Analysis of the Zn-Binding Domains of TRIM32, the E3 Ubiquitin Ligase Mutated in Limb Girdle Muscular Dystrophy 2H
Abstract
1. Introduction
2. Materials and Methods
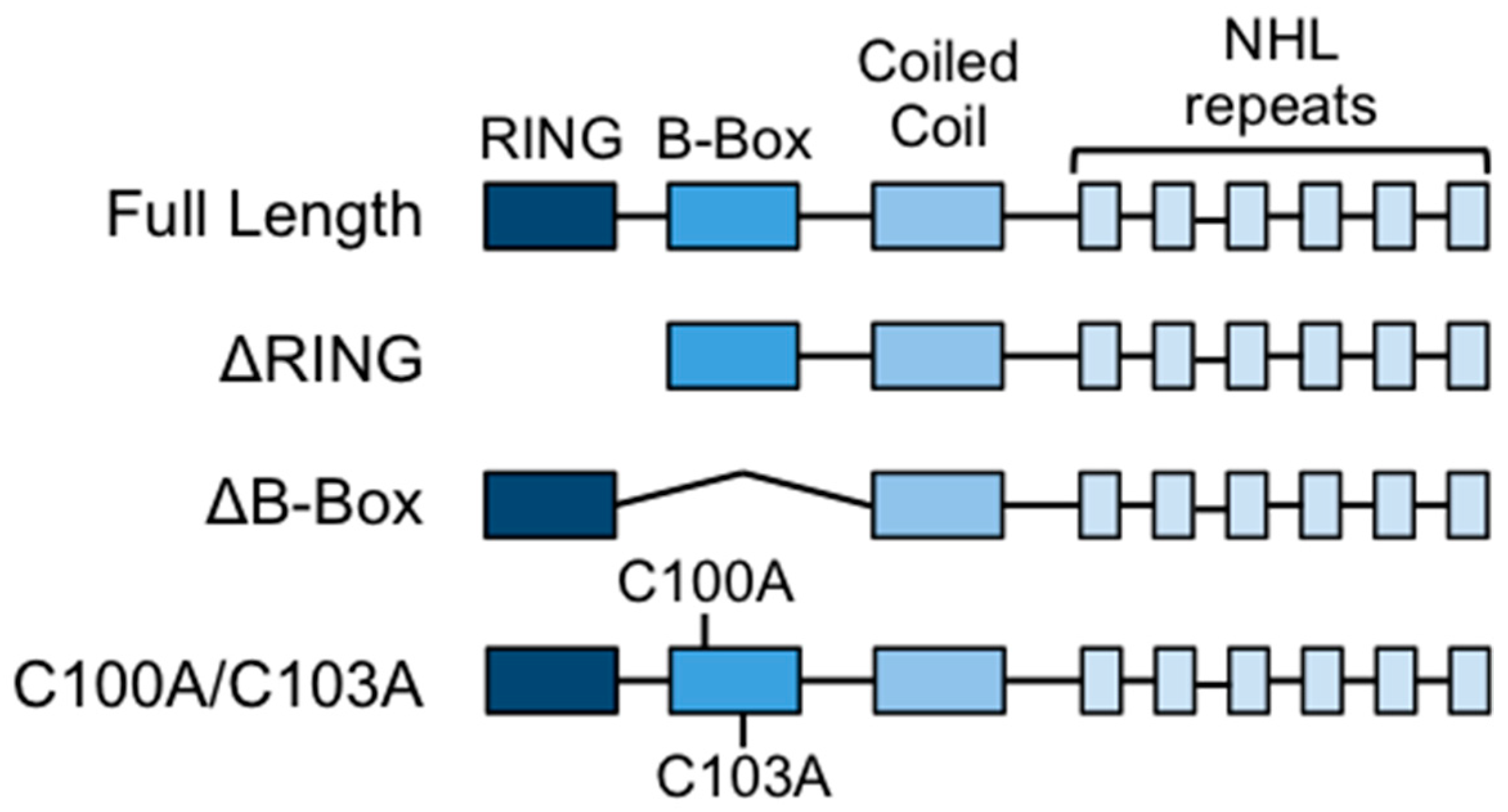
2.1. Constructs and Mutagenesis
2.2. Protein Expression and Purification
2.3. In Vitro Ubiquitination
2.4. Cell Culture and Transfection
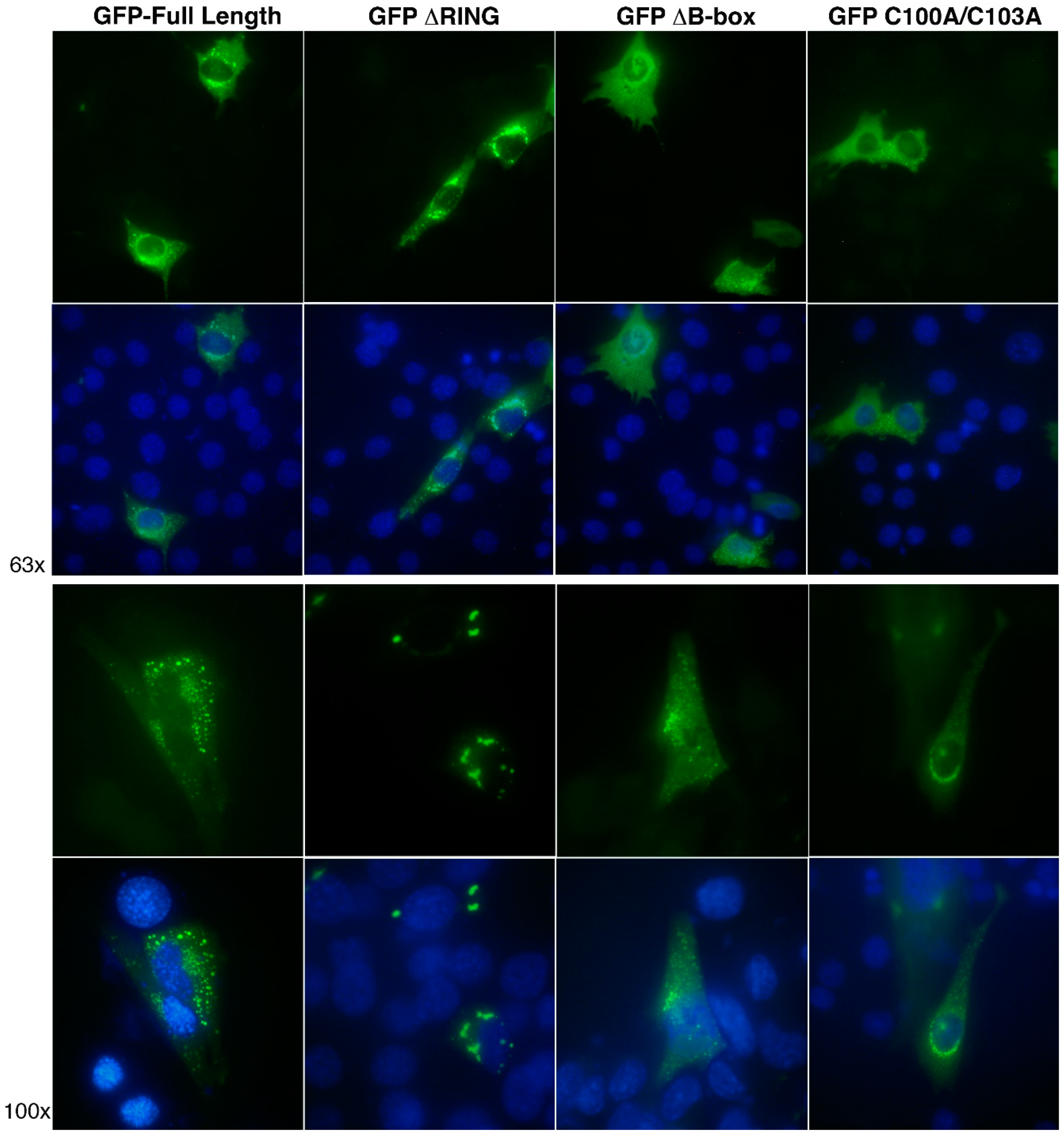
2.5. Immunofluorescence
2.6. Antibodies and Immunoblot Analysis
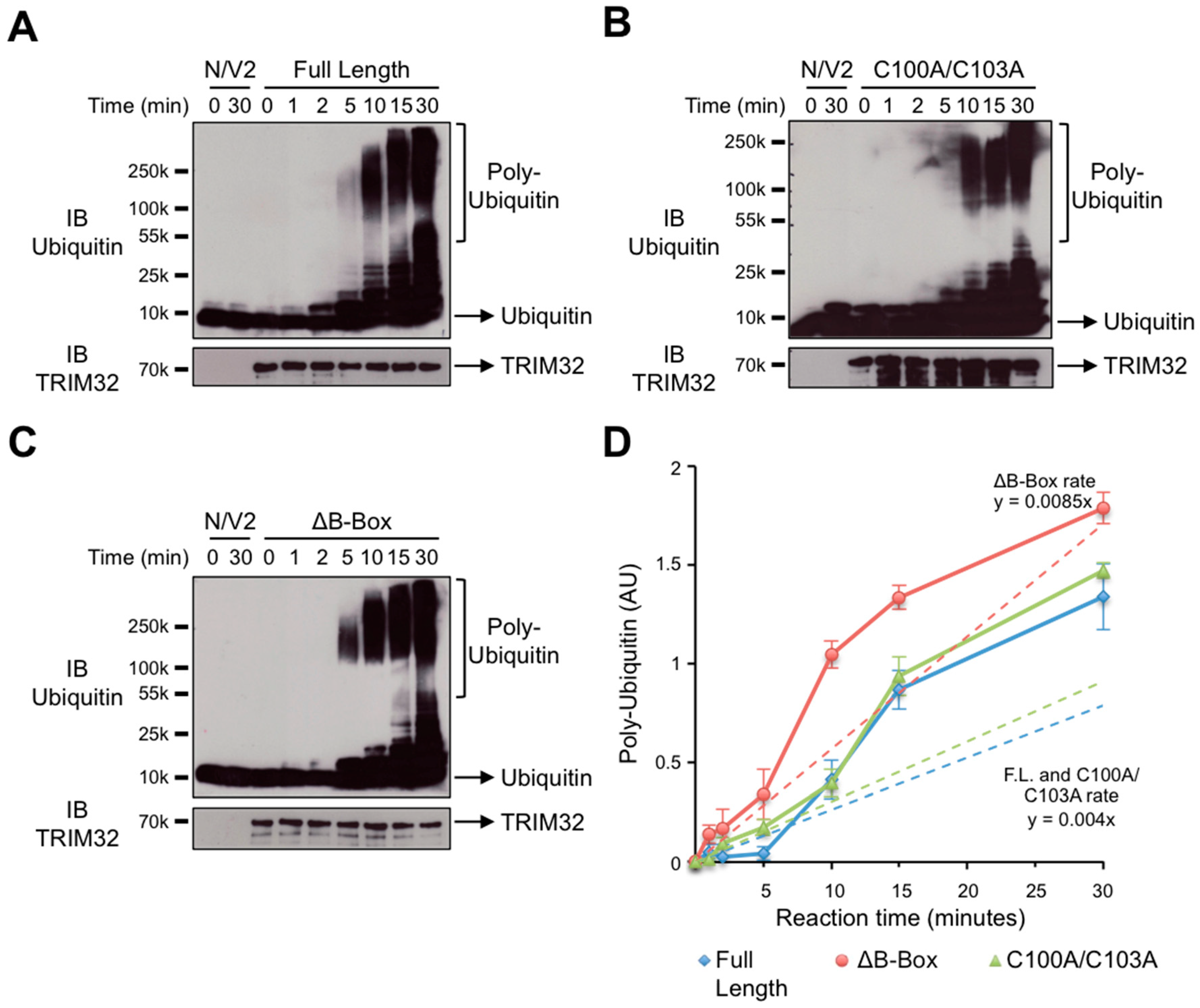
2.7. Quantification of In Vitro Ubiquitination Activity
3. Results
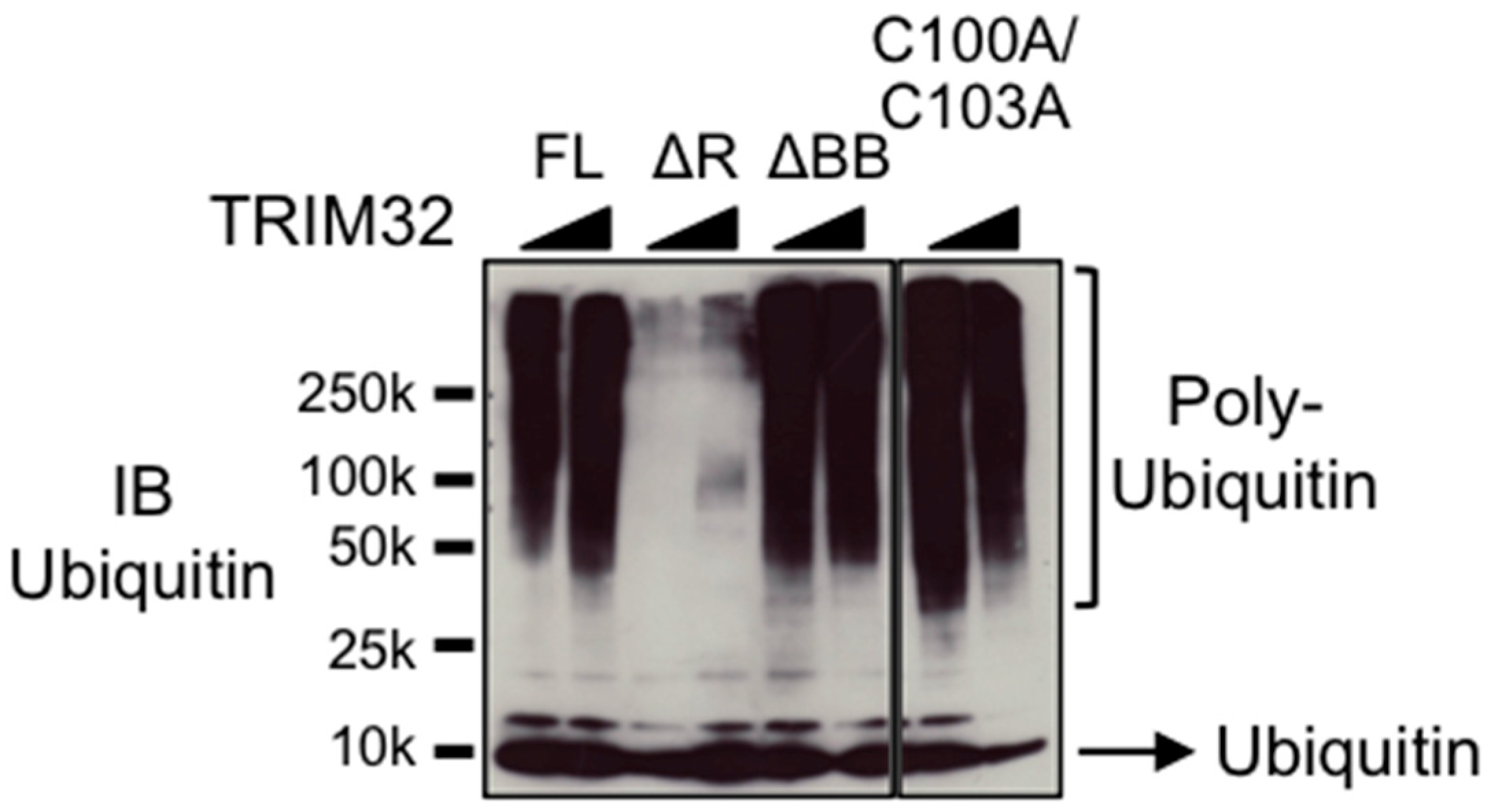
3.1. Deletion or Mutation of the B-box Domain Does Not Prevent TRIM32 E3 Ligase Catalytic Activity
3.2. The B-box Domain Plays a Role in Modulating TRIM32 E3 Ligase Reaction Rate
3.3. TRIM32 E2 Specificity Is Not Determined by the B-box
3.4. The RING Domain Is Involved in Higher-Order Self-Association of TRIM32
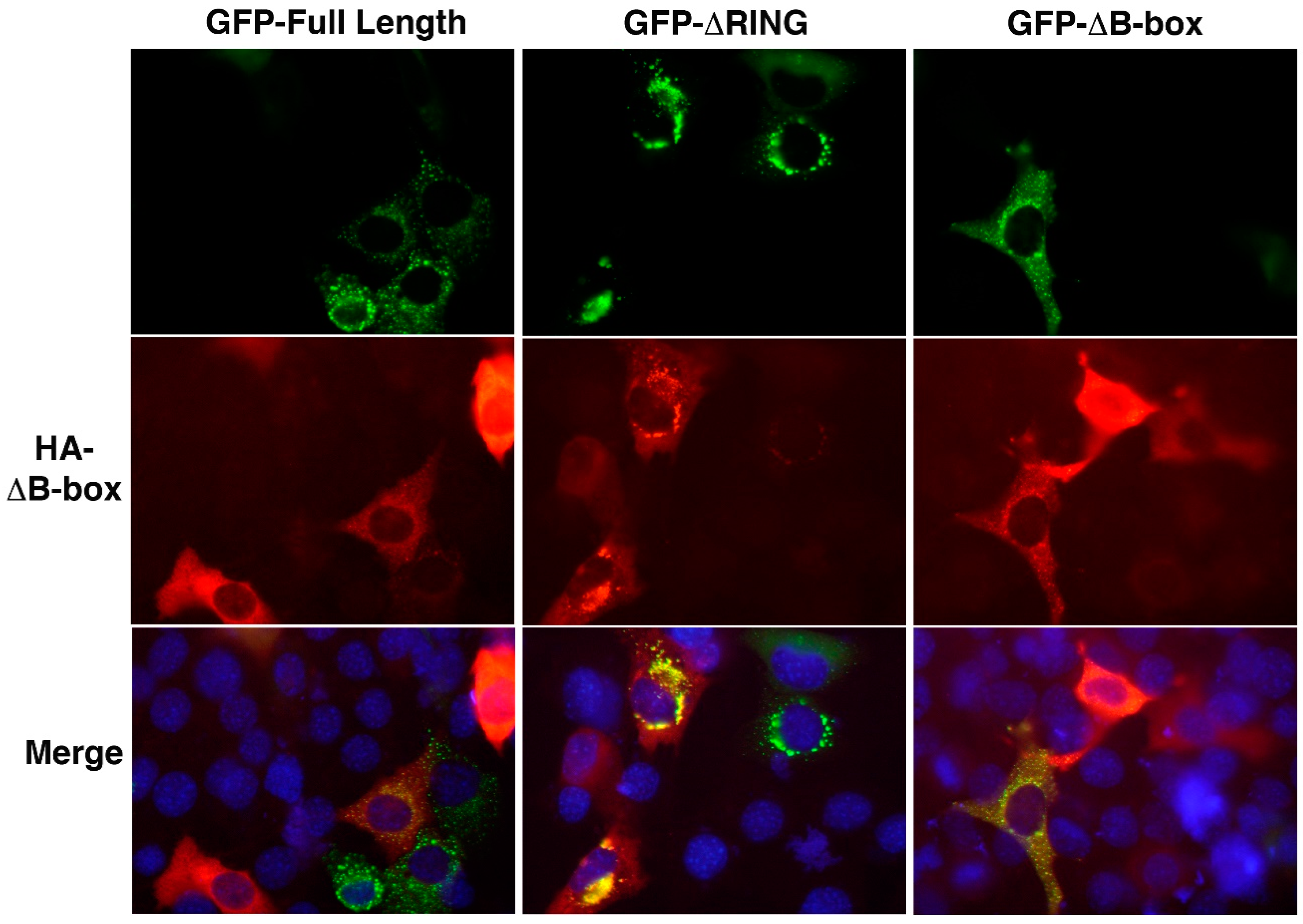
3.5. The RING and B-box Domains Contribute to Specify TRIM32 Subcellular Localization
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Komander, D.; Rape, M. The ubiquitin code. Ann. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Deshaies, R.J.; Joazeiro, C.A. RING domain E3 ubiquitin ligases. Ann. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef]
- Reymond, A.; Meroni, G.; Fantozzi, A.; Merla, G.; Cairo, S.; Luzi, L.; Riganelli, D.; Zanaria, E.; Messali, S.; Cainarca, S.; et al. The tripartite motif family identifies cell compartments. EMBO J. 2001, 20, 2140–2151. [Google Scholar] [CrossRef] [PubMed]
- Meroni, G.; Diez-Roux, G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays 2005, 27, 1147–1157. [Google Scholar] [CrossRef] [PubMed]
