TTYH1 and TTYH2 Serve as LRRC8A-Independent Volume-Regulated Anion Channels in Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Microarray and Analysis
2.3. Immunocytochemistry
2.4. shRNA Construction and Validation
2.5. Western Blot
2.6. RT-PCR and Real-Time PCR
2.7. Electrophysiology
2.8. Statistical Analysis
3. Results
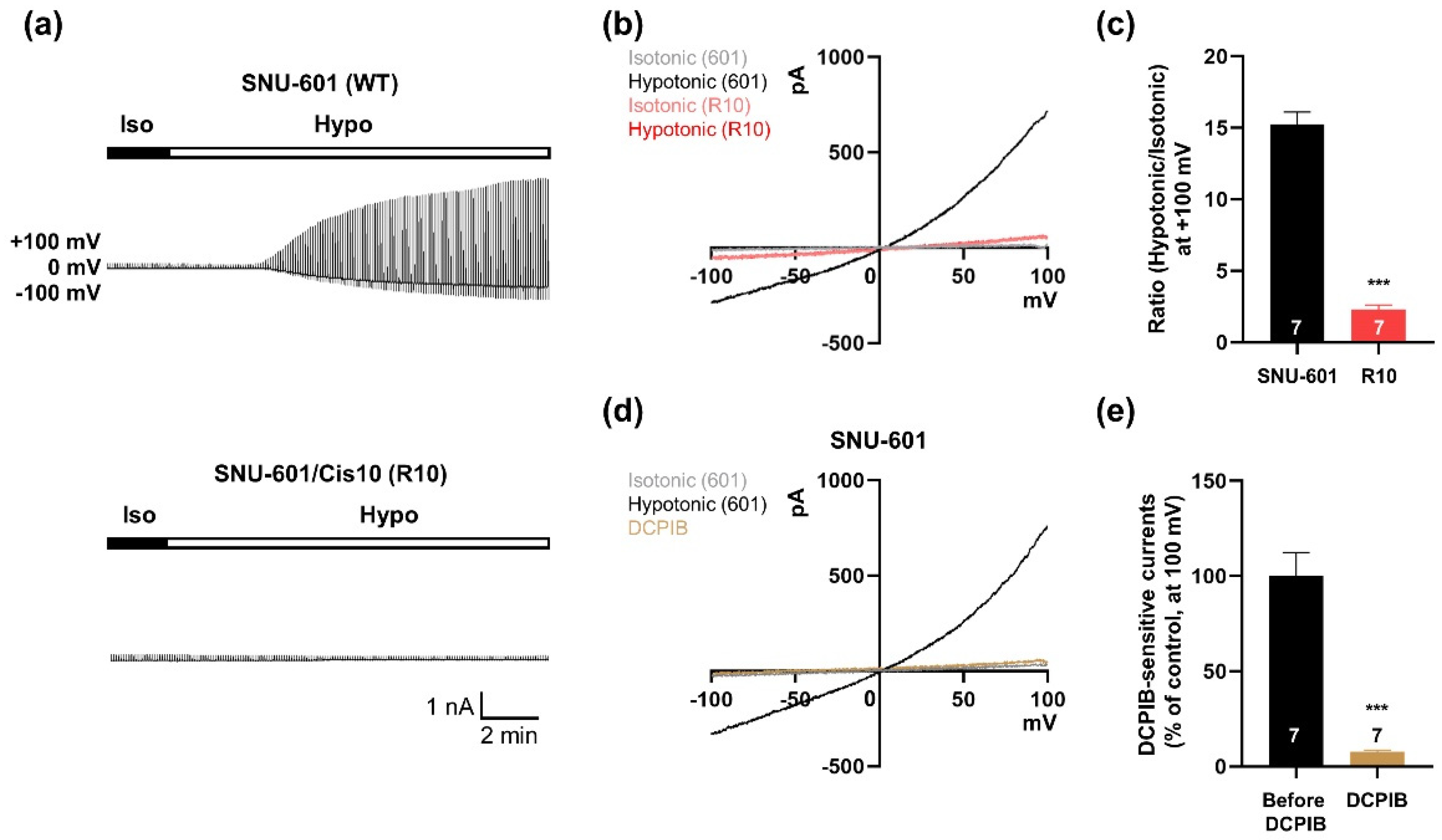
3.1. VRAC Currents are Shown in SNU-601 Cells but not in Cisplatin-Resistant R10 cells
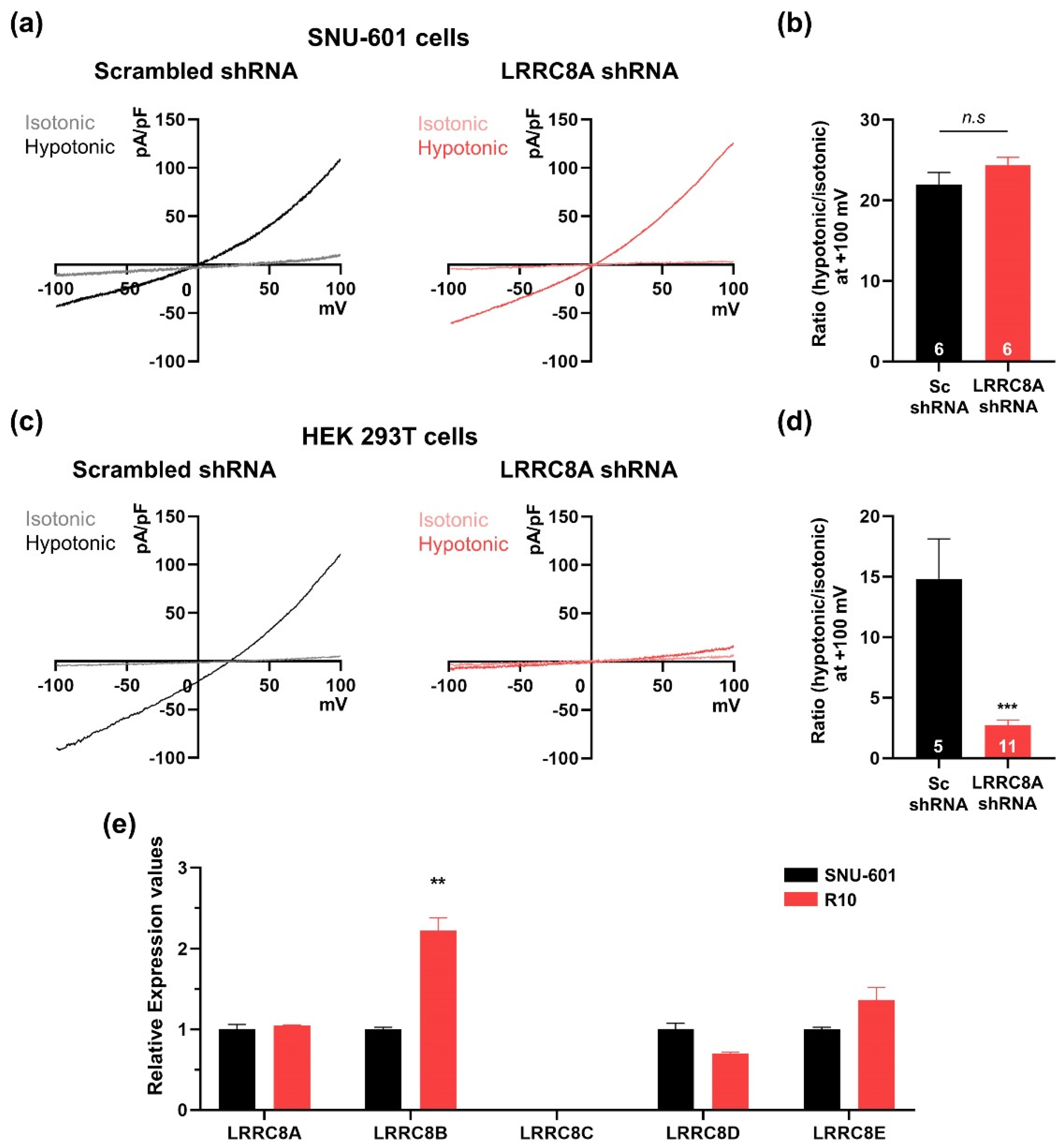
3.2. SNU-601 Cells Have LRRC8A-Independent VRAC Currents
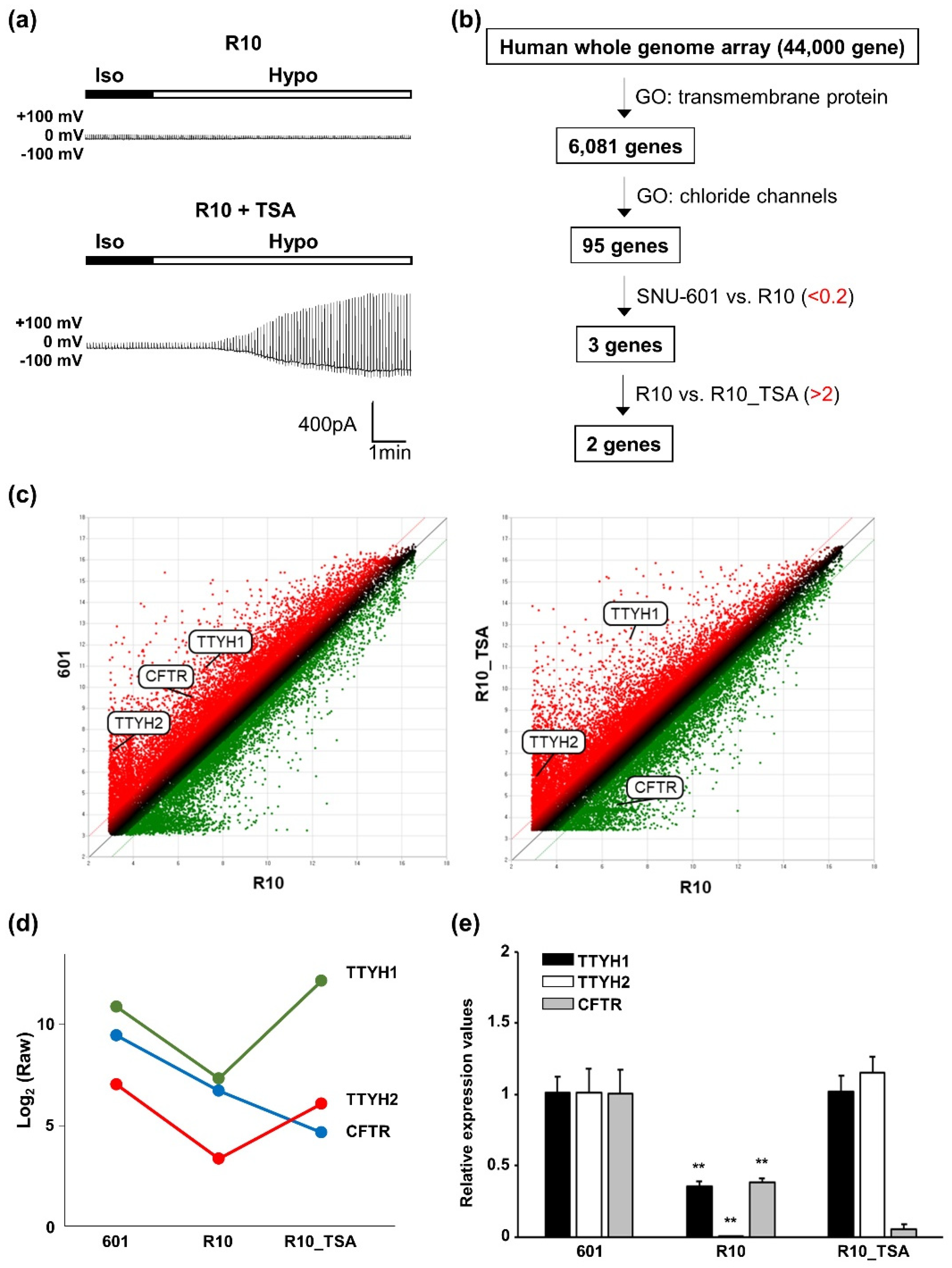
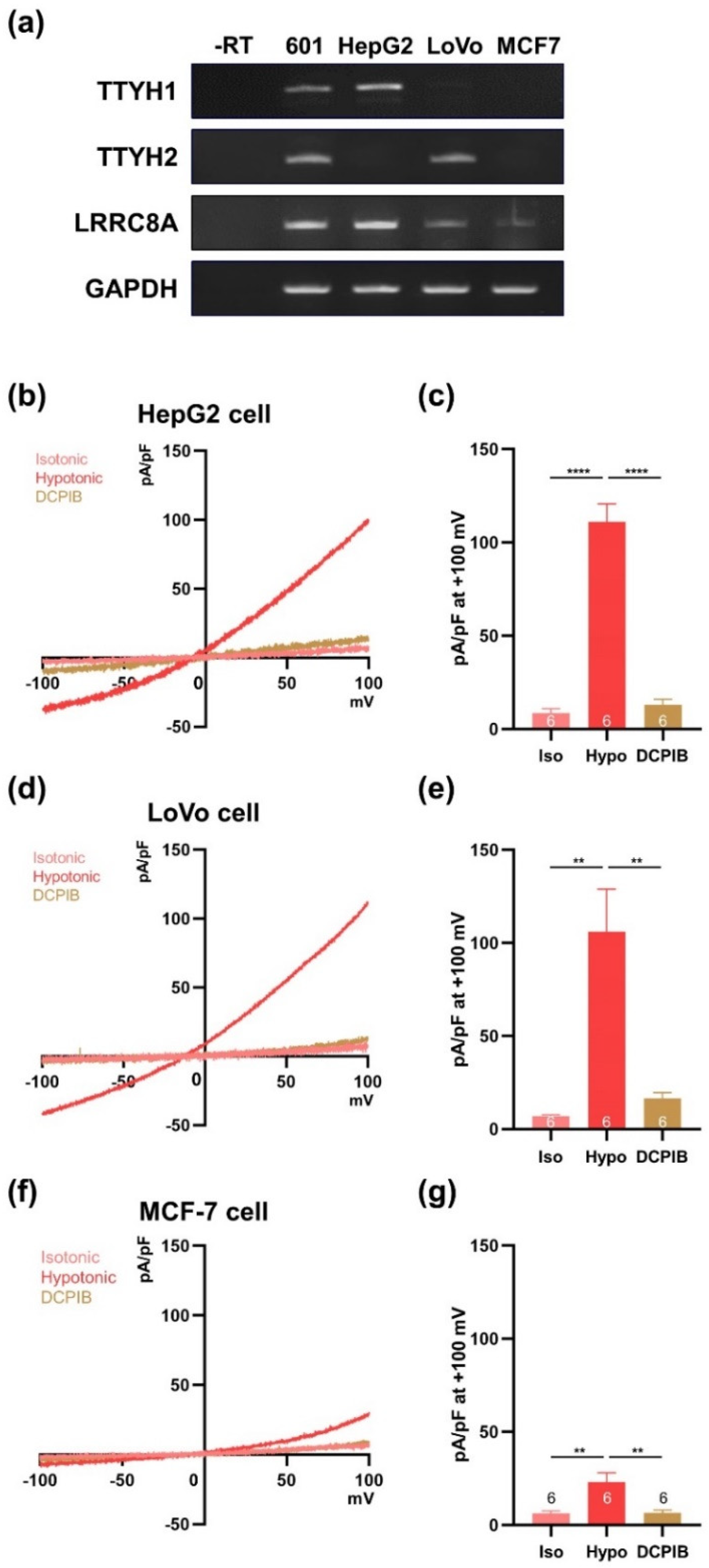
3.3. Whole-Genome Screening Identifies TTYH1 and TTYH2 as VRAC Candidates
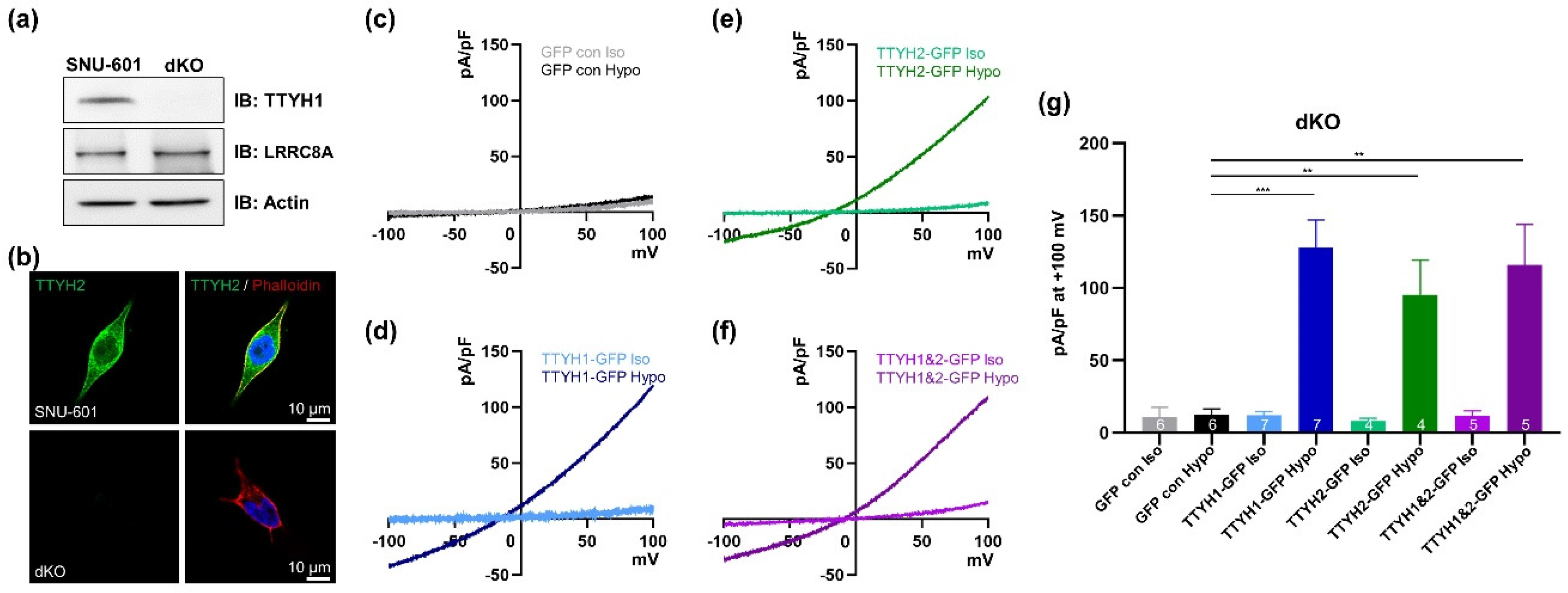
