcription (RT)-PCR and Quantitative (q)PCR Analyses
2.6. Immunoblotting Assay
2.7. Oil-Red-O (ORO) Staining
2.8. Mitochondrial Analysis
2.9. Mitochondrial Respiration Assay
2.10. Bioinformatic Prediction of miRNA-RBM4a Heteroduplex
2.11. In Vitro Luciferase Reporter Assay
2.12. Statistical Analyses
3. Results
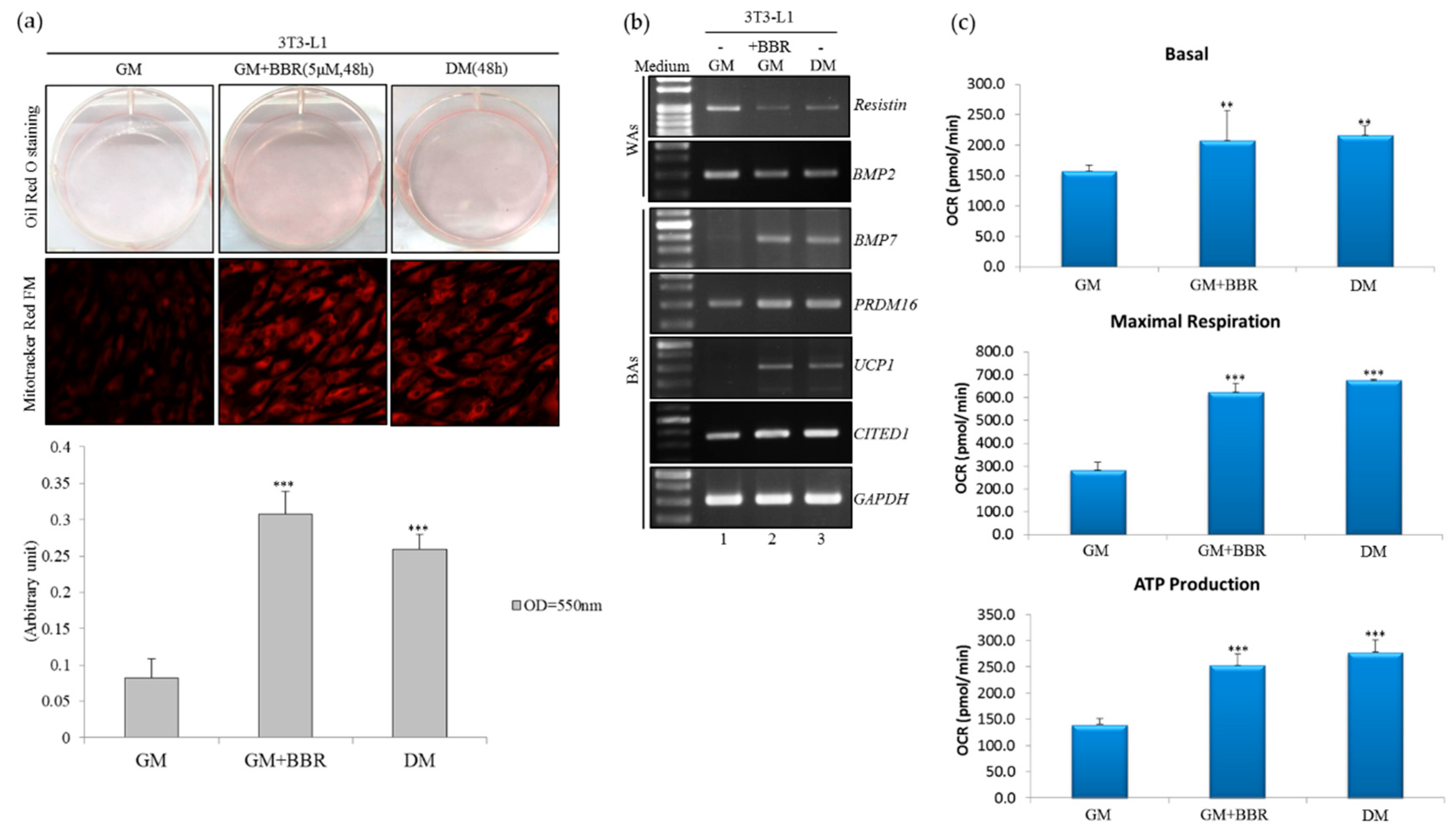
3.1. BBR Induces Brown-Adipogenic Signatures of 3T3-L1 Cells
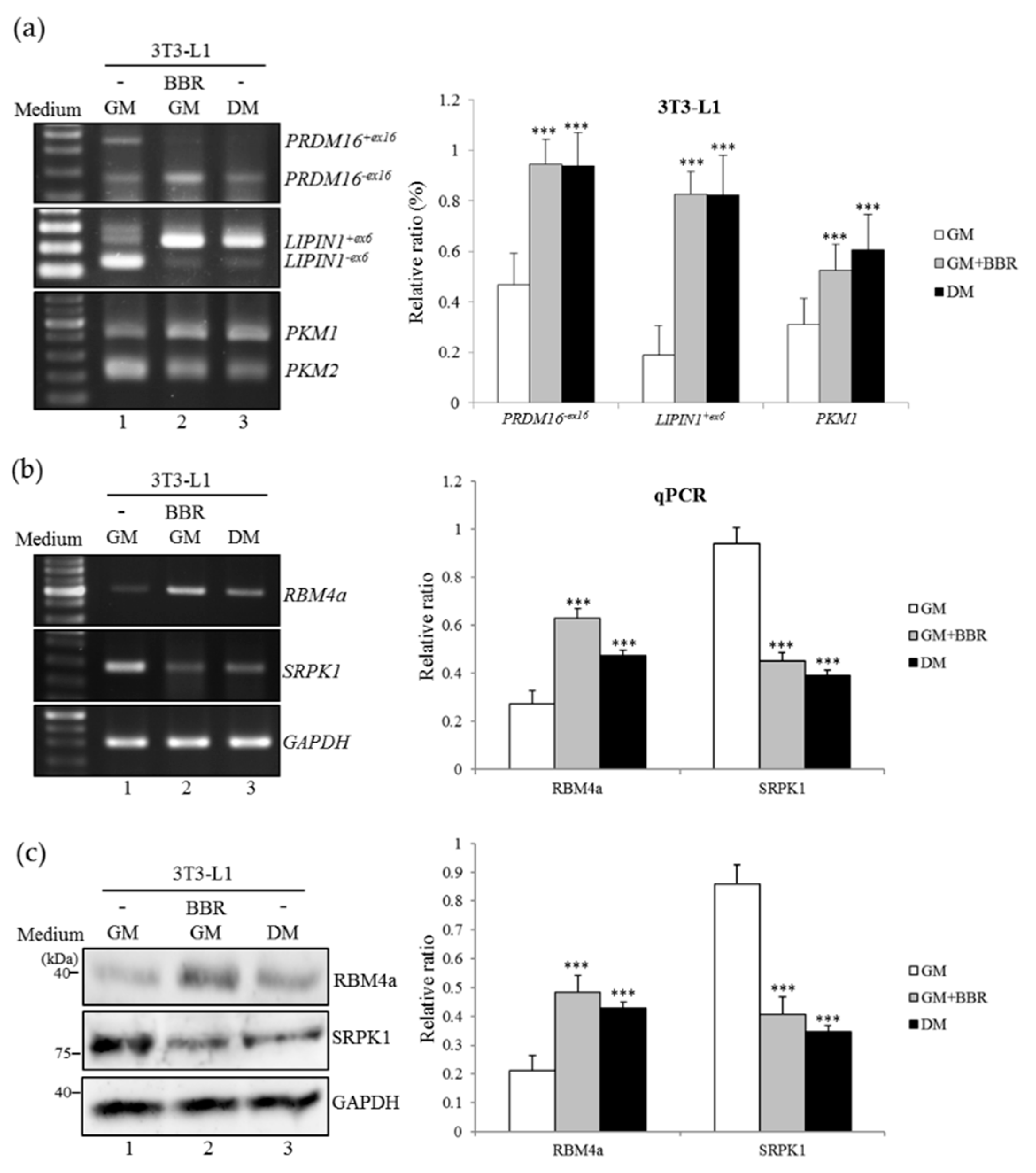
3.2. BBR-Enhanced BA-Related Splicing Events in 3T3-L1 Cells
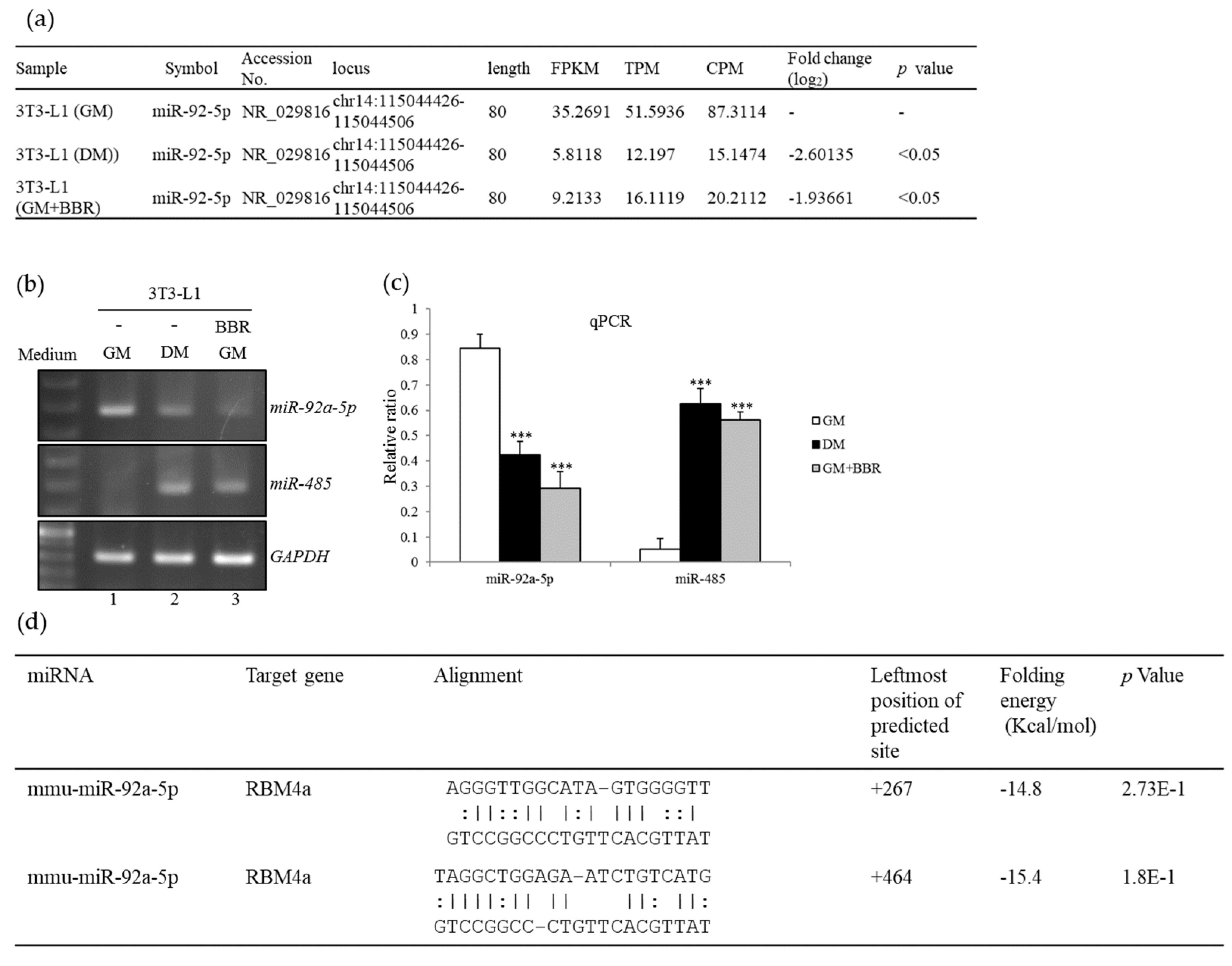
3.3. BBR Treatment Modulates the Expression Profiles of miR-92a and miR-485
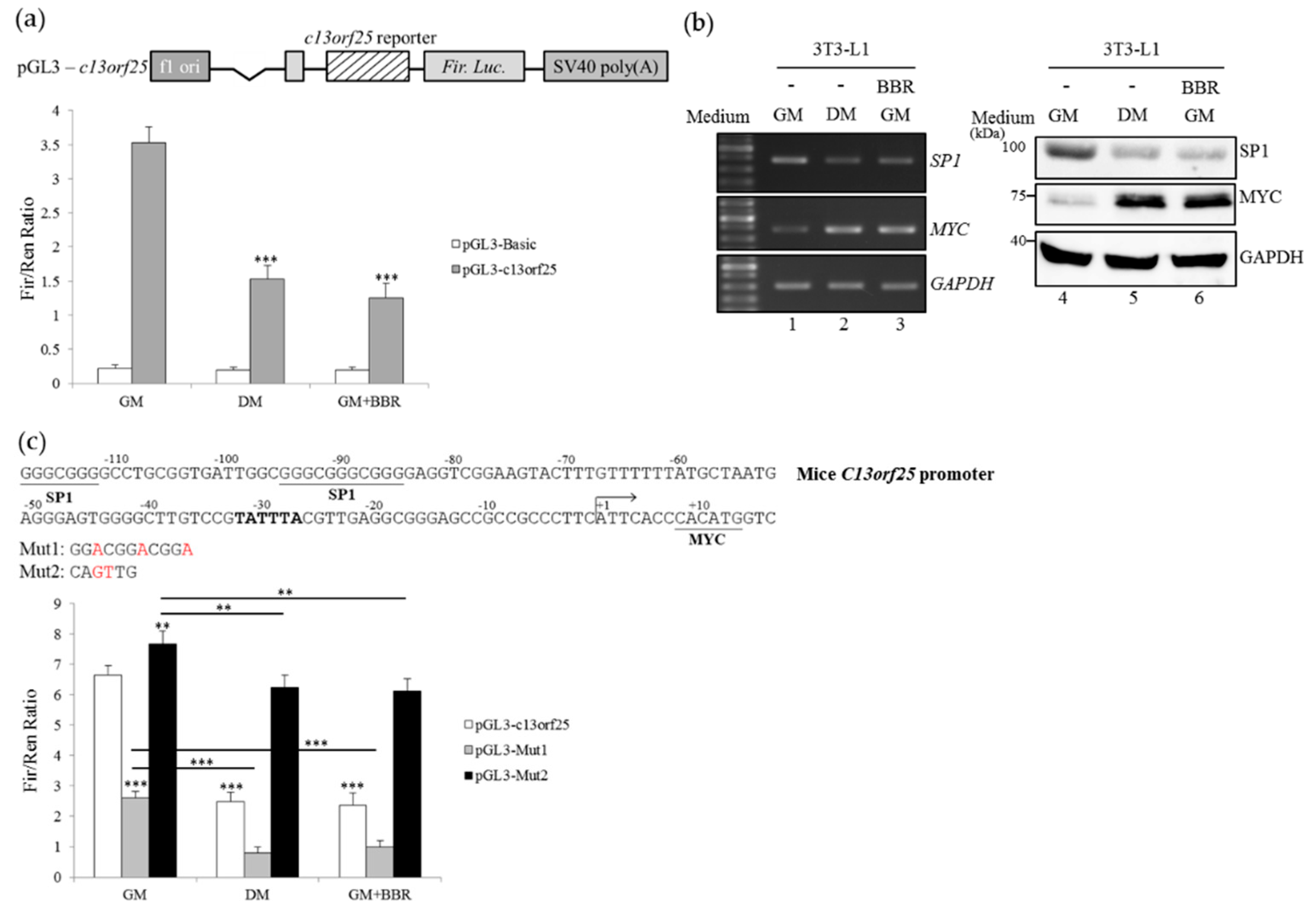
3.4. BBR-Modulated Transcriptional Regulation Reduces miR-92a Expression
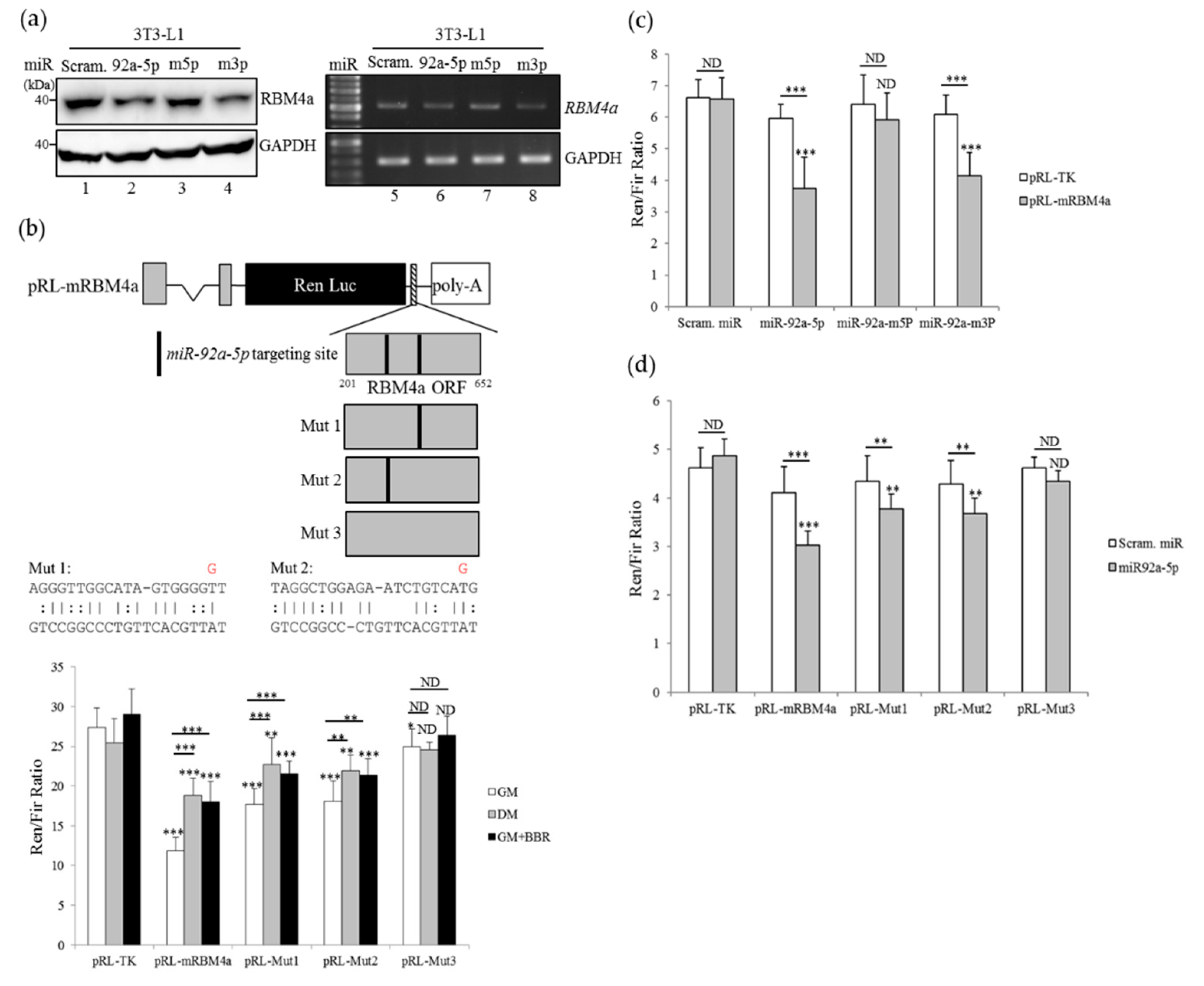
3.5. Overexpression of miR-92a-5p Leads to a Decrease in RBM4a Expression
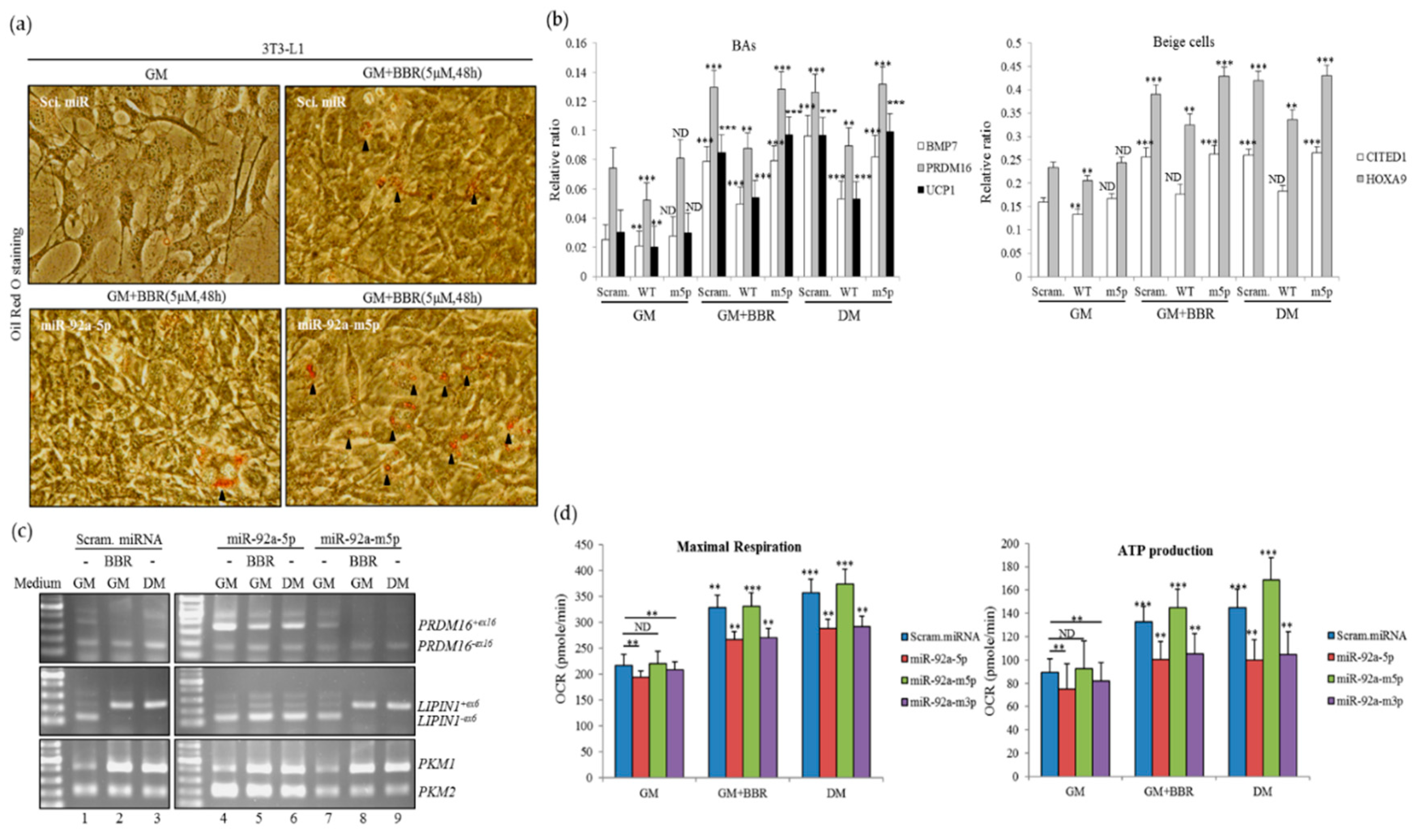
3.6. Overexpression of miR-92a-5p Interferes with the Influence of BBR on Beige Adipogenesis
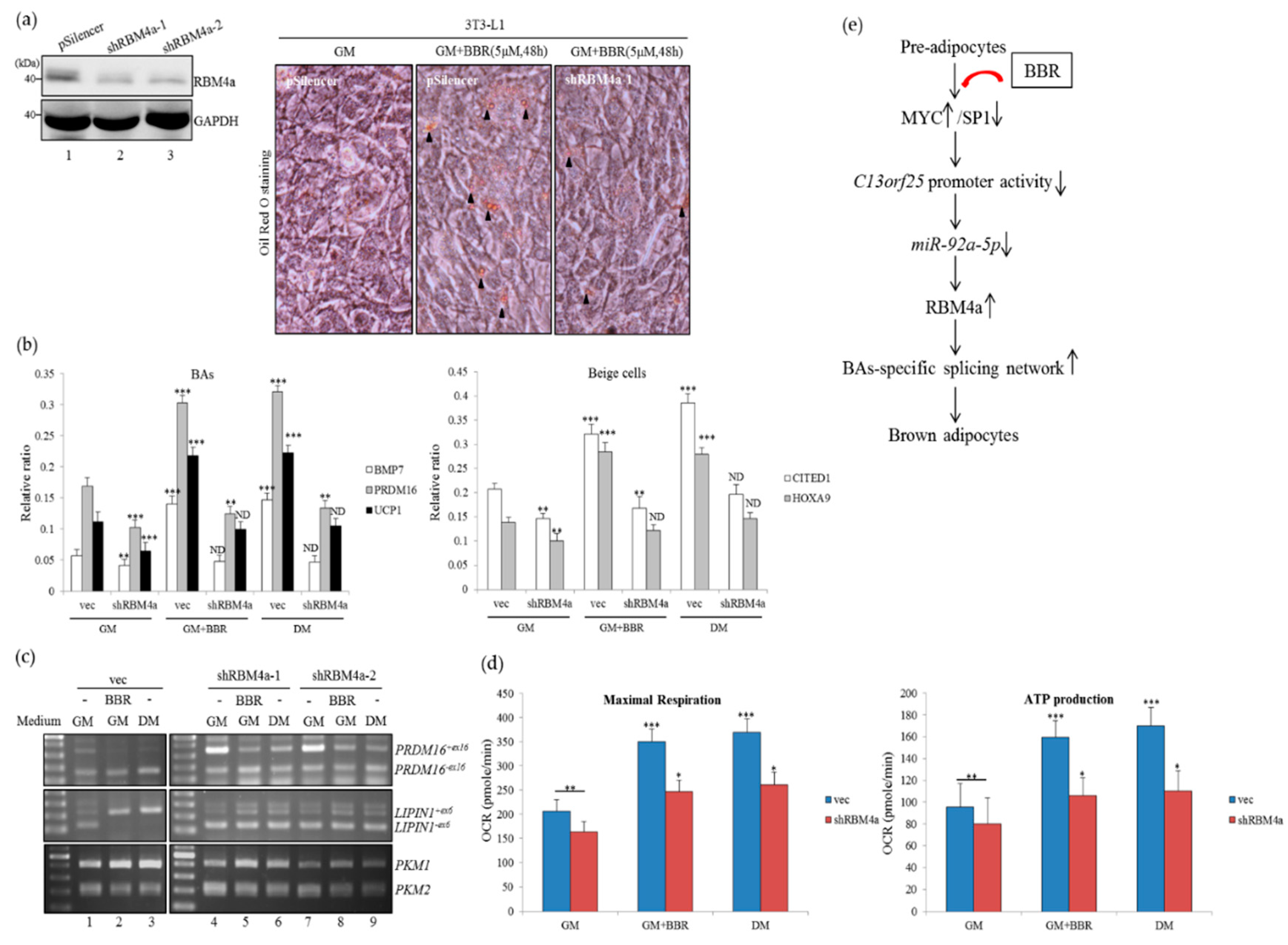
3.7. Depletion of RBM4a Diminishes the Effect of BBR on Brown Adipogenesis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kittiskulnam, P.; Thokanit, N.S.; Katavetin, P.; Susanthitaphong, P.; Srisawat, N.; Praditpornsilpa, K.; Tungsanga, K.; Eiam-Ong, S. The magnitude of obesity and metabolic syndrome among diabetic chronic kidney disease population: A nationwide study. PLoS ONE 2018, 13, e0196332. