Viral Appropriation: Laying Claim to Host Nuclear Transport Machinery
Abstract
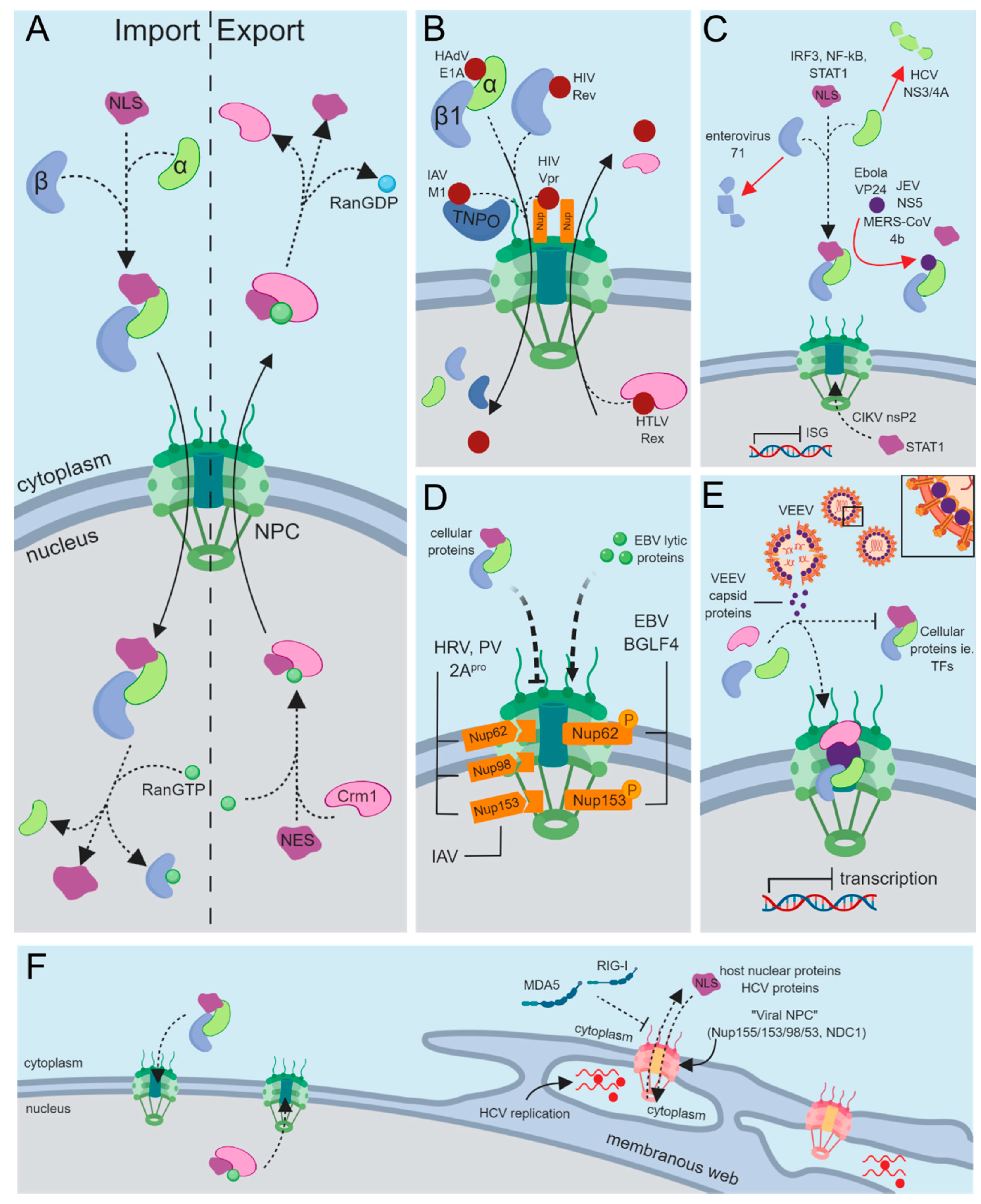
1. The Separation of Cytoplasmic and Nuclear Compartments
2. The Nuclear Envelope Can Represent an Obstacle for Viral Infection
3. Nucleocytoplasmic Transport of Cellular Proteins
4. Targeting Nuclear Import Machinery through Viral Mimicry
4.1. Interactions with the Nuclear Pore Complex (NPC)
4.2. Interactions with Importins
4.3. Nuclear Export and Nucleocytoplasmic Shuttling
5. Viral Disruption of Cellular Protein Trafficking
5.1. Subverting Antiviral Immunity through Nucleocytoplasmic Transport
5.2. General Disruption of Host Nucleocytoplasmic Transport
6. Viruses as a Tool to Discover Mechanisms of Nuclear Transport
7. Targeting Nuclear Transport to Control Viral Replication
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Martin, W.; Koonin, E.V. Introns and the origin of nucleus-cytosol compartmentalization. Nature 2006, 440, 41–45. [Google Scholar] [CrossRef]
- Devos, D.P.; Graf, R.; Field, M.C. Evolution of the nucleus. Curr. Opin. Cell Biol. 2014, 28, 8–15. [Google Scholar] [CrossRef]
- Schwartz, T.U. The Structure Inventory of the Nuclear Pore Complex. J. Mol. Biol. 2016, 428, 1986–2000. [Google Scholar] [CrossRef]
- Beck, M.; Hurt, E. The nuclear pore complex: Understanding its function through structural insight. Nat. Rev. Mol. Cell Biol. 2017, 18, 73–89. [Google Scholar] [CrossRef]
- Cautain, B.; Hill, R.; de Pedro, N.; Link, W. Components and regulation of nuclear transport processes. FEBS J. 2015, 282, 445–462. [Google Scholar] [CrossRef]
- Goldberg, M.W. Nuclear pore complex tethers to the cytoskeleton. Semin. Cell Dev. Biol. 2017, 68, 52–58. [Google Scholar] [CrossRef]
- Ptak, C.; Aitchison, J.D.; Wozniak, R.W. The multifunctional nuclear pore complex: A platform for controlling gene expression. Curr. Opin. Cell Biol. 2014, 28, 46–53. [Google Scholar] [CrossRef]
- Frey, S.; Gorlich, D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell 2007, 130, 512–523. [Google Scholar] [CrossRef]
- Aramburu, I.V.; Lemke, E.A. Floppy but not sloppy: Interaction mechanism of FG-nucleoporins and nuclear transport receptors. Semin. Cell Dev. Biol. 2017, 68, 34–41. [Google Scholar] [CrossRef]
- Frey, S.; Rees, R.; Schunemann, J.; Ng, S.C.; Funfgeld, K.; Huyton, T.; Gorlich, D. Surface Properties Determining Passage Rates of Proteins through Nuclear Pores. Cell 2018, 174, 202–217.e9. [Google Scholar] [CrossRef]
- Timney, B.L.; Raveh, B.; Mironska, R.; Trivedi, J.M.; Kim, S.J.; Russel, D.; Wente, S.R.; Sali, A.; Rout, M.P. Simple rules for passive diffusion through the nuclear pore complex. J. Cell Biol. 2016, 215, 57–76. [Google Scholar] [CrossRef] [PubMed]
- Chook, Y.M.; Blobel, G. Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 2001, 11, 703–715. [Google Scholar] [CrossRef]
- Radu, A.; Moore, M.S.; Blobel, G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell 1995, 81, 215–222. [Google Scholar] [CrossRef]
- Fay, N.; Pante, N. Nuclear entry of DNA viruses. Front. Microbiol. 2015, 6, 467. [Google Scholar] [CrossRef] [PubMed]
- Kobiler, O.; Drayman, N.; Butin-Israeli, V.; Oppenheim, A. Virus strategies for passing the nuclear envelope barrier. Nucleus 2012, 3, 526–539. [Google Scholar] [CrossRef] [PubMed]
- Kuss, S.K.; Mata, M.A.; Zhang, L.; Fontoura, B.M.A. Nuclear imprisonment: Viral strategies to arrest host mRNA nuclear export. Viruses 2013, 5, 1824–1849. [Google Scholar] [CrossRef] [PubMed]
- Yarbrough, M.L.; Mata, M.A.; Sakthivel, R.; Fontoura, B.M.A. Viral subversion of nucleocytoplasmic trafficking. Traffic 2014, 15, 127–140. [Google Scholar] [CrossRef] [PubMed]
- Harel, A.; Forbes, D.J. Importin beta: Conducting a much larger cellular symphony. Mol. Cell 2004, 16, 319–330. [Google Scholar] [CrossRef]
- Xu, D.; Farmer, A.; Chook, Y.M. Recognition of nuclear targeting signals by Karyopherin-beta proteins. Curr. Opin. Struct. Biol. 2010, 20, 782–790. [Google Scholar] [CrossRef] [PubMed]
- Goldfarb, D.S.; Corbett, A.H.; Mason, D.A.; Harreman, M.T.; Adam, S.A. Importin alpha: A multipurpose nuclear-transport receptor. Trends Cell Biol. 2004, 14, 505–514. [Google Scholar] [CrossRef]
- Cavazza, T.; Vernos, I. The RanGTP Pathway: From Nucleo-Cytoplasmic Transport to Spindle Assembly and Beyond. Front. Cell Dev. Biol. 2015, 3, 82. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 2007, 8, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Lott, K.; Cingolani, G. The importin beta binding domain as a master regulator of nucleocytoplasmic transport. Biochim. Biophys. Acta 2011, 1813, 1578–1592. [Google Scholar] [CrossRef] [PubMed]
- Riddick, G.; Macara, I.G. The adapter importin-alpha provides flexible control of nuclear import at the expense of efficiency. Mol. Syst. Biol. 2007, 3, 118. [Google Scholar] [CrossRef] [PubMed]
- Pumroy, R.A.; Cingolani, G. Diversification of importin-alpha isoforms in cellular trafficking and disease states. Biochem. J. 2015, 466, 13–28. [Google Scholar] [CrossRef]
- Kalderon, D.; Roberts, B.L.; Richardson, W.D.; Smith, A.E.; Hill, M. A Short Amino Acid Sequence Able to Specify Nuclear Location. Cell 1984, 39, 499–509. [Google Scholar] [CrossRef]
- Robbins, J.; Dilworth, S.M.; Laskey, R.A.; Dingwall, C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: Identification of a class of bipartite nuclear targeting sequence. Cell 1991, 64, 615–623. [Google Scholar] [CrossRef]
- Marfori, M.; Mynott, A.; Ellis, J.J.; Mehdi, A.M.; Saunders, N.F.W.; Curmi, P.M.; Forwood, J.K.; Bodén, M.; Kobe, B. Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim. Biophys. Acta 2011, 1813, 1562–1577. [Google Scholar] [CrossRef] [PubMed]
