1. Introduction
A comprehensive characterization of subcellular structures remains a challenge for biological research. The conventional optical imaging methods have advanced to achieve cell images at nano-resolution, but are limited to some complexities, such as the fluorophore localization and fitting algorithms to correct for sample drift during acquisition [
1]. Scanning probe microscopy has advanced the nanoscale imaging for bio-samples due to its strong capabilities of mapping the morphological and mechanical features of subcellular structures with extreme high spatial resolution [
2]. In particular, atomic force microscopy (AFM) can acquire topographical surface images as well as the mechanical properties of biological samples at nanoscale resolution by measuring the forces acting between surface chemical groups [
3,
4,
5,
6]. Various improvements have been made to the AFM platform to acquire more quantitative, dynamic and chemical information, which can be accurately extracted from the acquired images [
7,
8,
9,
10,
11,
12]. Another powerful approach for nanoscale imaging, scanning electron microscope (SEM), provides methods to characterize the inner architecture of a single soft material nanoparticle in addition to its size and shape [
13]. Transmission electron microscopy can be used to visualize some virus particles and cells at nano-resolution [
14]. Soft X-ray tomography can also be used for the quantitative imaging of organelle structure and distribution in whole cells [
15]. Although those techniques have achieved nanometer resolution on cell imaging, they both have their own limitations. For instance, AFM images have insoluble artifacts caused by the abrupt height of the samples and can only obtain the surface or sub-surface structure [
7,
8,
9,
10,
11,
12]; SEM and TEM have special requirements for sample preparation [
13,
14]; and soft X-ray tomography has potential radiation safety limitations [
15].
Among all biomedical imaging techniques, ultrasound has played a significant role in the assessment, diagnosis and imaging of organs and tissues due to its noninvasive natures. The use of ultrasonic waves to explore the inner structures of materials emerged with the introduction of scanning near-field ultrasound holography (SNFUH) [
16]. This technique utilized the acoustic resonance to provide lateral as well as depth-based imaging of nanostructures including alveolar macrophages and peripheral erythrocytes [
17,
18,
19,
20]. However, fast energy attenuation of standing waves limited the application of ultrasound waves to image thin samples tens of microns thick.
Recently, AFAM was developed by combining ultrasound imaging with the near-field AFM imaging. In contrast to SNFUH, in the transmission mode of AFAM, the single frequency acoustic wave penetrates through the specimens and is detected by the high-frequency vibration of the probe. Hence, the transmission mode of AFAM allows deeper imaging while still maintaining nanoscale resolution. In addition, atomic force acoustic microscopy can not only characterize the surface information of various cells at the submicron scale but also image interior substructures by measuring changes in the phase and amplitudes of acoustic waves. The acoustic wave phase represents the position of a wave at a particular time in its cycle, and the relative phase difference indicates that there are certain substances in the path to accelerate or decelerate the propagation of sound waves. The acoustic wave amplitude reflects the wave attenuation along the propagating direction due to the energy dissipation. Hence, the changes of amplitude and phase can explore the internal structure information.