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.; Bronec, C.; Napoli, E.; Graham, J.; Stanhope, K.L.; Marsilio, I.; Giron, M.C.; Havel, P.J.; Giulivi, C. Adipose depot-specific effects of ileal interposition surgery in UCD-T2D rats: Unexpected implications for obesity and diabetes. Biochem. J. 2018, 475, 649–662. [Google Scholar] [CrossRef] [PubMed]
- Banfi, S.; Gusarova, V.; Gromada, J.; Cohen, J.C.; Hobbs, H.H. Increased thermogenesis by a noncanonical pathway in ANGPTL3/8-deficient mice. Proc. Natl. Acad. Sci. USA 2018, 115, E1249–E1258. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Huard, C.; Vernochet, C.; Ziemek, D.; Knowlton, K.M.; Tyminski, E.; Paradis, T.; Zhang, Y.; Jones, J.E.; von Schack, D.; et al. Brown fat determination and development from muscle precursor cells by novel action of bone morphogenetic protein 6. PLoS ONE 2014, 9, e92608. [Google Scholar] [CrossRef] [PubMed]
- An, Y.; Wang, G.; Diao, Y.; Long, Y.; Fu, X.; Weng, M.; Zhou, L.; Sun, K.; Cheung, T.H.; Ip, N.Y.; et al. A molecular switch regulating cell fate choice between muscle progenitor cells and brown adipocytes. Dev. Cell 2017, 41, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Berry, D.C.; Graff, J.M. Distinct cellular and molecular mechanisms for β3 adrenergic receptor-induced beige adipocyte formation. eLife 2017, 6, e30329. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Yun, J.W. Chrysin induces brown fat-like phenotype and enhances lipid metabolism in 3T3-L1 adipocytes. Nutrition 2016, 32, 1002–1010. [Google Scholar] [CrossRef]