- Conti, E.; Uy, M.; Leighton, L.; Blobel, G.; Kuriyan, J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell 1998, 94, 193–204. [Google Scholar] [CrossRef]
- Dingwall, C.; Laskey, R.A. Nuclear import: A tale of two sites. Curr. Biol. 1998, 8, R922–R924. [Google Scholar] [CrossRef]
- Kimura, M.; Imamoto, N. Biological significance of the importin-β family-dependent nucleocytoplasmic transport pathways. Traffic 2014, 15, 727–748. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.J.; Cansizoglu, A.E.; Suel, K.E.; Louis, T.H.; Zhang, Z.; Chook, Y.M. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell 2006, 126, 543–558. [Google Scholar] [CrossRef] [PubMed]
- Soniat, M.; Chook, Y.M. Karyopherin-beta2 Recognition of a PY-NLS Variant that Lacks the Proline-Tyrosine Motif. Structure 2016, 24, 1802–1809. [Google Scholar] [CrossRef] [PubMed]
- Suel, K.E.; Gu, H.; Chook, Y.M. Modular organization and combinatorial energetics of proline-tyrosine nuclear localization signals. PLoS Biol. 2008, 6, e137. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Twyffels, L.; Gueydan, C.; Kruys, V. Transportin-1 and Transportin-2: Protein nuclear import and beyond. FEBS Lett. 2014, 588, 1857–1868. [Google Scholar] [CrossRef] [PubMed]
- Fornerod, M.; Ohno, M.; Yoshida, M.; Mattaj, I.W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 1997, 90, 1051–1060. [Google Scholar] [CrossRef]
- Xu, D.; Grishin, N.V.; Chook, Y.M. NESdb: A database of NES-containing CRM1 cargoes. Mol. Biol. Cell 2012, 23, 3673–3676. [Google Scholar] [CrossRef] [PubMed]
- Meyer, B.E.; Meinkoth, J.L.; Malim, M.H. Nuclear transport of human immunodeficiency virus type 1, visna virus, and equine infectious anemia virus Rev proteins: Identification of a family of transferable nuclear export signals. J. Virol. 1996, 70, 2350–2359. [Google Scholar]
- Fung, H.Y.J.; Fu, S.-C.; Chook, Y.M. Nuclear export receptor CRM1 recognizes diverse conformations in nuclear export signals. eLife 2017, 6. [Google Scholar] [CrossRef]
- Fung, H.Y.J.; Fu, S.-C.; Brautigam, C.A.; Chook, Y.M. Structural determinants of nuclear export signal orientation in binding to exportin CRM1. eLife 2015, 4. [Google Scholar] [CrossRef]
- Williams, T.; Ngo, L.H.; Wickramasinghe, V.O. Nuclear export of RNA: Different sizes, shapes and functions. Semin. Cell Dev. Biol. 2018, 75, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Kose, S.; Furuta, M.; Imamoto, N. Hikeshi, a nuclear import carrier for Hsp70s, protects cells from heat shock-induced nuclear damage. Cell 2012, 149, 578–589. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Zak, J.; Chen, S.; Sanchez-Pulido, L.; Severson, D.T.; Endicott, J.; Ponting, C.P.; Schofield, C.J.; Lu, X. A code for RanGDP binding in ankyrin repeats defines a nuclear import pathway. Cell 2014, 157, 1130–1145. [Google Scholar] [CrossRef] [PubMed]
- Copeland, A.M.; Newcomb, W.W.; Brown, J.C. Herpes simplex virus replication: Roles of viral proteins and nucleoporins in capsid-nucleus attachment. J. Virol. 2009, 83, 1660–1668. [Google Scholar] [CrossRef] [PubMed]
- Matreyek, K.A.; Yucel, S.S.; Li, X.; Engelman, A. Nucleoporin NUP153 phenylalanine-glycine motifs engage a common binding pocket within the HIV-1 capsid protein to mediate lentiviral infectivity. PLoS Pathog. 2013, 9, e1003693. [Google Scholar] [CrossRef]
- Strunze, S.; Engelke, M.F.; Wang, I.-H.; Puntener, D.; Boucke, K.; Schleich, S.; Way, M.; Schoenenberger, P.; Burckhardt, C.J.; Greber, U.F. Kinesin-1-mediated capsid disassembly and disruption of the nuclear pore complex promote virus infection. Cell Host Microbe 2011, 10, 210–223. [Google Scholar] [CrossRef]
- Le Sage, V.; Mouland, A.J. Viral subversion of the nuclear pore complex. Viruses 2013, 5, 2019–2042. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-W.; Lee, C.-P.; Huang, Y.-H.; Yang, P.-W.; Wang, J.-T.; Chen, M.-R. Epstein-Barr virus protein kinase BGLF4 targets the nucleus through interaction with nucleoporins. J. Virol. 2012, 86, 8072–8085. [Google Scholar] [CrossRef]
- Fouchier, R.A.; Meyer, B.E.; Simon, J.H.; Fischer, U.; Albright, A.V.; Gonzalez-Scarano, F.; Malim, M.H. Interaction of the human immunodeficiency virus type 1 Vpr protein with the nuclear pore complex. J. Virol. 1998, 72, 6004–6013. [Google Scholar]
- Jenkins, Y.; McEntee, M.; Weis, K.; Greene, W.C. Characterization of HIV-1 vpr nuclear import: Analysis of signals and pathways. J. Cell Biol. 1998, 143, 875–885. [Google Scholar] [CrossRef]
- Zachariae, U.; Grubmuller, H. Importin-beta: Structural and dynamic determinants of a molecular spring. Structure 2008, 16, 906–915. [Google Scholar] [CrossRef] [PubMed]
- Le Rouzic, E.; Mousnier, A.; Rustum, C.; Stutz, F.; Hallberg, E.; Dargemont, C.; Benichou, S. Docking of HIV-1 Vpr to the nuclear envelope is mediated by the interaction with the nucleoporin hCG1. J. Biol. Chem. 2002, 277, 45091–45098. [Google Scholar] [CrossRef] [PubMed]
- Bukrinsky, M.I.; Sharova, N.; Dempsey, M.P.; Stanwick, T.L.; Bukrinskaya, A.G.; Haggerty, S.; Stevenson, M. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc. Natl. Acad. Sci. USA 1992, 89, 6580–6584. [Google Scholar] [CrossRef] [PubMed]
- Kamata, M.; Nitahara-Kasahara, Y.; Miyamoto, Y.; Yoneda, Y.; Aida, Y. Importin-alpha promotes passage through the nuclear pore complex of human immunodeficiency virus type 1 Vpr. J. Virol. 2005, 79, 3557–3564. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nitahara-Kasahara, Y.; Kamata, M.; Yamamoto, T.; Zhang, X.; Miyamoto, Y.; Muneta, K.; Iijima, S.; Yoneda, Y.; Tsunetsugu-Yokota, Y.; Aida, Y. Novel nuclear import of Vpr promoted by importin alpha is crucial for human immunodeficiency virus type 1 replication in macrophages. J. Virol. 2007, 81, 5284–5293. [Google Scholar] [CrossRef] [PubMed]
