A Critical E-box in Barhl1 3′ Enhancer Is Essential for Auditory Hair Cell Differentiation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmid Construction and Site-Directed Mutagenesis
2.2. Reporter Assay
2.3. mESC Cell Culture and Differentiation
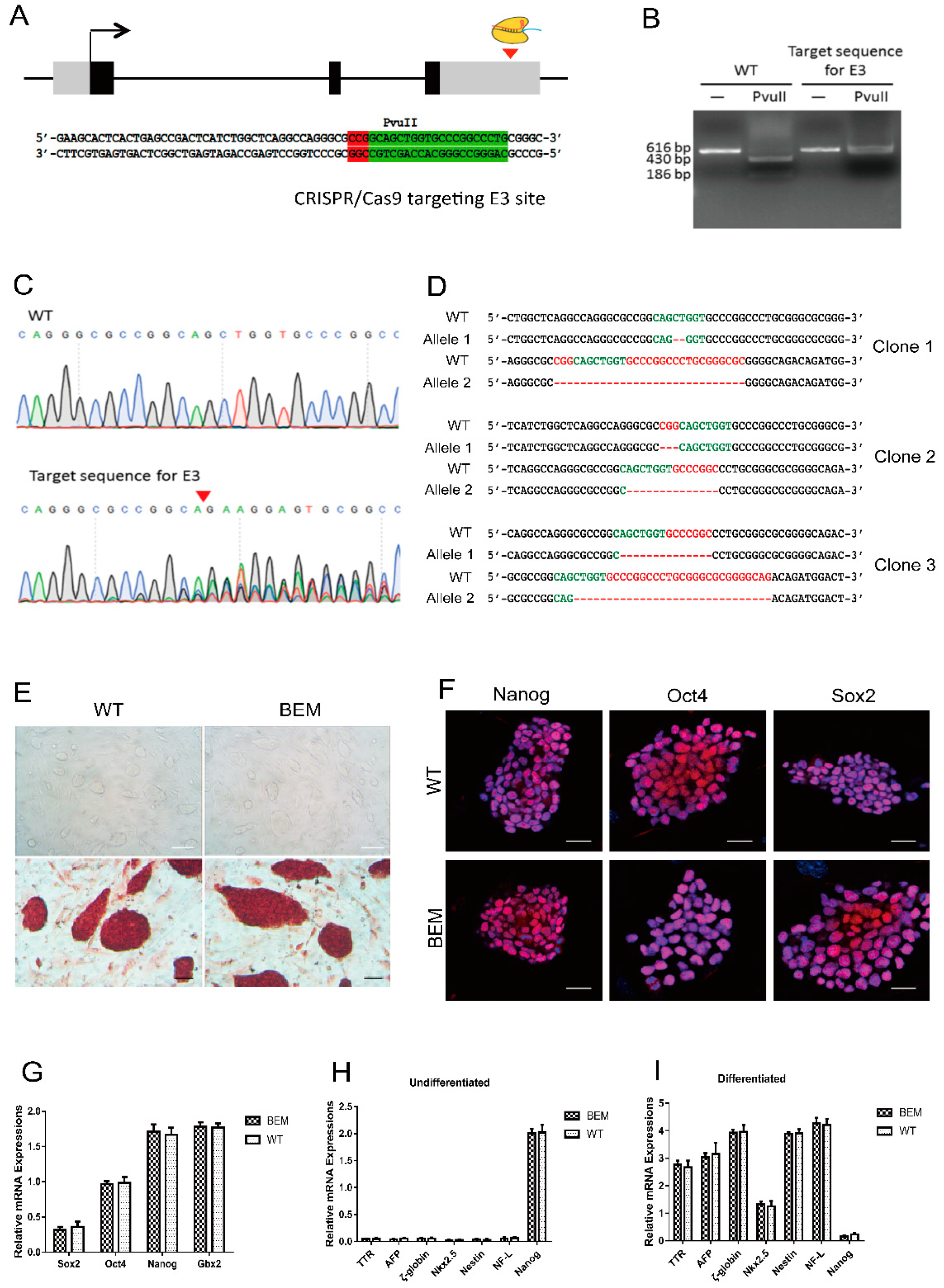
2.4. Generation of BEM mESC Lines
2.5. RNA Extraction and Real-Time Quantitative PCR (qPCR)
2.6. Western Blot
2.7. Alkaline Phosphatase (AP) and Immunofluorescence Staining
2.8. FM1-43FX Uptake Studies
2.9. RNA-Sequencing (RNA-Seq) and Data Analysis
2.10. Statistical Analysis
3. Results
3.1. Barhl1 Is a Direct Downstream Target of Atoh1 in the Inner Ear Hair Cells
3.2. Atoh1 Binds to an Essential E-box in Barhl1 3’-Flanking Enhancer
3.3. Establishment of BEM mESC Line Carrying the E3 Motif Mutant
3.4. Disruption of the Barhl1 E3 Site Does Not Affect the Pluripotency of mESCs
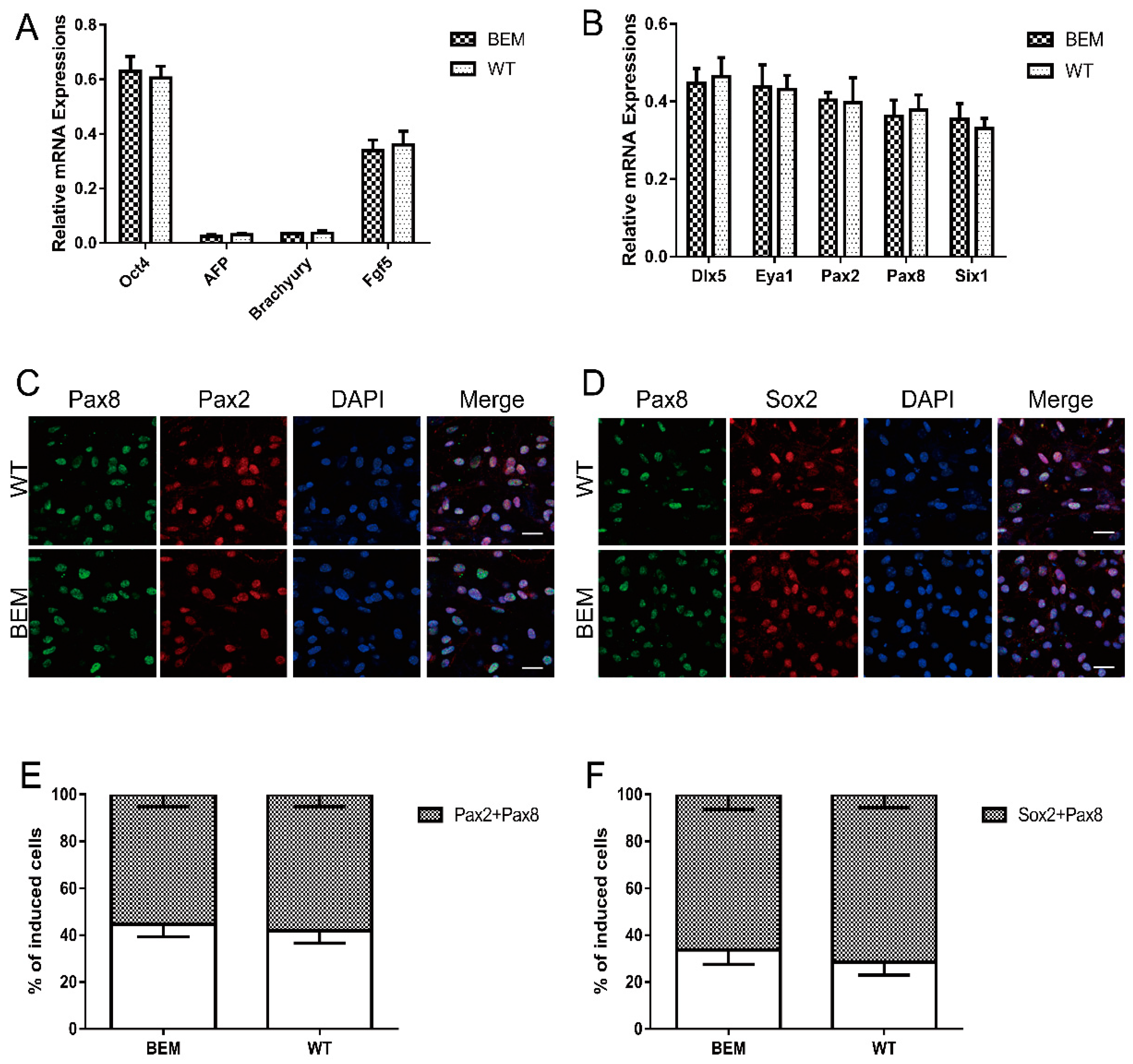
3.5. Disruption of the Barhl1 E3 Site Does Not Affect EPL Cell Induction and Otic Progenitor Differentiation
3.6. Disruption of the Barhl1 E3 Site Affects Auditory Hair Cell Differentiation
3.7. The E3 Box Has Similar Effects on Hair Cell Differentiation as Barhl1 Protein
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lentz, J.J.; Jodelka, F.M.; Hinrich, A.J.; McCaffrey, K.E.; Farris, H.E.; Spalitta, M.J.; Bazan, N.G.; Duelli, D.M.; Rigo, F.; Hastings, M.L. Rescue of hearing and vestibular function by antisense oligonucleotides in a mouse model of human deafness. Nat. Med. 2013, 19, 345–350. [Google Scholar] [CrossRef] [Green Version]
- Bramhall, N.F.; Shi, F.; Arnold, K.; Hochedlinger, K.; Edge, A.S. Lgr5-positive supporting cells generate new hair cells in the postnatal cochlea. Stem Cell Rep. 2014, 2, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Cox, B.C.; Chai, R.; Lenoir, A.; Liu, Z.; Zhang, L.; Nguyen, D.H.; Chalasani, K.; Steigelman, K.A.; Fang, J.; Rubel, E.W.; et al. Spontaneous hair cell regeneration in the neonatal mouse cochlea in vivo. Development 2014, 141, 816–829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bermingham, N.A.; Hassan, B.A.; Price, S.D.; Vollrath, M.A.; Ben-Arie, N.; Eatock, R.A.; Bellen, H.J.; Lysakowski, A.; Zoghbi, H.Y. Math1: An essential gene for the generation of inner ear hair cells. Science 1999, 284, 1837–1841. [Google Scholar] [PubMed]