- Sardiello, M.; Cairo, S.; Fontanella, B.; Ballabio, A.; Meroni, G. Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties. BMC Evol. Biol. 2008, 8, 225. [Google Scholar] [CrossRef] [PubMed]
- Short, K.M.; Cox, T.C. Sub-classification of the rbcc/trim superfamily reveals a novel motif necessary for microtubule binding. J. Biol. Chem. 2006, 281, 8970–8980. [Google Scholar] [CrossRef]
- Koliopoulos, M.G.; Esposito, D.; Christodoulou, E.; Taylor, I.A.; Rittinger, K. Functional role of TRIM E3 ligase oligomerization and regulation of catalytic activity. EMBO J. 2016, 35, 1204–1218. [Google Scholar] [CrossRef]
- Dawidziak, D.M.; Sanchez, J.G.; Wagner, J.M.; Ganser-Pornillos, B.K.; Pornillos, O. Structure and catalytic activation of the TRIM23 RING E3 ubiquitin ligase. Proteins 2017, 85, 1957–1961. [Google Scholar] [CrossRef]
- Plechanovova, A.; Jaffray, E.G.; Tatham, M.H.; Naismith, J.H.; Hay, R.T. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 2012, 489, 115–120. [Google Scholar] [CrossRef]
- Branigan, E.; Plechanovova, A.; Jaffray, E.G.; Naismith, J.H.; Hay, R.T. Structural basis for the RING-catalyzed synthesis of K63-linked ubiquitin chains. Nat. Struct. Mol. Biol. 2015, 22, 597–602. [Google Scholar] [CrossRef]
- Sanchez, J.G.; Chiang, J.J.; Sparrer, K.M.J.; Alam, S.L.; Chi, M.; Roganowicz, M.D.; Sankaran, B.; Gack, M.U.; Pornillos, O. Mechanism of TRIM25 Catalytic Activation in the Antiviral RIG-I Pathway. Cell Rep. 2016, 16, 1315–1325. [Google Scholar] [CrossRef] [PubMed]
- Massiah, M.A.; Simmons, B.N.; Short, K.M.; Cox, T.C. Solution Structure of the RBCC/TRIM B-box1 Domain of Human MID1: B-box with a RING. J. Mol. Biol. 2006, 358, 532–545. [Google Scholar] [CrossRef] [PubMed]