- Vodicka, M.A.; Koepp, D.M.; Silver, P.A.; Emerman, M. HIV-1 Vpr interacts with the nuclear transport pathway to promote macrophage infection. Genes Dev. 1998, 12, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Johnson, M.; Brocardo, M.; Jamieson, C.; Henderson, B.R. Wnt signaling proteins associate with the nuclear pore complex: Implications for cancer. Adv. Exp. Med. Biol. 2014, 773, 353–372. [Google Scholar] [CrossRef]
- Hudjetz, B.; Gabriel, G. Human-like PB2 627K influenza virus polymerase activity is regulated by importin-alpha1 and -alpha7. PLoS Pathog. 2012, 8, e1002488. [Google Scholar] [CrossRef] [PubMed]
- Pumroy, R.A.; Ke, S.; Hart, D.J.; Zachariae, U.; Cingolani, G. Molecular determinants for nuclear import of influenza A PB2 by importin alpha isoforms 3 and 7. Structure 2015, 23, 374–384. [Google Scholar] [CrossRef]
- Ao, Z.; Danappa Jayappa, K.; Wang, B.; Zheng, Y.; Kung, S.; Rassart, E.; Depping, R.; Kohler, M.; Cohen, E.A.; Yao, X. Importin alpha3 interacts with HIV-1 integrase and contributes to HIV-1 nuclear import and replication. J. Virol. 2010, 84, 8650–8663. [Google Scholar] [CrossRef] [PubMed]
- Kohler, M.; Gorlich, D.; Hartmann, E.; Franke, J. Adenoviral E1A protein nuclear import is preferentially mediated by importin alpha3 in vitro. Virology 2001, 289, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.J.; King, C.R.; Dikeakos, J.D.; Mymryk, J.S. Functional analysis of the C-terminal region of human adenovirus E1A reveals a misidentified nuclear localization signal. Virology 2014, 468–470, 238–243. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.-L.; Wilson, V.G. Common importin alpha specificity for papillomavirus E2 proteins. Virus Res. 2010, 150, 135–137. [Google Scholar] [CrossRef] [PubMed]
- Alvisi, G.; Musiani, D.; Jans, D.A.; Ripalti, A. An importin alpha/beta-recognized bipartite nuclear localization signal mediates targeting of the human herpes simplex virus type 1 DNA polymerase catalytic subunit pUL30 to the nucleus. Biochemistry 2007, 46, 9155–9163. [Google Scholar] [CrossRef] [PubMed]
- Van Roey, K.; Uyar, B.; Weatheritt, R.J.; Dinkel, H.; Seiler, M.; Budd, A.; Gibson, T.J.; Davey, N.E. Short linear motifs: Ubiquitous and functionally diverse protein interaction modules directing cell regulation. Chem. Rev. 2014, 114, 6733–6778. [Google Scholar] [CrossRef] [PubMed]
- Davey, N.E.; Cyert, M.S.; Moses, A.M. Short linear motifs - ex nihilo evolution of protein regulation. Cell Commun. Signal. 2015, 13, 43. [Google Scholar] [CrossRef]
- King, C.R.; Zhang, A.; Tessier, T.M.; Gameiro, S.F.; Mymryk, J.S. Hacking the Cell: Network Intrusion and Exploitation by Adenovirus E1A. mBio 2018, 9. [Google Scholar] [CrossRef]
- Davey, N.E.; Travé, G.; Gibson, T.J. How viruses hijack cell regulation. Trends Biochem. Sci. 2011, 36, 159–169. [Google Scholar] [CrossRef]
- Via, A.; Uyar, B.; Brun, C.; Zanzoni, A. How pathogens use linear motifs to perturb host cell networks. Trends Biochem. Sci. 2015, 40, 36–48. [Google Scholar] [CrossRef]
- Henderson, B.R.; Percipalle, P. Interactions between HIV Rev and nuclear import and export factors: The Rev nuclear localisation signal mediates specific binding to human importin-beta. J. Mol. Biol. 1997, 274, 693–707. [Google Scholar] [CrossRef]
- Truant, R.; Cullen, B. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin beta-dependent nuclear localization signals.pdf. Mol. Cell. Biol. 1999, 19, 1210–1217. [Google Scholar] [CrossRef] [PubMed]
- Palmeri, D.; Malim, M.H. Importin beta can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol. Cell. Biol. 1999, 19, 1218–1225. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Nath, A.; Hauber, J.; Kehlenbach, R.H. Multiple importins function as nuclear transport receptors for the Rev protein of human immunodeficiency virus type 1. J. Biol. Chem. 2006, 281, 20883–20890. [Google Scholar] [CrossRef] [PubMed]
- Ojala, P.M.; Sodeik, B.; Ebersold, M.W.; Kutay, U.; Helenius, A. Herpes simplex virus type 1 entry into host cells: Reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol. Cell. Biol. 2000, 20, 4922–4931. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wang, J.C.-Y.; Pierson, E.E.; Keifer, D.Z.; Delaleau, M.; Gallucci, L.; Cazenave, C.; Kann, M.; Jarrold, M.F.; Zlotnick, A. Importin beta Can Bind Hepatitis B Virus Core Protein and Empty Core-Like Particles and Induce Structural Changes. PLoS Pathog. 2016, 12, e1005802. [Google Scholar] [CrossRef] [PubMed]
- Wodrich, H.; Cassany, A.; D’Angelo, M.A.; Guan, T.; Nemerow, G.; Gerace, L. Adenovirus core protein pVII is translocated into the nucleus by multiple import receptor pathways. J. Virol. 2006, 80, 9608–9618. [Google Scholar] [CrossRef]
- Miyake, Y.; Keusch, J.J.; Decamps, L.; Ho-Xuan, H.; Iketani, S.; Gut, H.; Kutay, U.; Helenius, A.; Yamauchi, Y. Influenza virus uses transportin 1 for vRNP debundling during cell entry. Nat. Microbiol. 2019, 4, 578–586. [Google Scholar] [CrossRef]
- Wang, L.; Li, M.; Cai, M.; Xing, J.; Wang, S.; Zheng, C. A PY-nuclear localization signal is required for nuclear accumulation of HCMV UL79 protein. Med. Microbiol. Immunol. 2012, 201, 381–387. [Google Scholar] [CrossRef]
- Hindley, C.E.; Lawrence, F.J.; Matthews, D.A. A role for transportin in the nuclear import of adenovirus core proteins and DNA. Traffic 2007, 8, 1313–1322. [Google Scholar] [CrossRef]