- Velickovic, K.; Leija, H.A.L.; Bloor, I.; Law, J.; Sacks, H.; Symonds, M.; Sottile, V. Low temperature exposure induces browning of bone marrow stem cell derived adipocytes In Vitro. Sci. Rep. 2018, 8, 4974. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, H.; Li, B.; Meng, X.; Wang, J.; Zhang, Y.; Yao, S.; Ma, Q.; Jin, L.; Yang, J.; et al. Berberine activates thermogenesis in white and brown adipose tissue. Nat. Commun. 2014, 5, 5493. [Google Scholar] [CrossRef]
- Harms, M.; Seale, P. Brown and beige fat: Development, function and therapeutic potential. Nat. Med. 2013, 10, 1252–1263. [Google Scholar] [CrossRef]
- Wang, Y.; Tong, Q.; Shou, J.W.; Zhao, Z.X.; Li, X.Y.; Zhang, X.F.; Ma, S.R.; He, C.Y.; Lin, Y.; Wen, B.Y.; et al. Gut microbiota-mediated personalized treatment of hyperlipidemia using berberine. Theranostics 2017, 7, 2443–2451. [Google Scholar] [CrossRef] [PubMed]
- Nilsen, T.W.; Graveley, B.R. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010, 463, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Stelekati, E.; Kurachi, M.; Yu, S.; Cai, Z.; Manne, S.; Khan, O.; Yang, X.; Wherry, E.J. MiR-150 regulates memory CD8 T cell differentiation via c-Myb. Cell Rep. 2017, 20, 2584–2597. [Google Scholar] [CrossRef] [PubMed]
- Wittkopp, N.; Huntzinger, E.; Weiler, C.; Saulière, J.; Schmidt, S.; Sonawane, M.; Izaurralde, E. Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol. Cell. Biol. 2009, 29, 3517–3528. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Xu, S.; Kyaw, A.M.M.; Lim, Y.C.; Chia, S.Y.; Siang, D.T.C.; Alvarez-Dominguez, J.R.; Chen, P.; Leow, M.K.; Sun, L. RNA binding protein ybx2 regulates rna stability during cold-induced brown fat activation. Diabetes 2017, 66, 2987–3000. [Google Scholar] [CrossRef] [PubMed]