- Massiah, M.A.; Matts, J.A.; Short, K.M.; Simmons, B.N.; Singireddy, S.; Yi, Z.; Cox, T.C. Solution structure of the MID1 B-box2 CHC(D/C)C(2)H(2) zinc-binding domain: Insights into an evolutionarily conserved RING fold. J. Mol. Biol. 2007, 369, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Mrosek, M.; Meier, S.; Ucurum-Fotiadis, Z.; von Castelmur, E.; Hedbom, E.; Lustig, A.; Grzesiek, S.; Labeit, D.; Labeit, S.; Mayans, O. Structural analysis of B-Box 2 from MuRF1: Identification of a novel self-association pattern in a RING-like fold. Biochemistry 2008, 47, 10722–10730. [Google Scholar] [CrossRef] [PubMed]
- Goldstone, D.C.; Walker, P.A.; Calder, L.J.; Coombs, P.J.; Kirkpatrick, J.; Ball, N.J.; Hilditch, L.; Yap, M.W.; Rosenthal, P.B.; Stoye, J.P.; et al. Structural studies of postentry restriction factors reveal antiparallel dimers that enable avid binding to the HIV-1 capsid lattice. Proc. Natl. Acad. Sci. USA 2014, 111, 9609–9614. [Google Scholar] [CrossRef] [PubMed]
- Weinert, C.; Morger, D.; Djekic, A.; Grutter, M.G.; Mittl, P.R. Crystal structure of TRIM20 C-terminal coiled-coil/B30.2 fragment: Implications for the recognition of higher order oligomers. Sci. Rep. 2015, 5, 10819. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, J.G.; Okreglicka, K.; Chandrasekaran, V.; Welker, J.M.; Sundquist, W.I.; Pornillos, O. The tripartite motif coiled-coil is an elongated antiparallel hairpin dimer. Proc. Natl. Acad. Sci. USA 2014, 111, 2494–2499. [Google Scholar] [CrossRef] [PubMed]
- Napolitano, L.M.; Meroni, G. TRIM family: Pleiotropy and diversification through homomultimer and heteromultimer formation. IUBMB Life 2012, 64, 64–71. [Google Scholar] [CrossRef]
- Wagner, J.M.; Roganowicz, M.D.; Skorupka, K.; Alam, S.L.; Christensen, D.; Doss, G.; Wan, Y.; Frank, G.A.; Ganser-Pornillos, B.K.; Sundquist, W.I.; et al. Mechanism of B-box 2 domain-mediated higher-order assembly of the retroviral restriction factor TRIM5α. eLife 2016, 5, e16309. [Google Scholar] [CrossRef] [PubMed]