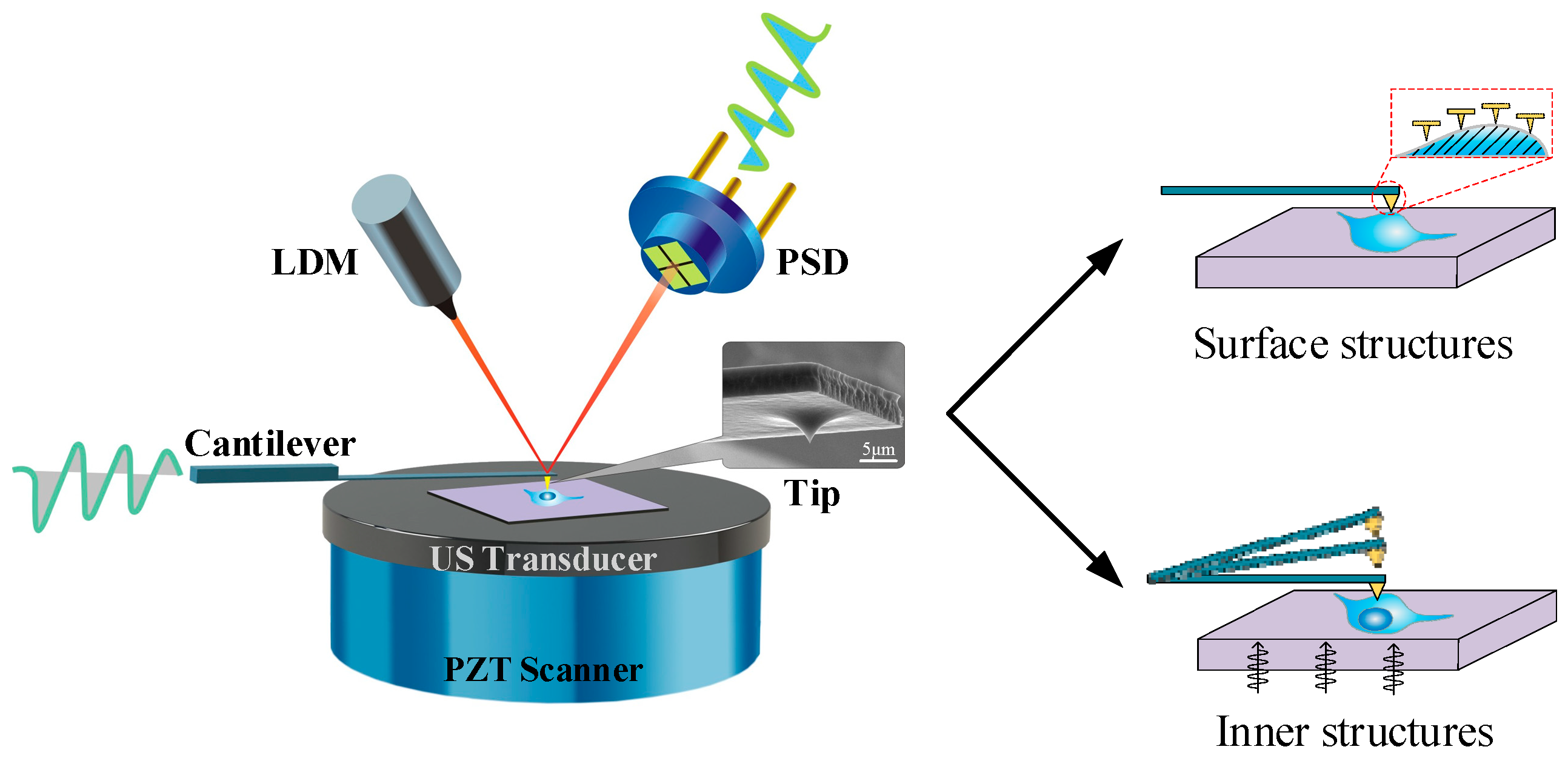
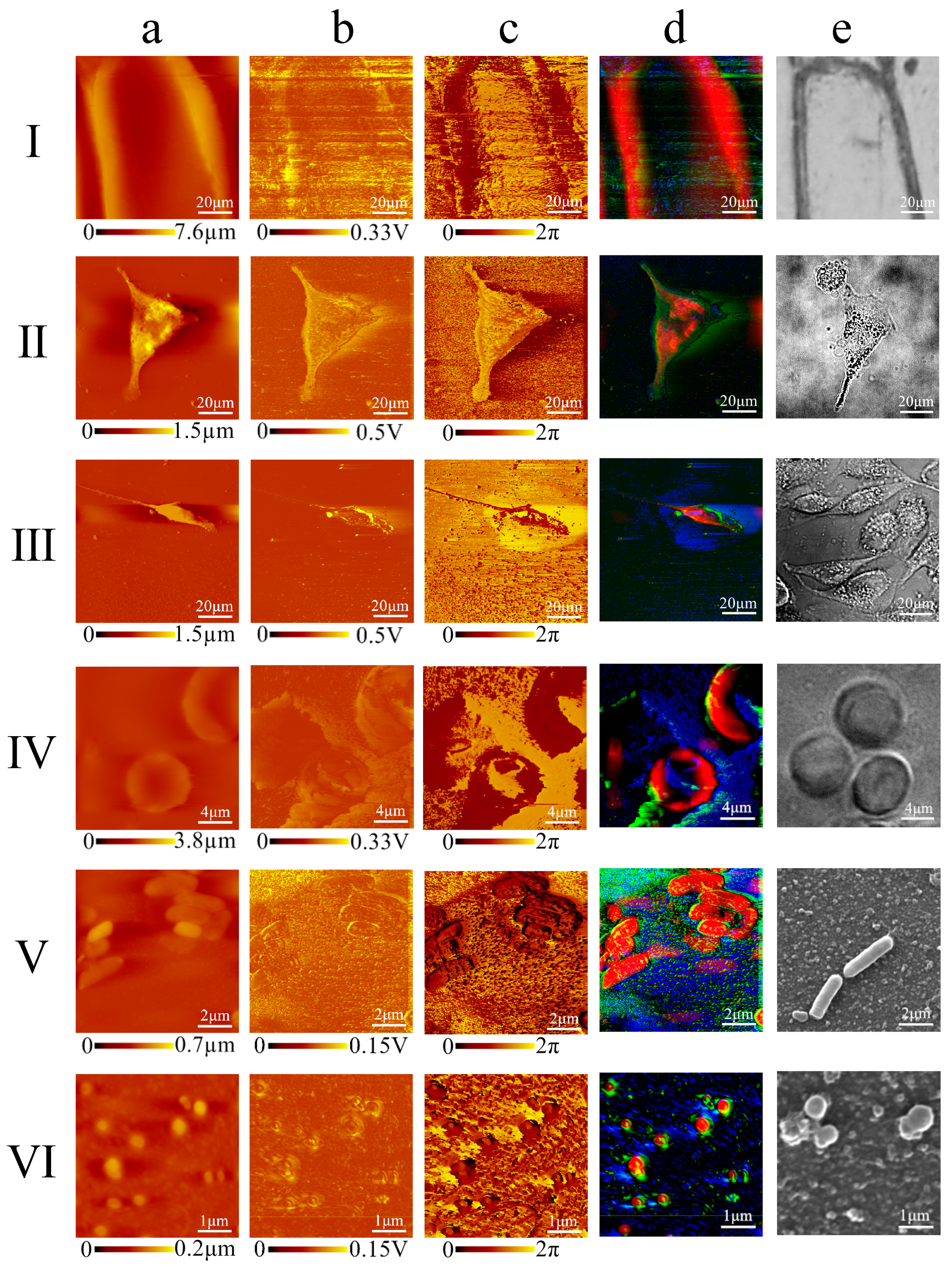
In this paper, we present a novel method to acquire cell images with nanoscale resolution in a non-invasive and easily handled manner (
Figure 1). According to the multiple repeated experiments, we optimized the scanning speed, feedback configurations and acoustic frequency of the AFAM to strengthen the acoustic signals and to stabilize the morphological signals. We also established an RGB-fusion model to illustrate the subcellular features by combining different information from the original images. Applying the optimized AFAM setup and an RGB-fusion algorithm, we obtained submicron-scale images of various types of cells, including
Escherichia coli (
E. coli),
Staphylococcus aureus (
S. aureus), onion epidermis, MCF7 breast cancer cells, MDA-MB-231 breast cancer cells and human erythrocytes, which proved the adaption capacity and robustness of this technique. These experiments revealed distinct morphology and indicated the internal substructures of both eukaryotes and prokaryotes. The fused images enabled better visual observations of morphology and subcellular structures.
2. Materials and Methods
AFAM imaging: CSPM5500, an open atomic force acoustic microscope, from Being Nano-Instruments Ltd., Guangzhou, China, was used as the measurement platform. The ultrasound transducer under the sample holder/plate was used to launch a single-frequency ultrasound wave and was coupled with a normal ultrasound coupling agent as a glue-like substance, to make the specimen contact the sample holder/plate closely. A scanning acoustic model, known as transmission mode, was applied, in which the acoustic wave penetrated through the samples. A contact probe, obtained from Budget Sensors, with a resonant frequency of 13 kHz and a force constant of 0.2 N/m, was used for imaging. The probe tip scanned across the surface of cells on a silicon (Si) wafer, while the acoustic images were acquired with the morphological images simultaneously. The low-frequency vibration represented the morphological information, and the relatively high-frequency vibration showed signals of the acoustic images. All the experiments were performed at room temperature (22 °C).