- Cai, T.; Jen, H.I.; Kang, H.; Klisch, T.J.; Zoghbi, H.Y.; Groves, A.K. Conditional deletion of Atoh1 reveals distinct critical periods for survival and function of hair cells in the organ of Corti. J. Neurosci. 2013, 33, 10110–10122. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Johnson, J.E.; Zoghbi, H.Y.; Segil, N. The role of Math1 in inner ear development: Uncoupling the establishment of the sensory primordium from hair cell fate determination. Development 2002, 29, 2495–2505. [Google Scholar]
- Woods, C.; Montcouquiol, M.; Kelley, M.W. Math1 regulates development of the sensory epithelium in the mammalian cochlea. Nat. Neurosci. 2004, 7, 1310–1318. [Google Scholar] [CrossRef] [PubMed]
- Gubbels, S.P.; Woessner, D.W.; Mitchell, J.C.; Ricci, A.J.; Brigande, J.V. Functional auditory hair cells produced in the mammalian cochlea by in utero gene transfer. Nature 2008, 455, 537–541. [Google Scholar] [CrossRef] [Green Version]
- Kelly, M.C.; Chang, Q.; Pan, A.; Lin, X.; Chen, P. Atoh1 directs the formation of sensory mosaics and induces cell proliferation in the postnatal mammalian cochlea in vivo. J. Neurosci. 2012, 32, 6699–6710. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.L.; Gao, W.Q. Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat. Neurosci 2000, 3, 580–586. [Google Scholar] [CrossRef] [PubMed]
- Izumikawa, M.; Minoda, R.; Kawamoto, K.; Abrashkin, K.A.; Swiderski, D.L.; Dolan, D.F.; Brough, D.E.; Raphael, Y. Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nat. Med. 2005, 11, 271–276. [Google Scholar] [CrossRef]
- Kawamoto, K.; Ishimoto, S.; Minoda, R.; Brough, D.E.; Raphael, Y. Math1 gene transfer generates new cochlear hair cells in mature guinea pigs in vivo. J. Neurosci. 2003, 23, 4395–4400. [Google Scholar] [CrossRef]
- Richardson, R.T.; Atkinson, P.J. Atoh1 gene therapy in the cochlea for hair cell regeneration. Expert. Opin. Biol. Ther. 2015, 15, 417–430. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Xie, X.; Deng, M.; Chen, X.; Gan, L. Generation and characterization of Atoh1-Cre knock-in mouse line. Genesis 2010, 480, 407–413. [Google Scholar] [CrossRef]