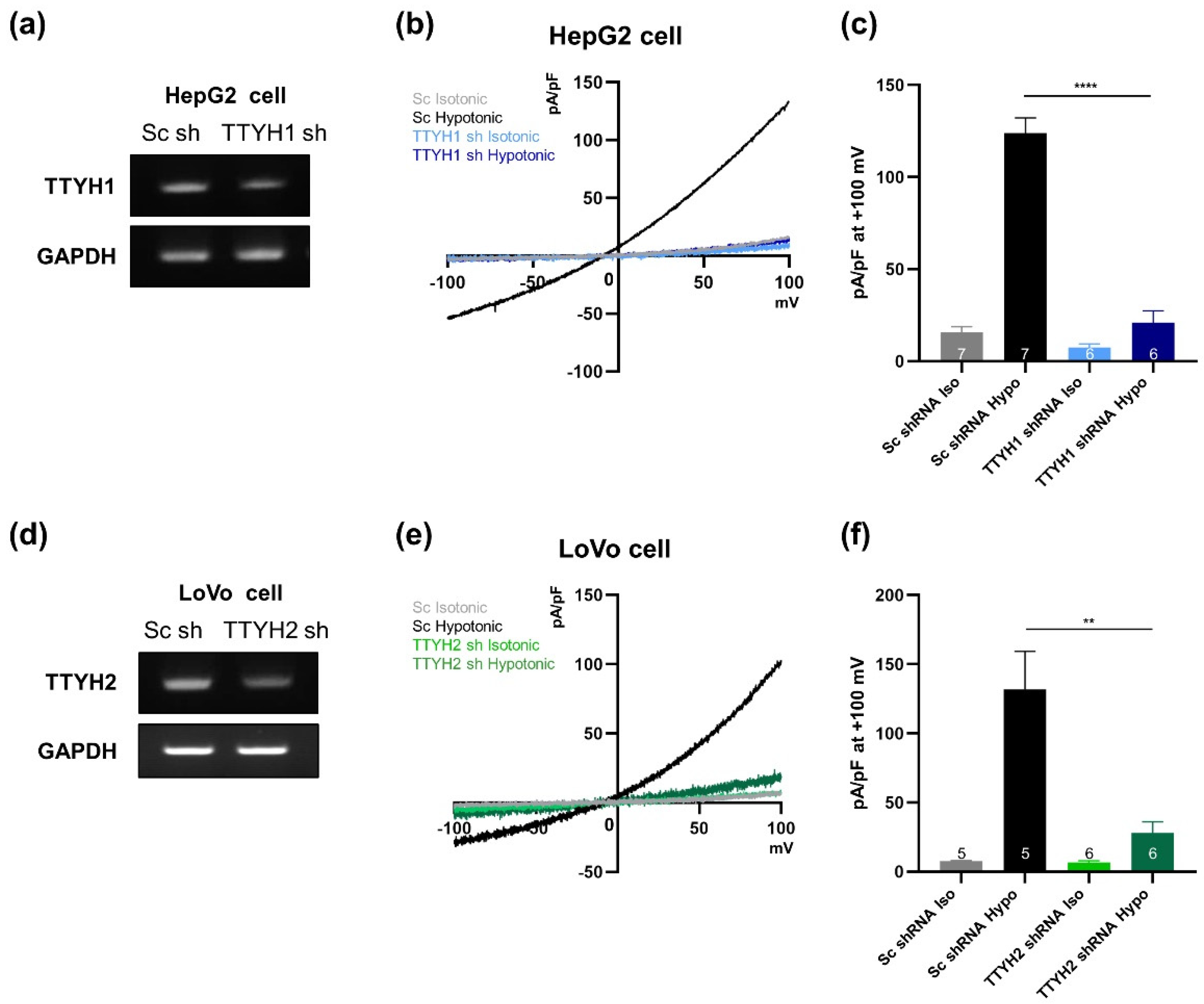
3.4. TTYH1/TTYH2 dKO Cells Lack VRAC Currents
3.5. TTYH1 and TTYH2 Have Independent VRAC Activity in Other Cancer Cell Lines
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pedersen, S.F.; Okada, Y.; Nilius, B. Biophysics and Physiology of the Volume-Regulated Anion Channel (VRAC)/Volume-Sensitive Outwardly Rectifying Anion Channel (VSOR). Pflug. Arch. Eur. J. Physiol. 2016, 468, 371–383. [Google Scholar] [CrossRef] [PubMed]
- Jentsch, T.J. VRACs and other ion channels and transporters in the regulation of cell volume and beyond. Nat. Rev. Mol. Cell Biol. 2016, 17, 293–307. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Dubin, A.E.; Mathur, J.; Tu, B.; Reddy, K.; Miraglia, L.J.; Reinhardt, J.; Orth, A.P.; Patapoutian, A. SWELL1, a