- Koh, E.H.; Chen, Y.; Bader, D.A.; Hamilton, M.P.; He, B.; York, B.; Kajimura, S.; McGuire, S.E.; Hartig, S.M. Mitochondrial activity in human white adipocytes is regulated by the ubiquitin carrier protein 9/microrna-30a axis. J. Biol. Chem. 2016, 291, 24747–24755. [Google Scholar] [CrossRef]
- Chang, J.S.; Ha, K. An unexpected role for the transcriptional coactivator isoform NT-PGC-1α in the regulation of mitochondrial respiration in brown adipocytes. J. Biol. Chem. 2017, 292, 9958–9966. [Google Scholar] [CrossRef]
- Lin, J.C. Multi-posttranscriptional regulations lessen the repressive effect of SRPK1 on brown adipogenesis. Biochim. Biophys. Acta 2018, 1863, 503–514. [Google Scholar] [CrossRef]
- Lin, J.C.; Lu, Y.H.; Liu, Y.R.; Lin, Y.J. RBM4a-regulated splicing cascade modulates the differentiation and metabolic activities of brown adipocytes. Sci. Rep. 2016, 6, 20665. [Google Scholar] [CrossRef]
- Wang, W.; Kissig, M.; Rajakumari, S.; Huang, L.; Lim, H.W.; Won, K.J.; Seale, P. Ebf2 is a selective marker of brown and beige adipogenic precursor cells. Proc. Natl. Acad. Sci. USA 2014, 111, 14466–14471. [Google Scholar] [CrossRef]