- Keown, J.R.; Yang, J.X.; Douglas, J.; Goldstone, D.C. Characterisation of assembly and ubiquitylation by the RBCC motif of Trim5α. Sci. Rep. 2016, 6, 26837. [Google Scholar] [CrossRef]
- Lazzari, E.; Meroni, G. TRIM32 ubiquitin E3 ligase, one enzyme for several pathologies: From muscular dystrophy to tumours. Int. J. Biochem. Cell Biol. 2016, 79, 469–477. [Google Scholar] [CrossRef]
- Frosk, P.; Weiler, T.; Nylen, E.; Sudha, T.; Greenberg, C.R.; Morgan, K.; Fujiwara, T.M.; Wrogemann, K. Limb-girdle muscular dystrophy type 2H associated with mutation in TRIM32, a putative E3-ubiquitin-ligase gene. Am. J. Hum. Genet. 2002, 70, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Muller-Felber, W.; Schlotter, B.; Topfer, M.; Ketelsen, U.P.; Muller-Hocker, J.; Pongratz, D. Phenotypic variability in two brothers with sarcotubular myopathy. J. Neurol. 1999, 246, 408–411. [Google Scholar] [CrossRef]
- Weiler, T.; Greenberg, C.R.; Zelinski, T.; Nylen, E.; Coghlan, G.; Crumley, M.J.; Fujiwara, T.M.; Morgan, K.; Wrogemann, K. A gene for autosomal recessive limb-girdle muscular dystrophy in Manitoba Hutterites maps to chromosome region 9q31–q33: Evidence for another limb-girdle muscular dystrophy locus. Am. J. Hum. Genet. 1998, 63, 140–147. [Google Scholar] [CrossRef]
- Kudryashova, E.; Kudryashov, D.; Kramerova, I.; Spencer, M.J. Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. J. Mol. Biol. 2005, 354, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Mokhonova, E.I.; Avliyakulov, N.K.; Kramerova, I.; Kudryashova, E.; Haykinson, M.J.; Spencer, M.J. The E3 ubiquitin ligase TRIM32 regulates myoblast proliferation by controlling turnover of NDRG2. Hum. Mol. Genet. 2015, 24, 2873–2883. [Google Scholar] [CrossRef]