- Darshan, M.S.; Lucchi, J.; Harding, E.; Moroianu, J. The l2 minor capsid protein of human papillomavirus type 16 interacts with a network of nuclear import receptors. J. Virol. 2004, 78, 12179–12188. [Google Scholar] [CrossRef]
- Le Roux, L.G.; Moroianu, J. Nuclear entry of high-risk human papillomavirus type 16 E6 oncoprotein occurs via several pathways. J. Virol. 2003, 77, 2330–2337. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.; Brooks, A.J.; Jans, D.A.; Vasudevan, S.G. A small region of the dengue virus-encoded RNA-dependent RNA polymerase, NS5, confers interaction with both the nuclear transport receptor importin-beta and the viral helicase, NS3. J. Gen. Virol. 2001, 82, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, E.C.; Orr, O.E.; Man Liu, S.; Engelhardt, O.G.; Fodor, E. Characterization of the interaction between the influenza A virus polymerase subunit PB1 and the host nuclear import factor Ran-binding protein 5. J. Gen. Virol. 2011, 92, 1859–1869. [Google Scholar] [CrossRef] [PubMed]
- Tarendeau, F.; Boudet, J.; Guilligay, D.; Mas, P.J.; Bougault, C.M.; Boulo, S.; Baudin, F.; Ruigrok, R.W.H.; Daigle, N.; Ellenberg, J.; et al. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat. Struct. Mol. Biol. 2007, 14, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.; Xu, H.; Ji, X.; Zhao, J.; Xu, H.; Hu, Y.; Deng, S.; Hu, S.; Liu, X. Importin alpha5 negatively regulates importin beta1-mediated nuclear import of Newcastle disease virus matrix protein and viral replication and pathogenicity in chicken fibroblasts. Virulence 2018, 9, 783–803. [Google Scholar] [CrossRef] [PubMed]
- Booth, D.S.; Cheng, Y.; Frankel, A.D. The export receptor Crm1 forms a dimer to promote nuclear export of HIV RNA. eLife 2014, 3, e04121. [Google Scholar] [CrossRef] [PubMed]
- Nakano, K.; Watanabe, T. HTLV-1 Rex Tunes the Cellular Environment Favorable for Viral Replication. Viruses 2016, 8, 58. [Google Scholar] [CrossRef]
- Pryor, M.J.; Rawlinson, S.M.; Wright, P.J.; Jans, D.A. CRM1-dependent nuclear export of dengue virus type 2 NS5. Novartis Found. Symp. 2006, 277, 143–149, 251–253. [Google Scholar]
- Sanchez, V.; Mahr, J.A.; Orazio, N.I.; Spector, D.H. Nuclear export of the human cytomegalovirus tegument protein pp65 requires cyclin-dependent kinase activity and the Crm1 exporter. J. Virol. 2007, 81, 11730–11736. [Google Scholar] [CrossRef]
- Ghildyal, R.; Ho, A.; Dias, M.; Soegiyono, L.; Bardin, P.G.; Tran, K.C.; Teng, M.N.; Jans, D.A. The respiratory syncytial virus matrix protein possesses a Crm1-mediated nuclear export mechanism. J. Virol. 2009, 83, 5353–5362. [Google Scholar] [CrossRef]
- Huang, S.; Chen, J.; Chen, Q.; Wang, H.; Yao, Y.; Chen, J.; Chen, Z. A second CRM1-dependent nuclear export signal in the influenza A virus NS2 protein contributes to the nuclear export of viral ribonucleoproteins. J. Virol. 2013, 87, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Chutiwitoonchai, N.; Aida, Y. NXT1, a Novel Influenza A NP Binding Protein, Promotes the Nuclear Export of NP via a CRM1-Dependent Pathway. Viruses 2016, 8, 209. [Google Scholar] [CrossRef] [PubMed]
- Kratzer, F.; Rosorius, O.; Heger, P.; Hirschmann, N.; Dobner, T.; Hauber, J.; Stauber, R.H. The adenovirus type 5 E1B-55K oncoprotein is a highly active shuttle protein and shuttling is independent of E4orf6, p53 and Mdm2. Oncogene 2000, 19, 850–857. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, Y.; Zhang, Z.; Zhao, X.; Wei, H.; Deng, J.; Cui, Z.; Zhang, X.-E. Human cytomegalovirus UL94 is a nucleocytoplasmic shuttling protein containing two NLSs and one NES. Virus Res. 2012, 166, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zheng, C. The first identified nucleocytoplasmic shuttling herpesviral capsid protein: Herpes simplex virus type 1 VP19C. PLoS ONE 2012, 7, e41825. [Google Scholar] [CrossRef]
- Jiang, H.; Olson, M.V.; Medrano, D.R.; Lee, O.-H.; Xu, J.; Piao, Y.; Alonso, M.M.; Gomez-Manzano, C.; Hung, M.-C.; Yung, W.K.A.; et al. A novel CRM1-dependent nuclear export signal in adenoviral E1A protein regulated by phosphorylation. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2006, 20, 2603–2605. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, L.; Wang, S.; Xing, J.; Zheng, C. Identification of a novel NLS of herpes simplex virus type 1 (HSV-1) VP19C and its nuclear localization is required for efficient production of HSV-1. J. Gen. Virol. 2012, 93, 1869–1875. [Google Scholar] [CrossRef]
- Adamson, W.E.; McNab, D.; Preston, V.G.; Rixon, F.J. Mutational analysis of the herpes simplex virus triplex protein VP19C. J. Virol. 2006, 80, 1537–1548. [Google Scholar] [CrossRef]
- Xing, J.; Wu, F.; Pan, W.; Zheng, C. Molecular anatomy of subcellular localization of HSV-1 tegument protein US11 in living cells. Virus Res. 2010, 153, 71–81. [Google Scholar] [CrossRef]
- Li, J.; Yu, M.; Zheng, W.; Liu, W. Nucleocytoplasmic shuttling of influenza A virus proteins. Viruses 2015, 7, 2668–2682. [Google Scholar] [CrossRef]
- Oksayan, S.; Wiltzer, L.; Rowe, C.L.; Blondel, D.; Jans, D.A.; Moseley, G.W. A novel nuclear trafficking module regulates the nucleocytoplasmic localization of the rabies virus interferon antagonist, P protein. J. Biol. Chem. 2012, 287, 28112–28121. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Denman, A.J.; Russo, A.; Wagstaff, K.M.; White, P.A.; Jans, D.A.; Mackenzie, J.M. Nucleocytoplasmic shuttling of the West Nile virus RNA-dependent RNA polymerase NS5 is critical to infection. Cell. Microbiol. 2018, 20, e12848. [Google Scholar] [CrossRef] [PubMed]