- Lone, J.; Choi, J.H.; Kim, S.W.; Yun, J.W. Curcumin induces brown fat-like phenotype in 3T3-L1 and primary white adipocytes. J. Nutr. Biochem. 2016, 27, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Lone, J.; Yun, J.W. Monoterpene limonene induces brown fat-like phenotype in 3T3-L1 white adipocytes. Life Sci. 2016, 153, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Asano, H.; Kanamori, Y.; Higurashi, S.; Nara, T.; Kato, K.; Matsui, T.; Funaba, M. Induction of beige-like adipocytes in 3T3-L1 cells. J. Vet. Med. Sci. 2014, 76, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Matsukawa, T.; Villareal, M.O.; Motojima, H.; Isoda, H. Increasing cAMP levels of preadipocytes by cyanidin-3-glucoside treatment induces the formation of beige phenotypes in 3T3-L1 adipocytes. J. Nutr. Biochem. 2017, 40, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Asai, Y.; Yamada, K.; Watanabe, T.; Keng, V.W.; Noguchi, T. Insulin stimulates expression of the pyruvate kinase M gene in 3T3-L1 adipocytes. Biosci. Biotechnol. Biochem. 2003, 67, 1272–1277. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Cheng, Y.; Wu, W.; Liu, Y.; Wei, N.; Feng, X.; Xie, Z.; Feng, Y. SRSF10 regulates alternative splicing and is required for adipocyte differentiation. Mol. Cell. Biol. 2014, 34, 2198–2207. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Buyel, J.J.; Hanssen, M.J.; Siegel, F.; Pan, R.; Naumann, J.; Schell, M.; van der Lans, A.; Schlein, C.; Froehlich, H.; et al. Exosomal microRNA miR-92a concentration in serum reflects human brown fat activity. Nat. Commun. 2016, 7, 11420. [Google Scholar] [CrossRef] [PubMed]