- Cohen, S.; Zhai, B.; Gygi, S.P.; Goldberg, A.L. Ubiquitylation by Trim32 causes coupled loss of desmin, Z-bands, and thin filaments in muscle atrophy. J. Cell Biol. 2012, 198, 575–589. [Google Scholar] [CrossRef] [PubMed]
- Nicklas, S.; Otto, A.; Wu, X.; Miller, P.; Stelzer, S.; Wen, Y.; Kuang, S.; Wrogemann, K.; Patel, K.; Ding, H.; et al. TRIM32 regulates skeletal muscle stem cell differentiation and is necessary for normal adult muscle regeneration. PLoS ONE 2012, 7, e30445. [Google Scholar] [CrossRef] [PubMed]
- Chiang, A.P.; Beck, J.S.; Yen, H.J.; Tayeh, M.K.; Scheetz, T.E.; Swiderski, R.E.; Nishimura, D.Y.; Braun, T.A.; Kim, K.Y.; Huang, J.; et al. Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet-Biedl syndrome gene (BBS11). Proc. Natl. Acad. Sci. USA 2006, 103, 6287–6292. [Google Scholar] [CrossRef]
- van den Ent, F.; Lowe, J. RF cloning: A restriction-free method for inserting target genes into plasmids. J. Biochem. Biophys. Methods 2006, 67, 67–74. [Google Scholar] [CrossRef]
- Streich, F.C., Jr.; Ronchi, V.P.; Connick, J.P.; Haas, A.L. Tripartite motif ligases catalyze polyubiquitin chain formation through a cooperative allosteric mechanism. J. Biol. Chem. 2013, 288, 8209–8221. [Google Scholar] [CrossRef] [PubMed]
- Napolitano, L.M.; Jaffray, E.G.; Hay, R.T.; Meroni, G. Functional interactions between ubiquitin E2 enzymes and TRIM proteins. Biochem. J. 2011, 434, 309–319. [Google Scholar] [CrossRef]
- Gack, M.U.; Shin, Y.C.; Joo, C.H.; Urano, T.; Liang, C.; Sun, L.; Takeuchi, O.; Akira, S.; Chen, Z.; Inoue, S.; et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 2007, 446, 916–920. [Google Scholar] [CrossRef]