- Seth, R.B.; Sun, L.; Chen, Z.J. Antiviral innate immunity pathways. Cell Res. 2006, 16, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Ivashkiv, L.B.; Donlin, L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014, 14, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-kappaB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; He, S.; Minassian, A.; Li, J.; Feng, P. Recent advances on viral manipulation of NF-kappaB signaling pathway. Curr. Opin. Virol. 2015, 15, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Morikawa, K.; Lange, C.M.; Gouttenoire, J.; Meylan, E.; Brass, V.; Penin, F.; Moradpour, D. Nonstructural protein 3-4A: The Swiss army knife of hepatitis C virus. J. Viral Hepat. 2011, 18, 305–315. [Google Scholar] [CrossRef]
- Gagne, B.; Tremblay, N.; Park, A.Y.; Baril, M.; Lamarre, D. Importin beta1 targeting by hepatitis C virus NS3/4A protein restricts IRF3 and NF-kappaB signaling of IFNB1 antiviral response. Traffic 2017, 18, 362–377. [Google Scholar] [CrossRef]
- Wang, C.; Sun, M.; Yuan, X.; Ji, L.; Jin, Y.; Cardona, C.J.; Xing, Z. Enterovirus 71 suppresses interferon responses by blocking Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling through inducing karyopherin-alpha1 degradation. J. Biol. Chem. 2017, 292, 10262–10274. [Google Scholar] [CrossRef]
- Deng, L.; Zeng, Q.; Wang, M.; Cheng, A.; Jia, R.; Chen, S.; Zhu, D.; Liu, M.; Yang, Q.; Wu, Y.; et al. Suppression of NF-kappaB Activity: A Viral Immune Evasion Mechanism. Viruses 2018, 10, 409. [Google Scholar] [CrossRef]
- Mateo, M.; Reid, S.P.; Leung, L.W.; Basler, C.F.; Volchkov, V.E. Ebolavirus VP24 binding to karyopherins is required for inhibition of interferon signaling. J. Virol. 2010, 84, 1169–1175. [Google Scholar] [CrossRef] [PubMed]
- Reid, S.P.; Valmas, C.; Martinez, O.; Sanchez, F.M.; Basler, C.F. Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin alpha proteins with activated STAT1. J. Virol. 2007, 81, 13469–13477. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Chen, Z.; Li, Y.; Zhao, Z.; He, W.; Zohaib, A.; Song, Y.; Deng, C.; Zhang, B.; Chen, H.; et al. Japanese Encephalitis Virus NS5 Inhibits Type I Interferon (IFN) Production by Blocking the Nuclear Translocation of IFN Regulatory Factor 3 and NF-kappaB. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Canton, J.; Fehr, A.R.; Fernandez-Delgado, R.; Gutierrez-Alvarez, F.J.; Sanchez-Aparicio, M.T.; Garcia-Sastre, A.; Perlman, S.; Enjuanes, L.; Sola, I. MERS-CoV 4b protein interferes with the NF-kappaB-dependent innate immune response during infection. PLoS Pathog. 2018, 14, e1006838. [Google Scholar] [CrossRef] [PubMed]
- Frieman, M.; Yount, B.; Heise, M.; Kopecky-Bromberg, S.A.; Palese, P.; Baric, R.S. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 2007, 81, 9812–9824. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sun, J.; Zhang, H.; Wang, M.; Gao, G.F.; Li, X. Ebola virus encodes a miR-155 analog to regulate importin-alpha5 expression. Cell. Mol. Life Sci. 2016, 73, 3733–3744. [Google Scholar] [CrossRef] [PubMed]
- Overby, A.K.; Weber, F. Hiding from intracellular pattern recognition receptors, a passive strategy of flavivirus immune evasion. Virulence 2011, 2, 238–240. [Google Scholar] [CrossRef]
- Paul, D.; Bartenschlager, R. Architecture and biogenesis of plus-strand RNA virus replication factories. World J. Virol. 2013, 2, 32–48. [Google Scholar] [CrossRef]
- Neufeldt, C.J.; Joyce, M.A.; Levin, A.; Steenbergen, R.H.; Pang, D.; Shields, J.; Tyrrell, D.L.J.; Wozniak, R.W. Hepatitis C virus-induced cytoplasmic organelles use the nuclear transport machinery to establish an environment conducive to virus replication. PLoS Pathog. 2013, 9, e1003744. [Google Scholar] [CrossRef]
- Neufeldt, C.J.; Joyce, M.A.; Van Buuren, N.; Levin, A.; Kirkegaard, K.; Gale, M.J.; Tyrrell, D.L.J.; Wozniak, R.W. The Hepatitis C Virus-Induced Membranous Web and Associated Nuclear Transport Machinery Limit Access of Pattern Recognition Receptors to Viral Replication Sites. PLoS Pathog. 2016, 12, e1005428. [Google Scholar] [CrossRef]
- Levin, A.; Neufeldt, C.J.; Pang, D.; Wilson, K.; Loewen-Dobler, D.; Joyce, M.A.; Wozniak, R.W.; Tyrrell, D.L.J. Functional characterization of nuclear localization and export signals in hepatitis C virus proteins and their role in the membranous web. PLoS ONE 2014, 9, e114629. [Google Scholar] [CrossRef] [PubMed]
- Weberruss, M.; Antonin, W. Perforating the nuclear boundary - how nuclear pore complexes assemble. J. Cell Sci. 2016, 129, 4439–4447. [Google Scholar] [CrossRef] [PubMed]
- Cordes, V.C.; Reidenbach, S.; Franke, W.W. Cytoplasmic annulate lamellae in cultured cells: Composition, distribution, and mitotic behavior. Cell Tissue Res. 1996, 284, 177–191. [Google Scholar] [CrossRef] [PubMed]
- Marshall, J.A.; Borg, J.; Coulepis, A.G.; Anderson, D.A. Annulate lamellae and lytic HAV infection in vitro. Tissue Cell 1996, 28, 205–214. [Google Scholar] [CrossRef]
- Wang, J.J.; Liao, C.L.; Chiou, Y.W.; Chiou, C.T.; Huang, Y.L.; Chen, L.K. Ultrastructure and localization of E proteins in cultured neuron cells infected with Japanese encephalitis virus. Virology 1997, 238, 30–39. [Google Scholar] [CrossRef][Green Version]
- Kim, K.S.; Boatman, E.S. Electron microscopy of monkey kidney cell cultures infected with rubella virus. J. Virol. 1967, 1, 205–214. [Google Scholar] [PubMed]
- Gustin, K.E.; Sarnow, P. Inhibition of nuclear import and alteration of nuclear pore complex composition by rhinovirus. J. Virol. 2002, 76, 8787–8796. [Google Scholar] [CrossRef]
- Gustin, K.E.; Sarnow, P. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J. 2001, 20, 240–249. [Google Scholar] [CrossRef]
- Context Affects Nuclear Protein Localization in Saccharomyces cerevisiae. Available online: http://mcb.asm.org/content/9/2/384.full.pdf (accessed on 2 March 2016).