All parameter-optimization experiments used the MDA-MB-231 cells. We first tested the imaging speed effects on MDA-MB-231 cell imaging, varying the scanning frequency from 0.1 to 2 Hz at 1024 data points per scanning line. Then, we changed the feedback configurations of the integral gain (100–300 times), proportional gain (100–300 times) and reference point value (0.10–0.20 V) to choose the most suitable feedback configurations. We selected 1.5 MHz and 30.5 kHz, which produced the best acoustic response, to illustrate the influence of the acoustic frequency.
Samples preparations for atomic force acoustic microscopy measurements: MDA-MB-231 and MCF7 cells, two classic types of breast cancer cells that play important roles in exploring the biochemical mechanisms of breast cancer, stored by the China Center for Type Culture Collection in Wuhan (Wuhan H22), were grown on a Si wafer in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 50 U/mL penicillin and 50 U/mL streptomycin at 37 °C in 95% humidified air with 5% CO
2. Before AFAM measurement, the cells on the Si wafer were treated with a 4% solution of paraformaldehyde for 15 min to fix the cells, which can maintain the cell morphology, make the cells adhere to the wafers more firmly to prevent them from being taken away by the probe during the imaging scan and further make the scanning process less affected by the sample shift. The fixation treatment with 4% solution of paraformaldehyde has been widely used in other optical imaging approaches, e.g. fluorescence imaging. There are no significant artifacts created in the procedure, which has been reported [
21]. After fixation, the cells were washed with ultrapure water to eliminate the salt crystal and air-dried naturally. Onion epidermis cells, widely used to observe the structures of plant cells under optical microscopes, were placed on a Si wafer after tearing off a small piece from the fresh root top of an onion. The morphology, structure and function of erythrocytes, or red blood cells, are active research areas because these cells carry oxygen to satisfy the respiration of various parts of the body. After resting at 4 °C for 4 h, fresh normal human erythrocytes extracted from human whole blood, which was generously provided by the volunteer in our laboratory, were spread on a Si wafer. Onion epidermis cells and human erythrocytes were applied to a Si wafer using micro pipette tips and dried naturally prior to AFAM measurement.
Escherichia coli (
E. coli) and
Staphylococcus aureus (
S. aureus) are two main species of contaminating bacteria that cause inflammatory reaction.
E. coli and
S. aureus, stored by the China Center for Type Culture Collection in Wuhan (Wuhan H22), were taken out of the −20 °C refrigerator. After cultured in Luria–Bertani (LB) media in an incubator shaker at 37 °C and 250 rpm for 12 h, the bacterial cells were in the log phase. A total of 50 μL medium of bacteria was added to a 1.5 mL EP tube and centrifuged for 4 min at 10,000 rpm/min and 4 °C. One milliliter of ultrapure water was added, and the solution was blended. The bacteria medium was applied to a Si wafer and air-dried naturally, and the Si wafers were exposed to an open flame for 0.2 s prior to AFAM measurement. We chose the six samples mentioned above to test the imaging capacity of our method, in which MDA-MB-231, MCF7 cells,
E. coli and
S. aureus were fixed, while the onion epidermis cells and erythrocytes were unfixed and native. The treatment with 4% solution of paraformaldehyde or the flame preprocessing aimed to fix the cells and keep their morphology during the imaging scan, which is a preprocessing of cells cultured in medium. In other words, this imaging approach does not require the cell fixation as a necessary process step, which we checked though a supplemented experiment. We fixed the cells to follow the common cell preparation, which kept the cell shape so that it can be easily compared with other imaging modality.