- Pan, N.; Jahan, I.; Kersigo, J.; Duncan, J.S.; Kopecky, B.; Fritzsch, B. A novel Atoh1 “self-terminating” mouse model reveals the necessity of proper Atoh1 level and duration for hair cell differentiation and viability. PLoS ONE 2012, 7, e30358. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Dearman, J.A.; Cox, B.C.; Walters, B.J.; Zhang, L.; Ayrault, O.; Zindy, F.; Gan, L.; Roussel, M.F.; Zuo, J. Age-dependent in vivo conversion of mouse cochlear pillar and Deiters’ cells to immature hair cells by Atoh1 ectopic expression. J. Neurosci. 2012, 32, 6600–6610. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.F.; Tong, M.; Edge, A.S. Destabilization of Atoh1 by E3 Ubiquitin Ligase Huwe1 and Casein Kinase 1 Is Essential for Normal Sensory Hair Cell Development. J. Biol. Chem. 2016, 291, 21096–21109. [Google Scholar]
- Shi, F.; Cheng, Y.F.; Wang, X.L.; Edge, A.S. Beta-catenin up-regulates Atoh1 expression in neural progenitor cells by interaction with an Atoh1 3’ enhancer. J. Biol. Chem. 2010, 285, 392–400. [Google Scholar] [CrossRef]
- Jansson, L.; Kim, G.S.; Cheng, A.G. Making sense of Wnt signaling-linking hair cell regeneration to development. Front. Cell. Neurosci. 2015, 9. [Google Scholar] [CrossRef]
- Zhao, X.; Heng, J.I.; Guardavaccaro, D.; Jiang, R.; Pagano, M.; Guillemot, F.; Iavarone, A.; Lasorella, A. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat. Cell Biol. 2008, 10, 643–653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; D’Arca, D.; Lim, W.K.; Brahmachary, M.; Carro, M.S.; Ludwig, T.; Cardo, C.C.; Guillemot, F.; Aldape, K.; Califano, A.; et al. The N-Myc-DLL3 cascade is suppressed by the ubiquitin ligase Huwe1 to inhibit proliferation and promote neurogenesis in the developing brain. Dev. Cell 2009, 17, 210–221. [Google Scholar] [Green Version]
- Forget, A.; Bihannic, L.; Cigna, S.M.; Lefevre, C.; Remke, M.; Barnat, M.; Dodier, S.; Shirvani, H.; Mercier, A.; Mensah, A.; et al. Shh signaling protects Atoh1 from degradation mediated by the E3 ubiquitin ligase Huwe1 in neural precursors. Dev. Cell 2014, 29, 649–661. [Google Scholar] [CrossRef] [PubMed]