- Watters, K.; Inankur, B.; Gardiner, J.C.; Warrick, J.; Sherer, N.M.; Yin, J.; Palmenberg, A.C. Differential Disruption of Nucleocytoplasmic Trafficking Pathways by Rhinovirus 2A Proteases. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Park, N.; Skern, T.; Gustin, K.E. Specific cleavage of the nuclear pore complex protein Nup62 by a viral protease. J. Biol. Chem. 2010, 285, 28796–28805. [Google Scholar] [CrossRef]
- Flather, D.; Nguyen, J.H.C.; Semler, B.L.; Gershon, P.D. Exploitation of nuclear functions by human rhinovirus, a cytoplasmic RNA virus. PLoS Pathog. 2018, 14, e1007277. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-W.; Lee, C.-P.; Su, M.-T.; Tsai, C.-H.; Chen, M.-R. BGLF4 kinase modulates the structure and transport preference of the nuclear pore complex to facilitate nuclear import of Epstein-Barr virus lytic proteins. J. Virol. 2015, 89, 1703–1718. [Google Scholar] [CrossRef] [PubMed]
- Muhlbauer, D.; Dzieciolowski, J.; Hardt, M.; Hocke, A.; Schierhorn, K.L.; Mostafa, A.; Muller, C.; Wisskirchen, C.; Herold, S.; Wolff, T.; et al. Influenza virus-induced caspase-dependent enlargement of nuclear pores promotes nuclear export of viral ribonucleoprotein complexes. J. Virol. 2015, 89, 6009–6021. [Google Scholar] [CrossRef] [PubMed]
- Atasheva, S.; Fish, A.; Fornerod, M.; Frolova, E.I. Venezuelan equine Encephalitis virus capsid protein forms a tetrameric complex with CRM1 and importin alpha/beta that obstructs nuclear pore complex function. J. Virol. 2010, 84, 4158–4171. [Google Scholar] [CrossRef] [PubMed]
- Atasheva, S.; Garmashova, N.; Frolov, I.; Frolova, E. Venezuelan equine encephalitis virus capsid protein inhibits nuclear import in Mammalian but not in mosquito cells. J. Virol. 2008, 82, 4028–4041. [Google Scholar] [CrossRef] [PubMed]
- King, C.R.; Cohen, M.J.; Fonseca, G.J.; Dirk, B.S.; Dikeakos, J.D.; Mymryk, J.S. Functional and Structural Mimicry of Cellular Protein Kinase A Anchoring Proteins by a Viral Oncoprotein. PLoS Pathog. 2016, 12, e1005621. [Google Scholar] [CrossRef] [PubMed]
- Fax, P.; Carlson, C.R.; Collas, P.; Tasken, K.; Esche, H.; Brockmann, D. Binding of PKA-RIIalpha to the Adenovirus E1A12S oncoprotein correlates with its nuclear translocation and an increase in PKA-dependent promoter activity. Virology 2001, 285, 30–41. [Google Scholar] [CrossRef]
- King, C.R.; Gameiro, S.F.; Tessier, T.M.; Zhang, A.; Mymryk, J.S. Mimicry of Cellular A Kinase-Anchoring Proteins Is a Conserved and Critical Function of E1A across Various Human Adenovirus Species. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef]
- Lange, A.; Mills, R.E.; Lange, C.J.; Stewart, M.; Devine, S.E.; Corbett, A.H. Classical Nuclear Localization Signals: Definition, Function, and Interaction with Importin. J. Biol. Chem. 2007, 282, 5101–5105. [Google Scholar] [CrossRef]
- Kosugi, S.; Hasebe, M.; Matsumura, N.; Takashima, H.; Miyamoto-Sato, E.; Tomita, M.; Yanagawa, H. Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J. Biol. Chem. 2009, 284, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, S.; Hasebe, M.; Entani, T.; Takayama, S.; Tomita, M.; Yanagawa, H. Design of peptide inhibitors for the importin alpha/beta nuclear import pathway by activity-based profiling. Chem. Biol. 2008, 15, 940–949. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, S.; Hasebe, M.; Tomita, M.; Yanagawa, H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA 2009, 106, 10171–10176. [Google Scholar] [CrossRef] [PubMed]
- Nguyen Ba, A.N.; Pogoutse, A.; Provart, N.; Moses, A.M. NLStradamus: A simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics 2009, 10, 202. [Google Scholar] [CrossRef] [PubMed]
- Nakada, R.; Hirano, H.; Matsuura, Y. Structure of importin-alpha bound to a non-classical nuclear localization signal of the influenza A virus nucleoprotein. Sci. Rep. 2015, 5, 15055. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Palese, P.; O’Neill, R.E. The NPI-1/NPI-3 (karyopherin alpha) binding site on the influenza a virus nucleoprotein NP is a nonconventional nuclear localization signal. J. Virol. 1997, 71, 1850–1856. [Google Scholar]
- Lott, K.; Bhardwaj, A.; Sims, P.J.; Cingolani, G. A minimal nuclear localization signal (NLS) in human phospholipid scramblase 4 that binds only the minor NLS-binding site of importin alpha1. J. Biol. Chem. 2011, 286, 28160–28169. [Google Scholar] [CrossRef]
- Giesecke, A.; Stewart, M. Novel binding of the mitotic regulator TPX2 (target protein for Xenopus kinesin-like protein 2) to importin-alpha. J. Biol. Chem. 2010, 285, 17628–17635. [Google Scholar] [CrossRef]
- Pang, X.; Zhou, H.-X. Design rules for selective binding of nuclear localization signals to minor site of importin alpha. PLoS ONE 2014, 9, e91025. [Google Scholar] [CrossRef]
- Wolff, T.; Unterstab, G.; Heins, G.; Richt, J.A.; Kann, M. Characterization of an unusual importin alpha binding motif in the borna disease virus p10 protein that directs nuclear import. J. Biol. Chem. 2002, 277, 12151–12157. [Google Scholar] [CrossRef]
- Wolff, T.; Pfleger, R.; Wehner, T.; Reinhardt, J.; Richt, J.A. A short leucine-rich sequence in the Borna disease virus p10 protein mediates association with the viral phospho- and nucleoproteins. J. Gen. Virol. 2000, 81, 939–947. [Google Scholar] [CrossRef] [PubMed]