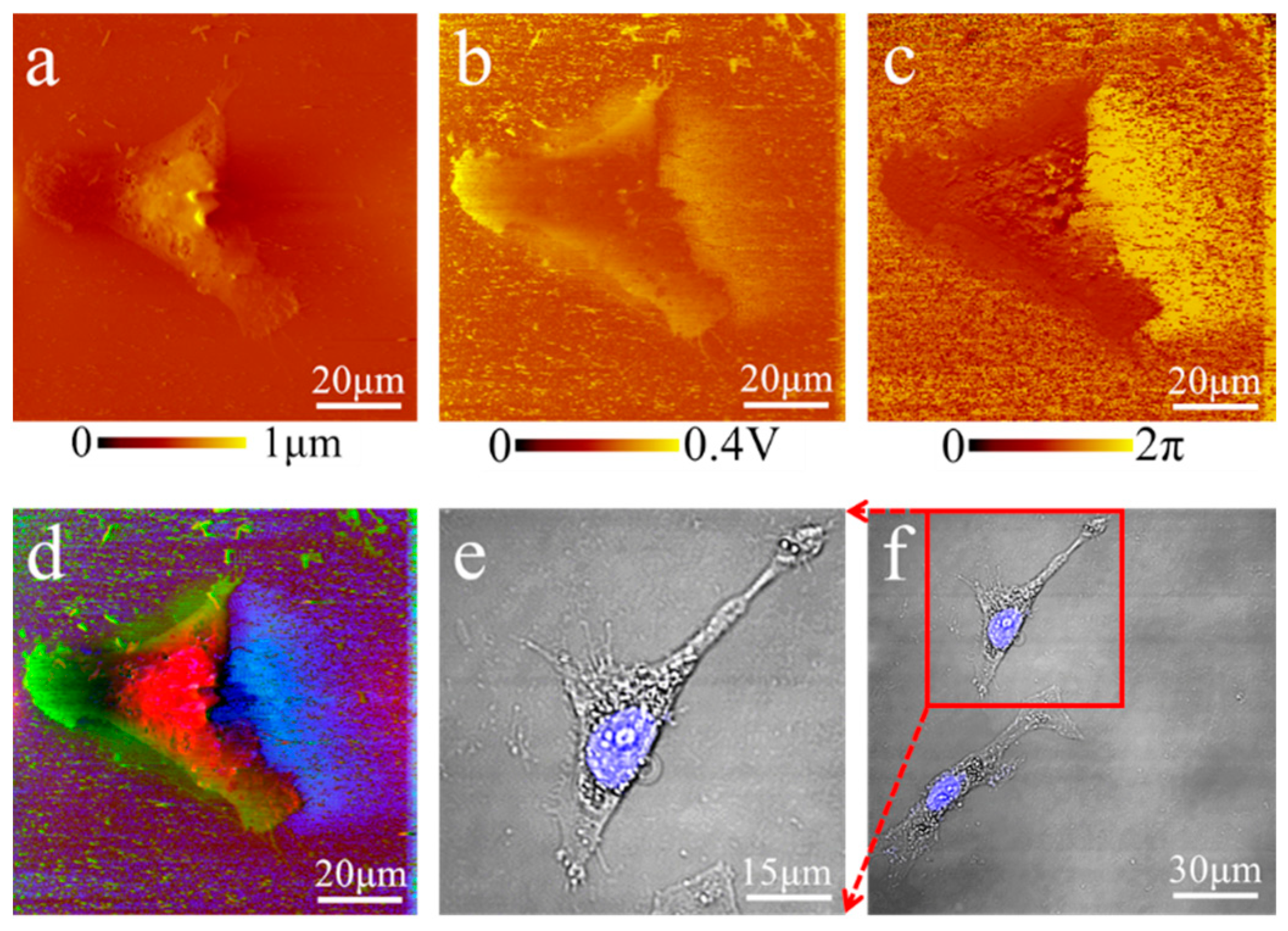
Samples preparations for laser scanning confocal microscope and scanning electron microscope measurements: Onion epidermis cells were placed on a microslide and photographed by an inverted fluorescence microscope (NIKON ECLIPSE Ti-S). MDA-MB-231 cells were first fixed by 4% solution of paraformaldehyde for 15 min, then stained by 4′,6-diamidino-2-phenylindole (DIPA) for 3 min, washed by phosphate buffer saline (PBS), and finally photographed by laser scanning confocal microscope (OLYMPUS FV1000). MCF7 cells were photographed by laser scanning confocal microscope (OLYMPUS FV1000) after fixation by 4% solution of paraformaldehyde for 15 min. Fresh normal human erythrocytes were first diluted 10 times, then applied to a microslide and photographed by laser scanning confocal microscope (OLYMPUS FV1000). We strictly followed the procedures described in Ref. [
18], to get the SEM images of
E. coli and
S. aureus [
22].