- Diaz-Griffero, F.; Li, X.; Javanbakht, H.; Song, B.; Welikala, S.; Stremlau, M.; Sodroski, J. Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology 2006, 349, 300–315. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Xu, J.; Xie, X.; Zhou, Y.; Tao, P.; Li, H.; Han, X.; Wang, C.; Liu, J.; Xu, P.; et al. Oligomerization-primed coiled-coil domain interaction with Ubc13 confers processivity to TRAF6 ubiquitin ligase activity. Nat. Commun. 2017, 8, 814. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Rape, M. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 2009, 10, 755–764. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.P.; Sun, L.; Chen, X.; Pineda, G.; Jiang, X.; Adhikari, A.; Zeng, W.; Chen, Z.J. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature 2009, 461, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Tatham, M.H.; Plechanovova, A.; Jaffray, E.G.; Salmen, H.; Hay, R.T. Ube2W conjugates ubiquitin to alpha-amino groups of protein N-termini. Biochem. J. 2013, 453, 137–145. [Google Scholar] [CrossRef]
- Schumacher, F.R.; Wilson, G.; Day, C.L. The N-terminal extension of UBE2E ubiquitin-conjugating enzymes limits chain assembly. J. Mol. Biol. 2013, 425, 4099–4111. [Google Scholar] [CrossRef]
- Locke, M.; Tinsley, C.L.; Benson, M.A.; Blake, D.J. TRIM32 is an E3 ubiquitin ligase for dysbindin. Hum. Mol. Genet. 2009, 18, 2344–2358. [Google Scholar] [CrossRef]
- Ryu, Y.S.; Lee, Y.; Lee, K.W.; Hwang, C.Y.; Maeng, J.S.; Kim, J.H.; Seo, Y.S.; You, K.H.; Song, B.; Kwon, K.S. TRIM32 protein sensitizes cells to tumor necrosis factor (TNFα)-induced apoptosis via its RING domain-dependent E3 ligase activity against X-linked inhibitor of apoptosis (XIAP). J. Biol. Chem. 2011, 286, 25729–25738. [Google Scholar] [CrossRef]
- Tatham, M.H.; Geoffroy, M.C.; Shen, L.; Plechanovova, A.; Hattersley, N.; Jaffray, E.G.; Palvimo, J.J.; Hay, R.T. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 2008, 10, 538–546. [Google Scholar] [CrossRef]