- Kenna, M.A.; Brachmann, C.B.; Devine, S.E.; Boeke, J.D. Invading the yeast nucleus: A nuclear localization signal at the C terminus of Ty1 integrase is required for transposition in vivo. Mol. Cell. Biol. 1998, 18, 1115–1124. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.P.; Rinckel, L.A.; Garfinkel, D.J. A Ty1 integrase nuclear localization signal required for retrotransposition. Mol. Cell. Biol. 1998, 18, 1105–1114. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lange, A.; McLane, L.M.; Mills, R.E.; Devine, S.E.; Corbett, A.H. Expanding the definition of the classical bipartite nuclear localization signal. Traffic 2010, 11, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Lyons, R.H.; Ferguson, B.Q.; Rosenberg, M. Pentapeptide nuclear localization signal in adenovirus E1a. Mol. Cell. Biol. 1987, 7, 2451–2456. [Google Scholar] [CrossRef]
- Cohen, M.J.; Yousef, A.F.; Massimi, P.; Fonseca, G.J.; Todorovic, B.; Pelka, P.; Turnell, A.S.; Banks, L.; Mymryk, J.S. Dissection of the C-terminal region of E1A redefines the roles of CtBP and other cellular targets in oncogenic transformation. J. Virol. 2013, 87, 10348–10355. [Google Scholar] [CrossRef] [PubMed]
- Atencio, D.; Barnes, C.; Duncan, T.M.; Willis, I.M.; Hanes, S.D. The yeast Ess1 prolyl isomerase controls Swi6 and Whi5 nuclear localization. G3 (Bethesda). 2014, 4, 523–537. [Google Scholar] [CrossRef] [PubMed]
- Biegalke, B.J. Nontraditional localization and retention signals localize human cytomegalovirus pUL34 to the nucleus. J. Virol. 2013, 87, 11939–11944. [Google Scholar] [CrossRef]
- Marshall, K.S.; Cohen, M.J.; Fonseca, G.J.; Todorovic, B.; King, C.R.; Yousef, A.F.; Zhang, Z.; Mymryk, J.S. Identification and characterization of multiple conserved nuclear localization signals within adenovirus E1A. Virology 2014, 454–455, 206–214. [Google Scholar] [CrossRef]
- De la Luna, S.; Burden, M.J.; Lee, C.W.; La Thangue, N.B. Nuclear accumulation of the E2F heterodimer regulated by subunit composition and alternative splicing of a nuclear localization signal. J. Cell Sci. 1996, 109 Pt 1, 2443–2452. [Google Scholar]
- Steidl, S.; Tuncher, A.; Goda, H.; Guder, C.; Papadopoulou, N.; Kobayashi, T.; Tsukagoshi, N.; Kato, M.; Brakhage, A.A. A single subunit of a heterotrimeric CCAAT-binding complex carries a nuclear localization signal: Piggy back transport of the pre-assembled complex to the nucleus. J. Mol. Biol. 2004, 342, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Prufer, K.; Racz, A.; Lin, G.C.; Barsony, J. Dimerization with retinoid X receptors promotes nuclear localization and subnuclear targeting of vitamin D receptors. J. Biol. Chem. 2000, 275, 41114–41123. [Google Scholar] [CrossRef] [PubMed]
- Asally, M.; Yoneda, Y. Beta-catenin can act as a nuclear import receptor for its partner transcription factor, lymphocyte enhancer factor-1 (lef-1). Exp. Cell Res. 2005, 308, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, K.M.; Jans, D.A. Importins and beyond: Non-conventional nuclear transport mechanisms. Traffic 2009, 10, 1188–1198. [Google Scholar] [CrossRef] [PubMed]
- Kosyna, F.K.; Depping, R. Controlling the Gatekeeper: Therapeutic Targeting of Nuclear Transport. Cells 2018, 7, 221. [Google Scholar] [CrossRef] [PubMed]
- Laing, R.; Gillan, V.; Devaney, E. Ivermectin - Old Drug, New Tricks? Trends Parasitol. 2017, 33, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Juarez, M.; Schcolnik-Cabrera, A.; Duenas-Gonzalez, A. The multitargeted drug ivermectin: From an antiparasitic agent to a repositioned cancer drug. Am. J. Cancer Res. 2018, 8, 317–331. [Google Scholar] [PubMed]
- Jin, L.; Wang, R.; Zhu, Y.; Zheng, W.; Han, Y.; Guo, F.; Ye, F.B.; Li, Y. Selective targeting of nuclear receptor FXR by avermectin analogues with therapeutic effects on nonalcoholic fatty liver disease. Sci. Rep. 2015, 5, 17288. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Gomez, G.; Chavez-Blanco, A.; Medina-Franco, J.L.; Saldivar-Gonzalez, F.; Flores-Torrontegui, Y.; Juarez, M.; Diaz-Chavez, J.; Gonzalez-Fierro, A.; Duenas-Gonzalez, A. Ivermectin as an inhibitor of cancer stemlike cells. Mol. Med. Rep. 2018, 17, 3397–3403. [Google Scholar] [CrossRef]
- Chaccour, C.; Rabinovich, N.R. Advancing the repurposing of ivermectin for malaria. Lancet 2019. [Google Scholar] [CrossRef]
- Foy, B.D.; Alout, H.; Seaman, J.A.; Rao, S.; Magalhaes, T.; Wade, M.; Parikh, S.; Soma, D.D.; Sagna, A.B.; Fournet, F.; et al. Efficacy and risk of harms of repeat ivermectin mass drug administrations for control of malaria (RIMDAMAL): A cluster-randomised trial. Lancet 2019. [Google Scholar] [CrossRef]
- Wagstaff, K.M.; Rawlinson, S.M.; Hearps, A.C.; Jans, D.A. An AlphaScreen(R)-based assay for high-throughput screening for specific inhibitors of nuclear import. J. Biomol. Screen. 2011, 16, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Tay, M.Y.F.; Fraser, J.E.; Chan, W.K.K.; Moreland, N.J.; Rathore, A.P.; Wang, C.; Vasudevan, S.G.; Jans, D.A. Nuclear localization of dengue virus (DENV) 1-4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin. Antiviral Res. 2013, 99, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Varghese, F.S.; Kaukinen, P.; Glasker, S.; Bespalov, M.; Hanski, L.; Wennerberg, K.; Kummerer, B.M.; Ahola, T. Discovery of berberine, abamectin and ivermectin as antivirals against chikungunya and other alphaviruses. Antiviral Res. 2016, 126, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, K.M.; Sivakumaran, H.; Heaton, S.M.; Harrich, D.; Jans, D.A. Ivermectin is a specific inhibitor of importin alpha/beta-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 2012, 443, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Guzman, M.G.; Harris, E. Dengue. Lancet 2015, 385, 453–465. [Google Scholar] [CrossRef]