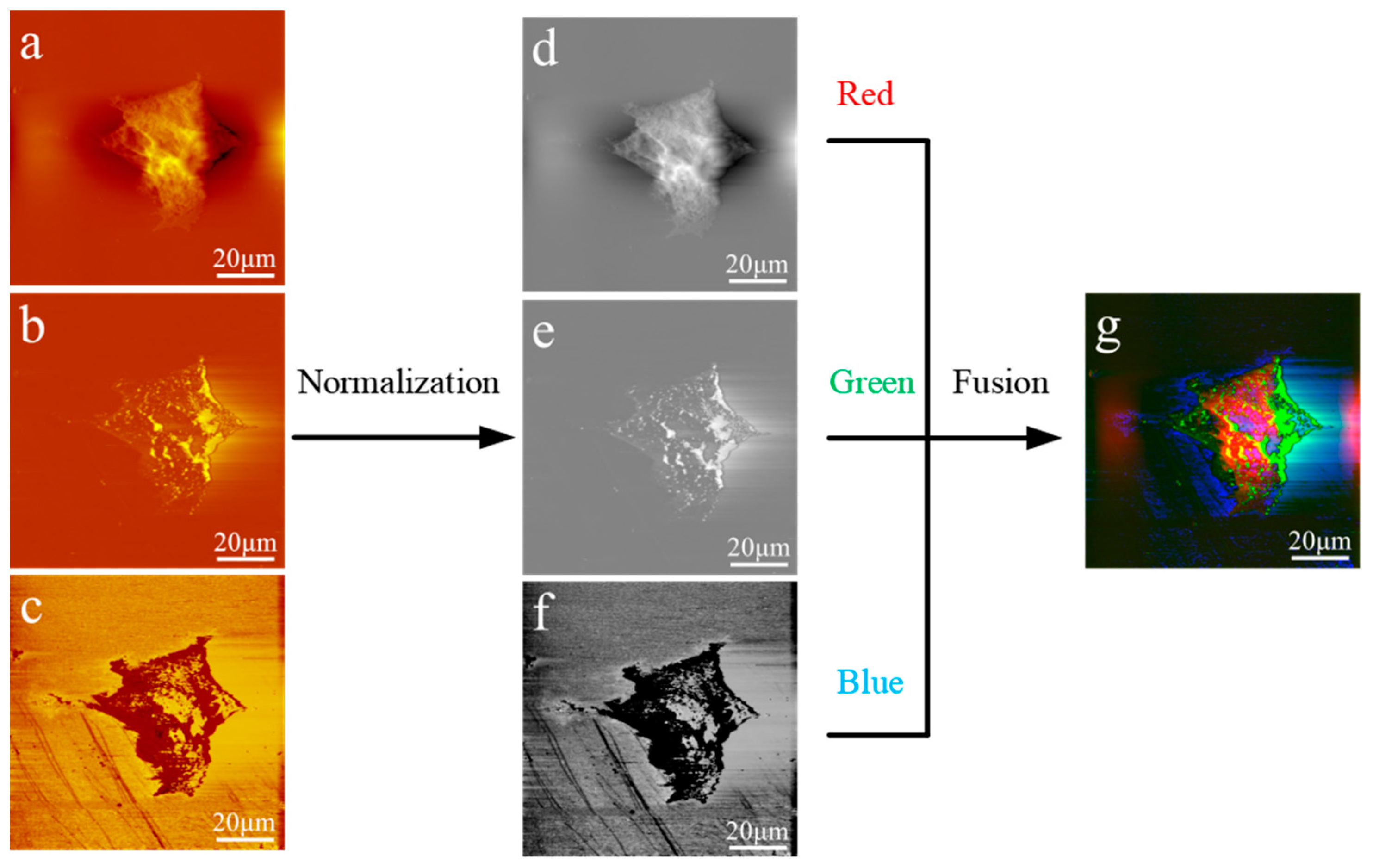
Data analysis: The original AFAM images were visualized with the software Imager in the AFAM system, which is called CSPM5500. Usually, each individual image of AFAM can only provide one restricted and specified view of the surface or the information of internal structures. In traditional AFAM experiments, the morphological and acoustic images are displayed separately, and it is difficult to distinguish structural information differences and overlaps among these obtained images. One possible solution is to integrate data from multiple images, creating a single fused image that retains all the features of the original sources. RGB (red, green and blue) color images are a special case of multispectral fused images that utilize the three basic colors of human vision. An image fusion algorithm based on RGB colors could be used to show the complementary information of all source images and provide a stereoscopic and colorful fused figure. The fusion of the RGB images comprised three steps. A normalization process was performed on three source images—the morphological image, phase image and amplitude image—to obtain gray images at the pixel level. A normalization process was performed using Equation (1),
where
i is the value of every pixel and max is the largest value of all pixels. Histogram modification was performed to maximize the image contrast using Equation (2),
where
N is the total number of pixels,
nk is the value of the pixel at the
kth level,
rk is the grayscale at kth level, and
pr (
ri) is the relative frequency at the
i level. To apply the RGB model, the morphological image was taken as the red channel, the phase image of the ultrasound as the green channel, and the amplitude image of the ultrasound as the blue image of the RGB color system to integrate the three images into one color image.
4. Discussion
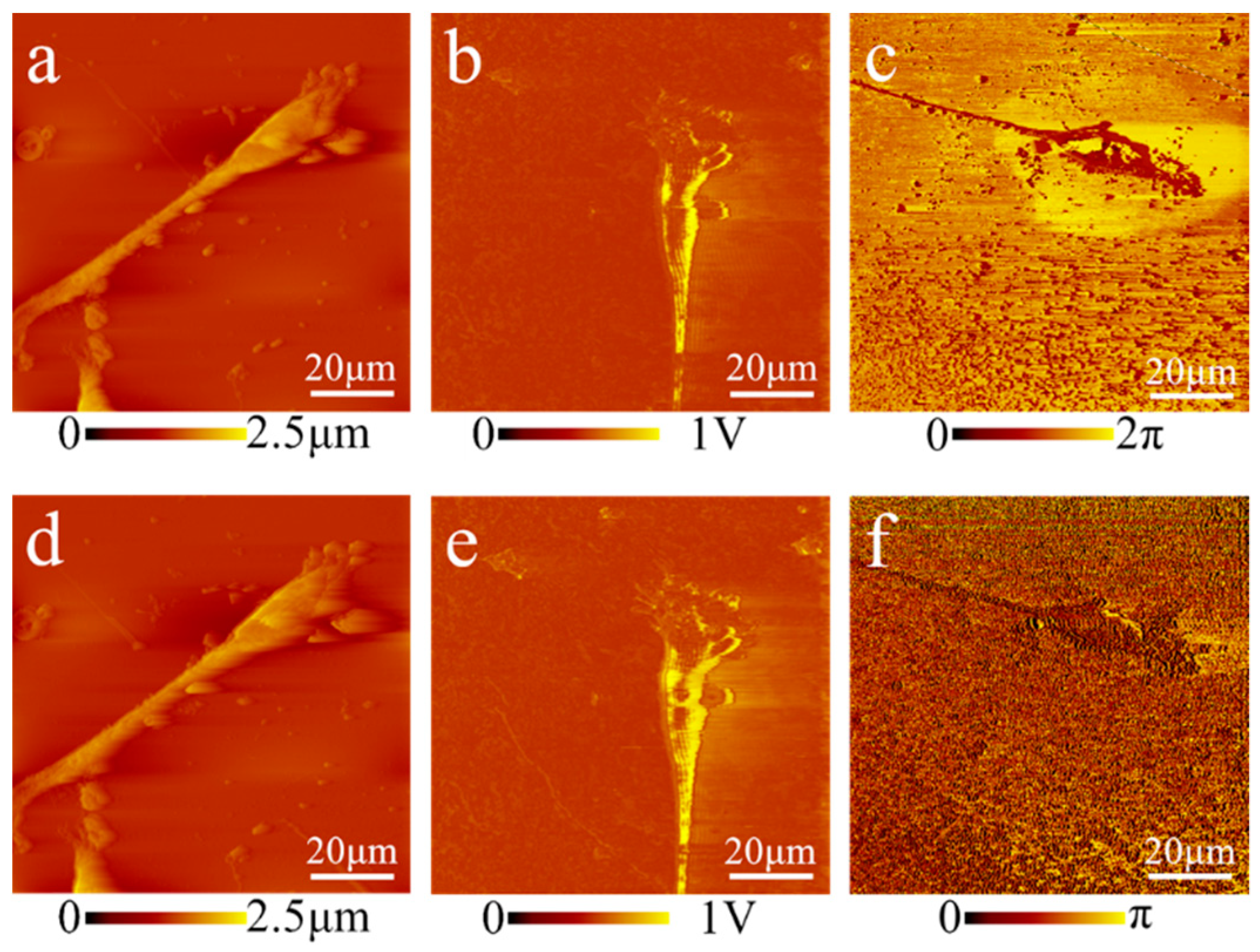