- Klisch, T.J.; Xi, Y.; Flora, A.; Wang, L.; Li, W.; Zoghbi, H.Y. In vivo Atoh1 targetome reveals how a proneural transcription factor regulates cerebellar development. Proc. Natl. Acad. Sci. USA 2011, 108, 3288–3293. [Google Scholar] [CrossRef]
- Bulfone, A.; Menguzzato, E.; Broccoli, V.; Marchitiello, A.; Gattuso, C.; Mariani, M.; Consalez, G.G.; Martinez, S.; Ballabio, A.; Banfi, S. Barhl1, a gene belonging to a new subfamily of mammalian homeobox genes, is expressed in migrating neurons of the CNS. Hum. Mol. Genet. 2000, 9, 1443–1452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Price, S.M.; Cahill, H.; Ryugo, D.K.; Shen, M.M.; Xiang, M. Hearing loss caused by progressive degeneration of cochlear hair cells in mice deficient for the Barhl1 homeobox gene. Development 2002, 129, 3523–3532. [Google Scholar]
- Zhong, C.; Chen, Z.; Luo, X.; Wang, C.; Jiang, H.; Shao, J.; Guan, M.; Huang, L.; Huang, X.; Wang, J. Barhl1 is required for the differentiation of inner ear hair cell-like cells from mouse embryonic stem cells. Int. J. Biochem. Cell. Biol. 2018, 96, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Chellappa, R.; Li, S.; Pauley, S.; Jahan, I.; Jin, K.; Xiang, M. Barhl1 regulatory sequences required for cell-specific gene expression and autoregulation in the inner ear and central nervous system. Mol. Cell Biol. 2008, 28, 1905–1914. [Google Scholar] [CrossRef] [PubMed]
- Chonko, K.T.; Jahan, I.; Stone, J.; Wright, M.C.; Fujiyama, T.; Hoshino, M.; Fritzsch, B.; Maricich, S.M. Atoh1 directs hair cell differentiation and survival in the late embryonic mouse inner ear. Dev. Biol. 2013, 381, 401–410. [Google Scholar] [CrossRef]
- Scheffer, D.I.; Shen, J.; Corey, D.P.; Chen, Z.Y. Gene Expression by Mouse Inner Ear Hair Cells during Development. J. Neurosci. 2015, 35, 6366–6380. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Cai, T.; Jen, H.I.; Kang, H.; Klisch, T.J.; Zoghbi, H.Y.; Groves, A.K. Characterization of the transcriptome of nascent hair cells and identification of direct targets of the Atoh1 transcription factor. J. Neurosci. 2015, 35, 5870–5883. [Google Scholar] [CrossRef] [PubMed]