- Pang, T.; Mak, T.K.; Gubler, D.J. Prevention and control of dengue-the light at the end of the tunnel. Lancet. Infect. Dis. 2017, 17, e79–e87. [Google Scholar] [CrossRef]
- World Health Organization. Global Strategy for Dengue Prevention and Control, 2012–2020; WHO: Geneva, Switzerland, 2012; ISBN 978-92-4-150403-4. [Google Scholar]
- Xu, T.-L.; Han, Y.; Liu, W.; Pang, X.-Y.; Zheng, B.; Zhang, Y.; Zhou, X.-N. Antivirus effectiveness of ivermectin on dengue virus type 2 in Aedes albopictus. PLoS Negl. Trop. Dis. 2018, 12, e0006934. [Google Scholar] [CrossRef]
- Barrows, N.J.; Campos, R.K.; Powell, S.T.; Prasanth, K.R.; Schott-Lerner, G.; Soto-Acosta, R.; Galarza-Munoz, G.; McGrath, E.L.; Urrabaz-Garza, R.; Gao, J.; et al. A Screen of FDA-Approved Drugs for Inhibitors of Zika Virus Infection. Cell Host Microbe 2016, 20, 259–270. [Google Scholar] [CrossRef]
- Lundberg, L.; Pinkham, C.; Baer, A.; Amaya, M.; Narayanan, A.; Wagstaff, K.M.; Jans, D.A.; Kehn-Hall, K. Nuclear import and export inhibitors alter capsid protein distribution in mammalian cells and reduce Venezuelan Equine Encephalitis Virus replication. Antiviral Res. 2013, 100, 662–672. [Google Scholar] [CrossRef]
- Marrugal-Lorenzo, J.A.; Serna-Gallego, A.; Gonzalez-Gonzalez, L.; Bunuales, M.; Poutou, J.; Pachon, J.; Gonzalez-Aparicio, M.; Hernandez-Alcoceba, R.; Sanchez-Cespedes, J. Inhibition of adenovirus infection by mifepristone. Antiviral Res. 2018, 159, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.R.; Lundberg, L.; Pinkham, C.; Shechter, S.; DeBono, A.; Baell, J.; Wagstaff, K.M.; Hick, C.A.; Kehn-Hall, K.; Jans, D.A. Identification of novel antivirals inhibiting recognition of Venezuelan equine encephalitis virus capsid protein by the Importin alpha/beta1 heterodimer through high-throughput screening. Antiviral Res. 2018, 151, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Wolff, B.; Sanglier, J.J.; Wang, Y. Leptomycin B is an inhibitor of nuclear export: Inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 1997, 4, 139–147. [Google Scholar] [CrossRef]
- Newlands, E.S.; Rustin, G.J.; Brampton, M.H. Phase I trial of elactocin. Br. J. Cancer 1996, 74, 648–649. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, M.; Asano, S.; Nakamura, T.; Adachi, M.; Yoshida, M.; Yanagida, M.; Nishida, E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 1997, 390, 308–311. [Google Scholar] [CrossRef]
- Mathew, C.; Ghildyal, R. CRM1 Inhibitors for Antiviral Therapy. Front. Microbiol. 2017, 8, 1171. [Google Scholar] [CrossRef]
- Parikh, K.; Cang, S.; Sekhri, A.; Liu, D. Selective inhibitors of nuclear export (SINE)—A novel class of anti-cancer agents. J. Hematol. Oncol. 2014, 7, 78. [Google Scholar] [CrossRef]
- Pickens, J.A.; Tripp, R.A. Verdinexor Targeting of CRM1 is a Promising Therapeutic Approach against RSV and Influenza Viruses. Viruses 2018, 10, 48. [Google Scholar] [CrossRef]
- Jorquera, P.A.; Mathew, C.; Pickens, J.; Williams, C.; Luczo, J.M.; Tamir, S.; Ghildyal, R.; Tripp, R.A. Verdinexor (KPT-335), a Selective Inhibitor of Nuclear Export, Reduces Respiratory Syncytial Virus Replication In Vitro. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Lundberg, L.; Pinkham, C.; de la Fuente, C.; Brahms, A.; Shafagati, N.; Wagstaff, K.M.; Jans, D.A.; Tamir, S.; Kehn-Hall, K. Selective Inhibitor of Nuclear Export (SINE) Compounds Alter New World Alphavirus Capsid Localization and Reduce Viral Replication in Mammalian Cells. PLoS Negl. Trop. Dis. 2016, 10, e0005122. [Google Scholar] [CrossRef]
- Perwitasari, O.; Johnson, S.; Yan, X.; Howerth, E.; Shacham, S.; Landesman, Y.; Baloglu, E.; McCauley, D.; Tamir, S.; Tompkins, S.M.; et al. Verdinexor, a novel selective inhibitor of nuclear export, reduces influenza a virus replication in vitro and in vivo. J. Virol. 2014, 88, 10228–10243. [Google Scholar] [CrossRef] [PubMed]
- Widman, D.G.; Gornisiewicz, S.; Shacham, S.; Tamir, S. In vitro toxicity and efficacy of verdinexor, an exportin 1 inhibitor, on opportunistic viruses affecting immunocompromised individuals. PLoS ONE 2018, 13, e0200043. [Google Scholar] [CrossRef] [PubMed]
- Perwitasari, O.; Johnson, S.; Yan, X.; Register, E.; Crabtree, J.; Gabbard, J.; Howerth, E.; Shacham, S.; Carlson, R.; Tamir, S.; et al. Antiviral Efficacy of Verdinexor In Vivo in Two Animal Models of Influenza A Virus Infection. PLoS ONE 2016, 11, e0167221. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tessier, T.M.; Dodge, M.J.; Prusinkiewicz, M.A.; Mymryk, J.S. Viral Appropriation: Laying Claim to Host Nuclear Transport Machinery. Cells 2019, 8, 559. https://doi.org/10.3390/cells8060559
Tessier TM, Dodge MJ, Prusinkiewicz MA, Mymryk JS. Viral Appropriation: Laying Claim to Host Nuclear Transport Machinery. Cells. 2019; 8(6):559. https://doi.org/10.3390/cells8060559
Chicago/Turabian StyleTessier, Tanner M., Mackenzie J. Dodge, Martin A. Prusinkiewicz, and Joe S. Mymryk. 2019. "Viral Appropriation: Laying Claim to Host Nuclear Transport Machinery" Cells 8, no. 6: 559. https://doi.org/10.3390/cells8060559
APA StyleTessier, T. M., Dodge, M. J., Prusinkiewicz, M. A., & Mymryk, J. S. (2019). Viral Appropriation: Laying Claim to Host Nuclear Transport Machinery. Cells, 8(6), 559. https://doi.org/10.3390/cells8060559