Technology to non-invasively acquire cell images at nanoscale resolution is essential to meet the demand for characterizing subcellular structures. AFAM can extensively reduce the artifacts and avoid complex sample preparation associated with SEM and AFM. The acoustic images of prokaryotes and eukaryotes acquired by AFAM showed clearer exterior contours than the morphological images compared to the alternative techniques (
Figure 6). Moreover, the samples for AFAM measurements are easy to prepare. For instance,
E. coli samples for AFAM only required a few simple processing steps such as placing a droplet of bacterial suspension on a silicon (Si) wafer and waiting for the samples to dry naturally, whereas, for SEM, the sample processing steps include cleaning, chemical fixation, drying and depositing noble metal onto the cell surface.
Comprehensive, accurate and thorough analysis of acoustic images is also a challenge. Usually, acoustic images in AFAM are separated from the morphological image, and it is difficult to identify signals which are different from surface structures. The morphology images acquired by AFM can highlight cytoplasmic regions, and the acoustic phase and amplitude images may suggest internal structures, similar to the optical fluorescence system with nuclear and cytoplasmic regions separated. Notably, combined AFAM images with RGB-colors presentation could demonstrate detailed, accurate topological information of the physical features, and reduce the image artifacts in the morphology in a more visual and colored manner. For the artifacts caused by diversity of samples’ height in the morphological images, the cell boundary could be clearly outlined by the acoustic phase and amplitude images, while each single color represented one type of image. The multicolor image highlighted subcellular edges by superimposed color from artifacts and noise, while separated color represented extraneous information which showed outside the protruding border. With the fusion method, the atlas images showed distinct morphology and suggested internal structure information in a color perspective.