plasma membrane protein, is an essential component of volume-regulated anion channel. Cell 2014, 157, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Voss, F.K.; Ullrich, F.; Münch, J.; Lazarow, K.; Lutter, D.; Mah, N.; Andrade-Navarro, M.A.; von Kries, J.P.; Stauber, T.; Jentsch, T.J. Identification of LRRC8 heteromers as an essential component of the volume-regulated anion channel VRAC. Science 2014, 344, 634–638. [Google Scholar] [CrossRef]
- Sirianant, L.; Wanitchakool, P.; Ousingsawat, J.; Benedetto, R.; Zormpa, A.; Cabrita, I.; Schreiber, R.; Kunzelmann, K. Non-essential contribution of LRRC8A to volume regulation. Pflug. Arch. Eur. J. Physiol. 2016, 468, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Okada, T.; Islam, M.R.; Tsiferova, N.A.; Okada, Y.; Sabirov, R.Z. Specific and essential but not sufficient roles of LRRC8A in the activity of volume-sensitive outwardly rectifying anion channel (VSOR). Channels 2017, 11, 109–120. [Google Scholar] [CrossRef]
- Milenkovic, A.; Brandl, C.; Milenkovic, V.M.; Jendryke, T.; Sirianant, L.; Wanitchakool, P.; Zimmermann, S.; Reiff, C.M.; Horling, F.; Schrewe, H.; et al. Bestrophin 1 is indispensable for volume regulation in human retinal pigment epithelium cells. Proc. Natl. Acad. Sci. USA 2015, 112, E2630–E2639. [Google Scholar] [CrossRef] [Green Version]
- Campbell, H.D.; Schimansky, T.; Claudianos, C.; Ozsarac, N.; Kasprzak, A.B.; Cotsell, J.N.; Young, I.G.; de Couet, H.G.; Miklos, G.L. The Drosophila melanogaster