- Fang, S.; Jensen, J.P.; Ludwig, R.L.; Vousden, K.H.; Weissman, A.M. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 2000, 275, 8945–8951. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.L.; Malyukova, A.; Holien, J.K.; Koach, J.; Parker, M.W.; Kavallaris, M.; Marshall, G.M.; Cheung, B.B. TRIM16 acts as an E3 ubiquitin ligase and can heterodimerize with other TRIM family members. PLoS ONE 2012, 7, e37470. [Google Scholar] [CrossRef]
- Wada, K.; Kamitani, T. Autoantigen Ro52 is an E3 ubiquitin ligase. Biochem. Biophys. Res. Commun. 2006, 339, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.; Gao, B.; Xu, W.; Xiong, S. Identification of TRIM22 as a RING finger E3 ubiquitin ligase. Biochem. Biophys. Res. Commun. 2008, 374, 502–506. [Google Scholar] [CrossRef] [PubMed]
- Tao, H.; Simmons, B.N.; Singireddy, S.; Jakkidi, M.; Short, K.M.; Cox, T.C.; Massiah, M.A. Structure of the MID1 tandem B-boxes reveals an interaction reminiscent of intermolecular ring heterodimers. Biochemistry 2008, 47, 2450–2457. [Google Scholar] [CrossRef]
- Saccone, V.; Palmieri, M.; Passamano, L.; Piluso, G.; Meroni, G.; Politano, L.; Nigro, V. Mutations that impair interaction properties of TRIM32 associated with limb-girdle muscular dystrophy 2H. Hum. Mutat. 2008, 29, 240–247. [Google Scholar] [CrossRef]
- Joazeiro, C.A.; Weissman, A.M. RING finger proteins: Mediators of ubiquitin ligase activity. Cell 2000, 102, 549–552. [Google Scholar] [CrossRef]
- Short, K.M.; Hopwood, B.; Yi, Z.; Cox, T.C. MID1 and MID2 homo- and heterodimerise to tether the rapamycin- sensitive PP2A regulatory subunit, Alpha 4, to microtubules: Implications for the clinical variability of X-linked Opitz GBBB syndrome and other developmental disorders. BMC Cell Biol. 2002, 3, 1. [Google Scholar] [CrossRef]