Ultrasound is rarely used to image nanoscale structures and is more commonly used to image organs deep within the body or within tissues, with the aid of an ultrasound contrast agent. Although SNFUH was developed to detect inner structures at nanoscale resolution in 2005, no prototype that can be widely used in scientific research has been developed, due to difficult maneuverability, e.g., the hard formation of the standing wave [
15,
16,
17,
18,
19]. Different from the resonance mode of SNFUH, transmission mode is easy to operate and suitable for large-scale use. In transmission AFAM, acoustic images exhibited enhanced edges of morphological information and fewer artifacts of the morphology, highlighted the faint structures in the morphology and suggested different internal areas that will be explored in detail in the future. As a commercialized instrument, AFAM is mature and stable in operating transmission mode. AFAM has been used to image chemical molecular structures, and our group began the research applying transmission AFAM to image cells. As cell imaging using transmission AFAM achieved notable results, this novel approach could be advanced as a general method to experimentally obtaining insight into subcellular mechanisms in a nondestructive and facile manner at a nanoscale resolution.
Recently, the Human Cell Atlas attracts great interests to advance better understanding of human evolution, and help diagnose human diseases by comparing cell types [
24,
25,
26,
27]. With extensive imaging range, easy sample preparation, multi-color visualization and nondestructive imaging, the AFAM could advance cell imaging, which would improve the Human Cell Atlas project. Hence, the future applications of AFAM on biological research are promising.