flightless-I gene involved in gastrulation and muscle degeneration encodes gelsolin-like and leucine-rich repeat domains and is conserved in Caenorhabditis elegans and humans. Proc. Natl. Acad. Sci. USA 1993, 90, 11386–11390. [Google Scholar] [CrossRef]
- Campbell, H.D.; Kamei, M.; Claudianos, C.; Woollatt, E.; Sutherland, G.R.; Suzuki, Y.; Hida, M.; Sugano, S.; Young, I.G. Human and mouse homologues of the Drosophila melanogaster tweety (tty) gene: A novel gene family encoding predicted transmembrane proteins. Genomics 2000, 68, 89–92. [Google Scholar] [CrossRef]
- Suzuki, M.; Mizuno, A. A Novel Human Cl- Channel Family Related to Drosophila flightless Locus. J. Biol. Chem. 2004, 279, 22461–22468. [Google Scholar] [CrossRef]
- Lee, E.L.; Shimizu, T.; Ise, T.; Numata, T.; Kohno, K.; Okada, Y. Impaired activity of volume-sensitive Cl- channel is involved in cisplatin resistance of cancer cells. J. Cell. Physiol. 2007, 211, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Min, X.J.; Li, H.; Hou, S.C.; He, W.; Liu, J.; Hu, B.; Wang, J. Dysfunction of volume-sensitive chloride channels contributes to cisplatin resistance in human lung adenocarcinoma cells. Exp. Biol. Med. 2011, 236, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.; Kim, D.-G.; Lee, Y.-S.; Bae, Y.; Kim, A.; Park, N.; Hwang, E.M.; Park, J.-Y. Surface expression of TTYH2 is attenuated by direct interaction with β-COP. BMB Rep. 2019, 4311. Available online: https://www.ncbi.nlm.nih.gov/pubmed/30670146 (accessed on 23 January 2019).