- Ji, M.; Rao, E.; Ramachandrareddy, H.; Shen, Y.; Jiang, C.; Chen, J.; Hu, Y.; Rizzino, A.; Chan, W.C.; Fu, K.; et al. The miR-17-92 microRNA cluster is regulated by multiple mechanisms in B-cell malignancies. Am. J. Pathol. 2011, 179, 1645–1656. [Google Scholar] [CrossRef]
- Hepler, C.; Shao, M.; Xia, J.Y.; Ghaben, A.L.; Pearson, M.J.; Vishvanath, L.; Sharma, A.X.; Morley, T.S.; Holland, W.L.; Gupta, R.K. Directing visceral white adipocyte precursors to a thermogenic adipocyte fate improves insulin sensitivity in obese mice. eLife 2017, 6, e27669. [Google Scholar] [CrossRef]
- Zhang, S.; Cao, H.; Li, Y.; Jing, Y.; Liu, S.; Ye, C.; Wang, H.; Yu, S.; Peng, C.; Hui, L.; et al. Metabolic benefits of inhibition of p38α in white adipose tissue in obesity. PLoS Biol. 2018, 16, e2004225. [Google Scholar] [CrossRef]
- Xiong, Y.; Yue, F.; Jia, Z.; Gao, Y.; Jin, W.; Hu, K.; Zhang, Y.; Zhu, D.; Yang, G.; Kuang, S. A novel brown adipocyte-enriched long non-coding RNA that is required for brown adipocyte differentiation and sufficient to drive thermogenic gene program in white adipocytes. Biochim. Biophys. Acta 2018, 1863, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Kiefer, F.W. The significance of beige and brown fat in humans. Endocr. Connect. 2017, 6, R70–R79. [Google Scholar] [CrossRef] [PubMed]
- Okla, M.; Kim, J.; Koehler, K.; Chung, S. Dietary factors promoting brown and beige fat development and thermogenesis. Adv. Nutr. 2017, 8, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Azhar, Y.; Parmar, A.; Miller, C.N.; Samuels, J.S.; Rayalam, S. Phytochemicals as novel agents for the induction of browning in white adipose tissue. Nutr. Metab. 