- Helms, A.W.; Abney, A.L.; Ben-Arie, N.; Zoghbi, H.Y.; Johnson, J.E. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development 2000, 127, 1185–1196. [Google Scholar] [PubMed]
- Nakano, Y.; Jahan, I.; Bonde, G.; Sun, X.; Hildebrand, M.S.; Engelhardt, J.F.; Smith, R.J.; Cornell, R.A.; Fritzsch, B.; Banfi, B. A mutation in the Srrm4 gene causes alternative splicing defects and deafness in the Bronx waltzer mouse. PLoS Genet. 2012, 8, e1002966. [Google Scholar] [CrossRef]
- Oshima, K.; Shin, K.; Diensthuber, M.; Peng, A.W.; Ricci, A.J.; Heller, S. Mechanosensitive hair cell-like cells from embryonic and induced pluripotent stem cells. Cell 2010, 141, 704–716. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.; Wong, E.Y.; Sun, J.; Xu, J.; Wang, F.; Xu, P.X. Eya1-Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2. Dev. Cell 2012, 22, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Roblin, G.; Liu, H.; Heller, S. Generation of hair cells by stepwise differentiation of embryonic stem cells. Proc. Natl. Acad. Sci. USA 2003, 100, 13495–13500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouji, Y.; Ishizaka, S.; Nakamura-Uchiyama, F.; Wanaka, A.; Yoshikawa, M. Induction of inner ear hair cell-like cells from Math1-transfected mouse ES cells. Cell. Death Dis. 2013, 4, e700. [Google Scholar] [CrossRef]
- Mulvaney, J.; Dabdoub, A. Atoh1, an essential transcription factor in neurogenesis and intestinal and inner ear development: Function, regulation, and context dependency. J. Assoc. Res. Otolaryngol. 2012, 13, 281–293. [Google Scholar] [CrossRef]
- Lai, H.C.; Klisch, T.J.; Roberts, R.; Zoghbi, H.Y.; Johnson, J.E. In vivo neuronal subtype-specific targets of Atoh1 (Math1) in dorsal spinal cord. J. Neurosci. 2011, 31, 10859–10871. [Google Scholar] [CrossRef] [PubMed]
- Flora, A.; Garcia, J.J.; Thaller, C.; Zoghbi, H.Y. The E-protein Tcf4 interacts with Math1 to regulate differentiation of a specific subset of neuronal progenitors. Proc. Natl. Acad. Sci. USA 2007, 104, 15382–15387. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Bermingham, N.A.; Finegold, M.J.; Zoghbi, H.Y. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science 2001, 294, 2155–2158. [Google Scholar] [CrossRef]