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Xu, H.; Choi, S.M.; An, C.S.; Min, Y.D.; Kim, K.C.; Kim, K.J.; Choi, C.H. Concentration-dependent collateral sensitivity of cisplatin-resistant gastric cancer cell sublines. Biochem. Biophys. Res. Commun. 2005, 328, 618–622. [Google Scholar] [CrossRef] [PubMed]
- Decher, N.; Lang, H.J.; Nilius, B.; Brüggemann, A.; Busch, A.E.; Steinmeyer, K. DCPIB is a novel selective blocker of I(Cl,swell) and prevents swelling-induced shortening of guinea-pig atrial action potential duration. Br. J. Pharmacol. 2001, 134, 1467–1479. [Google Scholar] [CrossRef] [PubMed]
- Best, L.; Yates, A.P.; Decher, N.; Steinmeyer, K.; Nilius, B. Inhibition of glucose-induced electrical activity in rat pancreatic beta-cells by DCPIB, a selective inhibitor of volume-sensitive anion currents. Eur. J. Pharmacol. 2004, 489, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Syeda, R.; Qiu, Z.; Dubin, A.E.; Murthy, S.E.; Maria, N.; Mason, D.E.; Mathur, J.; Cahalan, S.M.; Peters, E.C. LRRC8 Proteins Form Volume-Regulated Anion Channels that Sense Ionic Strength. Cell 2016, 164, 499–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, H.; Du, W.; Zhang, Y.-J.; Hong, J.; Su, W.-Y.; Tang, J.-T.; Wang, Y.-C.; Lu, R.; Fang, J.-Y. Trichostatin A, a histone deacetylase inhibitor, suppresses JAK2/STAT3 signaling via inducing the promoter-associated histone acetylation of SOCS1 and SOCS3 in human colorectal cancer cells. Mol. Carcinog. 2012, 51, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Planells-Cases, R.; Lutter, D.; Guyader, C.; Gerhards, N.M.; Ullrich, F.; Elger, D.A.; Kucukosmanoglu, A.; Xu, G.; Voss, F.K.; Reincke, S.M.; et al. Subunit composition of VRAC channels determines substrate specificity and cellular resistance to Pt-based anti-cancer drugs. EMBO J. 2015, 34, 2993–3008. [Google Scholar] [CrossRef] [PubMed]
- Cemazar, M.; Grosel, A.; Glavac, D.; Kotnik, V.; Skobrne, M.; Kranjc, S.; Mir, L.M.; Andre, F.; Opolon, P.; Sersa, G. Effects of Electrogenetherapy with p53wt Combined with Cisplatin on Survival of Human Tumor Cell Lines with Different p53 Status. DNA Cell Biol. 2003, 22, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Tardito, S.; Isella, C.; Medico, E.; Marchiò, L.; Bevilacqua, E.; Hatzoglou, M.; Bussolati, O.; Franchi-Gazzola, R. The thioxotriazole copper (II) complex A0 induces endoplasmic reticulum stress and paraptotic death in human cancer cells. J. Biol. Chem. 2009, 284, 24306–24319. [Google Scholar] [CrossRef] [PubMed]
- Jung, E.; Osswald, M.; Blaes, J.; Wiestler, B.; Sahm, F.; Schmenger, T.; Solecki, G.; Deumelandt, K.; Kurz, F.T.; Xie, R.; et al. Tweety-Homolog 1 Drives Brain Colonization of Gliomas. J. Neurosci. 2017, 37, 6837–6850. [Google Scholar] [CrossRef] [PubMed]