2016, 13, 89. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, X.; Feng, X.; Liu, X.; Deng, C.; Hu, C.H. Berberine alleviates olanzapine-induced adipogenesis via the AMPKα-SREBP Pathway in 3T3-L1 cells. Int. J. Mol. Sci. 2016, 17, 1865. [Google Scholar] [CrossRef] [PubMed]
- Hirai, T.; Mitani, Y.; Kurumisawa, K.; Nomura, K.; Wang, W.; Nakashima, K.I.; Inoue, M. Berberine stimulates fibroblast growth factor 21 by modulating the molecular clock component brain and muscle Arnt-like 1 in brown adipose tissue. Biochem. Pharmacol. 2019, 164, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Cuevas-Ramos, D.; Mehta, R.; Aguilar-Salinas, C.A. Fibroblast Growth Factor 21 and Browning of White Adipose Tissue. Front. Physiol. 2019, 10, 37. [Google Scholar] [CrossRef]
- Sheng, Z.; Sun, Y.; Zhu, R.; Jiao, N.; Tang, K.; Cao, Z.; Ma, C. Functional Cross-Talking between Differentially Expressed and Alternatively Spliced Genes in Human Liver Cancer Cells Treated with Berberine. PLoS ONE 2015, 10, e0143742. [Google Scholar] [CrossRef]
- Li, J.; Cao, B.; Liu, X.; Fu, X.; Xiong, Z.; Chen, L.; Sartor, O.; Dong, Y.; Zhang, H. Berberine suppresses androgen receptor signaling in prostate cancer. Mol. Cancer Ther. 2011, 10, 1346–1356. [Google Scholar] [CrossRef]
- Karbiener, M.; Pisani, D.F.; Frontini, A.; Oberreiter, L.M.; Lang, E.; Vegiopoulos, A.; Mössenböck, K.; Bernhardt, G.A.; Mayr, T.; Hildner, F.; et al. MicroRNA-26 family is required for human adipogenesis and drives characteristics of brown adipocytes. Stem Cells 2014, 32, 1578–1590. [Google Scholar] [CrossRef]