- Van Dussen, K.L.; Samuelson, L.C. Mouse atonal homolog 1 directs intestinal progenitors to secretory cell rather than absorptive cell fate. Dev. Biol. 2010, 346, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.H.; Tupal, S.; Huang, T.W.; Ward, C.S.; Neul, J.L.; Klisch, T.J.; Gray, P.A.; Zoghbi, H.Y. Atoh1 governs the migration of postmitotic neurons that shape respiratory effectiveness at birth and chemoresponsiveness in adulthood. Neuron 2012, 75, 799–809. [Google Scholar] [CrossRef] [PubMed]
- Miesegaes, G.R.; Klisch, T.J.; Thaller, C.; Ahmad, K.A.; Atkinson, R.C.; Zoghbi, H.Y. Identification and subclassification of new Atoh1 derived cell populations during mouse spinal cord development. Dev. Biol. 2009, 327, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Ruffault, P.L.; D’Autreaux, F.; Hayes, J.A.; Nomaksteinsky, M.; Autran, S.; Fujiyama, T.; Hoshino, M.; Hagglund, M.; Kiehn, O.; Brunet, J.F.; et al. The retrotrapezoid nucleus neurons expressing Atoh1 and Phox2b are essential for the respiratory response to CO(2). eLife 2015, 4, e07051. [Google Scholar] [CrossRef]
- Wang, V.Y.; Rose, M.F.; Zoghbi, H.Y. Math1 expression redefines the rhombic lip derivatives and reveals novel lineages within the brainstem and cerebellum. Neuron 2005, 48, 31–43. [Google Scholar] [CrossRef] [PubMed]
- Visel, A.; Prabhakar, S.; Akiyama, J.A.; Shoukry, M.; Lewis, K.D.; Holt, A.; Plajzer-Frick, I.; Afzal, V.; Rubin, E.M.; Pennacchio, L.A. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat. Genet. 2008, 40, 158–160. [Google Scholar] [CrossRef] [Green Version]
- Kawauchi, D.; Saito, T. Tra nscriptional cascade from Math1 to Mbh1 and Mbh2 is required for cerebellar granule cell differentiation. Dev. Biol. 2008, 322, 345–354. [Google Scholar] [CrossRef]
- Saba, R.; Johnson, J.E.; Saito, T. Commissural neuron identity is specified by a homeodomain protein, Mbh1, that is directly downstream of Math1. Development 2005, 132, 2147–2155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atchley, W.R.; Fitch, W.M. A natural classification of the basic helix-loop-helix class of transcription factors. Proc. Natl. Acad. Sci. USA 1997, 94, 5172–51726. [Google Scholar] [CrossRef]