- Toiyama, Y.; Mizoguchi, A.; Kimura, K.; Hiro, J.; Inoue, Y.; Tutumi, T.; Miki, C.; Kusunoki, M. TTYH2, a human homologue of the Drosophila melanogaster gene tweety, is up-regulated in colon carcinoma and involved in cell proliferation and cell aggregation. World J. Gastroenterol. 2007, 13, 2717–2721. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, B.H.; Nielsen, D.; Thorsteinsdottir, U.A.; Hoffmann, E.K.; Lambert, I.H. Downregulation of LRRC8A protects human ovarian and alveolar carcinoma cells against Cisplatin-induced expression of p53, MDM2, p21 Waf1/Cip1, and Caspase-9/-3 activation. Am. J. Physiol. Physiol. 2016, 310, C857–C873. [Google Scholar] [CrossRef]
- Kim, E.; Hwang, E.M.; Yarishkin, O.; Yoo, J.C.; Kim, D.; Park, N.; Cho, M.; Lee, Y.S.; Sun, C.H.; Yi, G.S.; et al. Enhancement of TREK1 channel surface expression by protein-protein interaction with β-COP. Biochem. Biophys. Res. Commun. 2010, 395, 244–250. [Google Scholar] [CrossRef]
- Lee, Y.S.; Bae, Y.; Park, N.; Yoo, J.C.; Cho, C.H.; Ryoo, K.; Hwang, E.M.; Park, J.Y. Surface expression of the Anoctamin-1 (ANO1) channel is suppressed by protein-protein interactions with β-COP. Biochem. Biophys. Res. Commun. 2016, 475, 216–222. [Google Scholar] [CrossRef]
- Kim, J.; Han, D.; Byun, S.; Kwon, M.; Cho, J.Y.; Pleasure, S.J.; Yoon, K. Ttyh1 regulates embryonic neural stem cell properties by enhancing the Notch signaling pathway. EMBO Rep. 2018, 19, e45472. [Google Scholar] [CrossRef]
- He, Y.; Hryciw, D.H.; Carroll, M.L.; Myers, S.A.; Whitbread, A.K.; Kumar, S.; Poronnik, P.; Hooper, J.D. The ubiquitin-protein ligase Nedd4-2 differentially interacts with and regulates members of the tweety family of chloride ion channels. J. Biol. Chem. 2008, 283, 24000–24010. [Google Scholar] [CrossRef]
- Benedetto, R.; Sirianant, L.; Pankonien, I.; Wanitchakool, P.; Ousingsawat, J.; Cabrita, I.; Schreiber, R.; Amaral, M.; Kunzelmann, K. Relationship between TMEM16A/anoctamin 1 and LRRC8A. Pflug. Arch. Eur. J. Physiol. 2016, 468, 1751–1763. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bae, Y.; Kim, A.; Cho, C.-H.; Kim, D.; Jung, H.-G.; Kim, S.-S.; Yoo, J.; Park, J.-Y.; Hwang, E.M. TTYH1 and TTYH2 Serve as LRRC8A-Independent Volume-Regulated Anion Channels in Cancer Cells. Cells 2019, 8, 562. https://doi.org/10.3390/cells8060562
Bae Y, Kim A, Cho C-H, Kim D, Jung H-G, Kim S-S, Yoo J, Park J-Y, Hwang EM. TTYH1 and TTYH2 Serve as LRRC8A-Independent Volume-Regulated Anion Channels in Cancer Cells. Cells. 2019; 8(6):562. https://doi.org/10.3390/cells8060562
Chicago/Turabian StyleBae, Yeonju, Ajung Kim, Chang-Hoon Cho, Donggyu Kim, Hyun-Gug Jung, Seong-Seop Kim, Jiyun Yoo, Jae-Yong Park, and Eun Mi Hwang. 2019. "TTYH1 and TTYH2 Serve as LRRC8A-Independent Volume-Regulated Anion Channels in Cancer Cells" Cells 8, no. 6: 562. https://doi.org/10.3390/cells8060562
APA StyleBae, Y., Kim, A., Cho, C.-H., Kim, D., Jung, H.-G., Kim, S.-S., Yoo, J., Park, J.-Y., & Hwang, E. M. (2019). TTYH1 and TTYH2 Serve as LRRC8A-Independent Volume-Regulated Anion Channels in Cancer Cells. Cells, 8(6), 562. https://doi.org/10.3390/cells8060562