- Kulyté, A.; Belarbi, Y.; Lorente-Cebrián, S.; Bambace, C.; Arner, E.; Daub, C.O.; Hedén, P.; Rydén, M.; Mejhert, N.; Arner, P. Additive effects of microRNAs and transcription factors on CCL2 production in human white adipose tissue. Diabetes 2014, 63, 1248–1258. [Google Scholar] [CrossRef] [PubMed]
- Roger, E.; Dumortier, O.; Pisani, D.F.; Gautier, N.; Obberghen, E.V.; Hinault, C. Argonaute-2 is associated to brown adipose tissue activation. Biochim. Biophys. Acta Mol. Basis Dis. 2019. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, Y.-C.; Lee, Y.-C.; Lin, Y.-J.; Lin, J.-C. Berberine Promotes Beige Adipogenic Signatures of 3T3-L1 Cells by Regulating Post-transcriptional Events. Cells 2019, 8, 632. https://doi.org/10.3390/cells8060632
Lin Y-C, Lee Y-C, Lin Y-J, Lin J-C. Berberine Promotes Beige Adipogenic Signatures of 3T3-L1 Cells by Regulating Post-transcriptional Events. Cells. 2019; 8(6):632. https://doi.org/10.3390/cells8060632
Chicago/Turabian StyleLin, Ying-Chin, Yuan-Chii Lee, Ying-Ju Lin, and Jung-Chun Lin. 2019. "Berberine Promotes Beige Adipogenic Signatures of 3T3-L1 Cells by Regulating Post-transcriptional Events" Cells 8, no. 6: 632. https://doi.org/10.3390/cells8060632
APA StyleLin, Y.-C., Lee, Y.-C., Lin, Y.-J., & Lin, J.-C. (2019). Berberine Promotes Beige Adipogenic Signatures of 3T3-L1 Cells by Regulating Post-transcriptional Events. Cells, 8(6), 632. https://doi.org/10.3390/cells8060632