- Rathjen, J.; Lake, J.A.; Bettess, M.D.; Washington, J.M.; Chapman, G.; Rathjen, P.D. Formation of a primitive ectoderm like cell population, EPL cells, from ES cells in response to biologically derived factors. J. Cell Sci. 1999, 112, 601–612. [Google Scholar] [PubMed]
- Sher, A.E. The embryonic and postnatal development of the inner ear of the mouse. Acta Otolaryngol. Suppl. 1971, 285, 1–77. [Google Scholar] [PubMed]
- Kelley, M.W. Cell adhesion molecules during inner ear and hair cell development, including notch and its ligands. Curr. Top. Dev. Biol. 2003, 57, 321–356. [Google Scholar] [PubMed]
- Kelley, M.W. Regulation of cell fate in the sensory epithelia of the inner ear. Nat. Rev. Neurosci. 2006, 7, 837–849. [Google Scholar] [CrossRef]
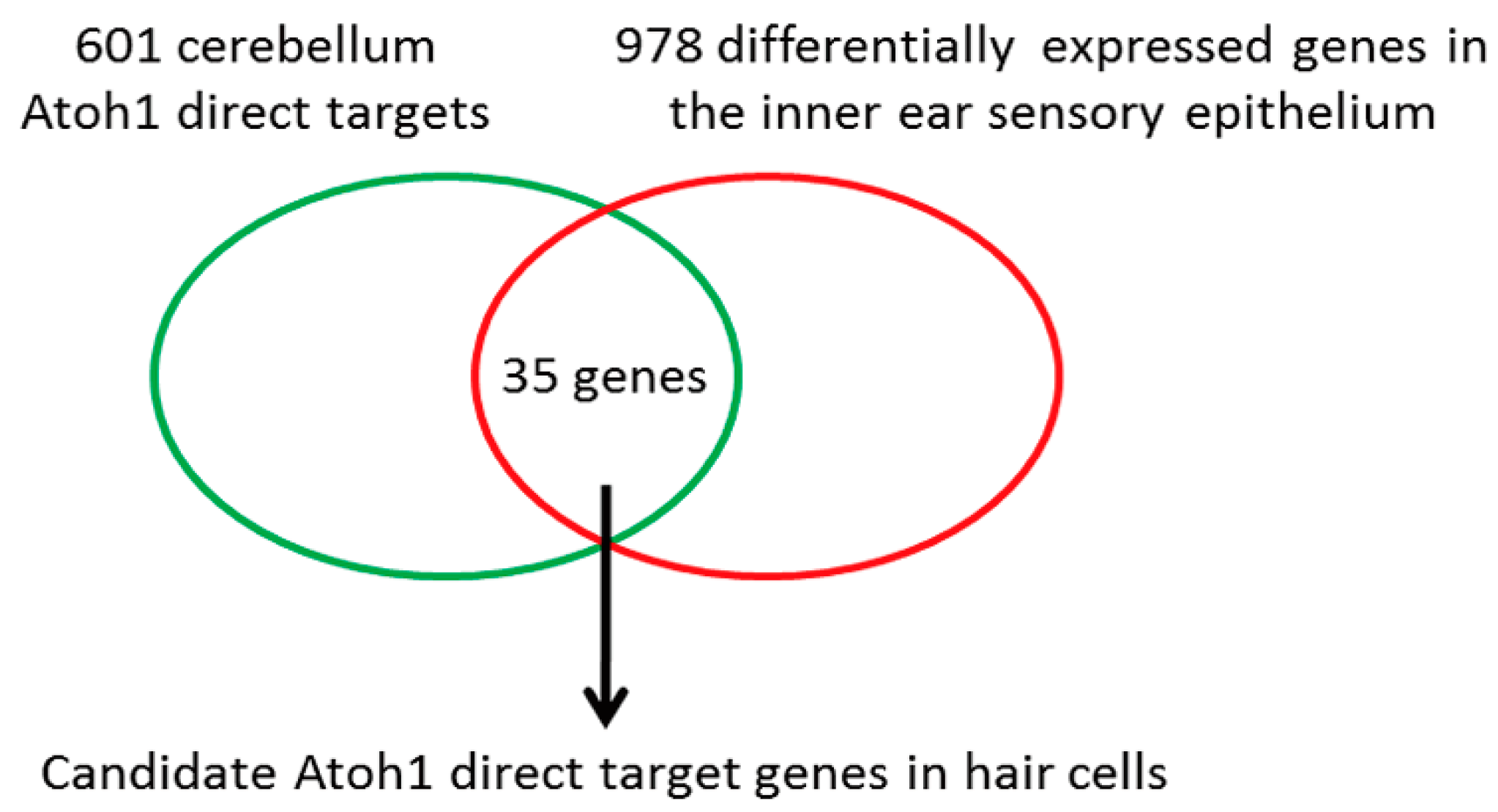




| Candidate Genes Upregulated in Hair Cells | Candidate Genes Downregulated in Hair Cells |
|---|---|
| Ano3 Arfgef3 Atoh1 Barhl1 * Cacng2 Chgb Htr3a Mfng Nceh1 Necab3 Nhlh1 Pacrg Pcsk9 Plch2 Rab36 Scg5 Sgpp2 Slc9a9 Srrm4 * Tmcc2 Ubash3b Wdr95 Zbtb18 | Adamts1 Arhgap23 Ccnd2 Cntn6 Eln Gatm Gm5089 Lypd6 Man1c1 Pcca Ppp1r14c Rgs2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, K.; Jiang, H.; Karim, M.R.; Zhong, C.; Xu, Z.; Liu, L.; Guan, M.; Shao, J.; Huang, X. A Critical E-box in Barhl1 3′ Enhancer Is Essential for Auditory Hair Cell Differentiation. Cells 2019, 8, 458. https://doi.org/10.3390/cells8050458
Hou K, Jiang H, Karim MR, Zhong C, Xu Z, Liu L, Guan M, Shao J, Huang X. A Critical E-box in Barhl1 3′ Enhancer Is Essential for Auditory Hair Cell Differentiation. Cells. 2019; 8(5):458. https://doi.org/10.3390/cells8050458
Chicago/Turabian StyleHou, Kun, Hui Jiang, Md. Rezaul Karim, Chao Zhong, Zhouwen Xu, Lin Liu, Minxin Guan, Jianzhong Shao, and Xiao Huang. 2019. "A Critical E-box in Barhl1 3′ Enhancer Is Essential for Auditory Hair Cell Differentiation" Cells 8, no. 5: 458. https://doi.org/10.3390/cells8050458
APA StyleHou, K., Jiang, H., Karim, M. R., Zhong, C., Xu, Z., Liu, L., Guan, M., Shao, J., & Huang, X. (2019). A Critical E-box in Barhl1 3′ Enhancer Is Essential for Auditory Hair Cell Differentiation. Cells, 8(5), 458. https://doi.org/10.3390/cells8050458