- Wagner, J.M.; Christensen, D.E.; Bhattacharya, A.; Dawidziak, D.M.; Roganowicz, M.D.; Wan, Y.; Pumroy, R.A.; Demeler, B.; Ivanov, D.N.; Ganser-Pornillos, B.K.; et al. General Model for Retroviral Capsid Pattern Recognition by TRIM5 Proteins. J. Virol. 2018, 92, e01563-17. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Lord, C.J.; Scheel, H.; Swift, S.; Hofmann, K.; Ashworth, A.; Barford, D. The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol. Cell 2008, 29, 451–464. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, T.; Taoka, M.; Shoji, I.; Kato, H.; Sato, T.; Hatakeyama, S.; Isobe, T.; Hachiya, N. 14-3-3 proteins sequester a pool of soluble TRIM32 ubiquitin ligase to repress autoubiquitylation and cytoplasmic body formation. J. Cell Sci. 2013, 126, 2014–2026. [Google Scholar] [CrossRef] [PubMed]


| TRIM32 | Forward (N1) | 5′-gagaatctttattttcagggcgccatggctgcagcagcagcttctcac-3′ |
| Reverse (C5) | 5′-caagcttgtcgacggagctcgaattcctatggggtggaatatcttctcagatggta-3′ | |
| TRIM32 ΔRING (Δ1–88) | Forward (N2) | 5′-gagaatctttattttcagggcgccgctgggctcagcgaggctgtggggc-3′ |
| Reverse (C5) | 5′-caagcttgtcgacggagctcgaattcctatggggtggaatatcttctcagatggta-3′ | |
| TRIM32 ΔB-box (Δ99–134) | Forward | 5’-agctgcttctttgacagggagtggcagcagccccacagcctcgctgag-3’ |
| Reverse | 5’-ctcagcgaggctgtggggctgctgccactccctgtcaaagaagcagct-3’ | |
| TRIM32 C100A/C103A | Forward | 5’-cagacgccgcccagcggaccgagccatgagcagcccc-3’ |
| Reverse | 5’-ggggctgctcatggctcggtccgctgggcggcgtctg-3’ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lazzari, E.; El-Halawany, M.S.; De March, M.; Valentino, F.; Cantatore, F.; Migliore, C.; Onesti, S.; Meroni, G. Analysis of the Zn-Binding Domains of TRIM32, the E3 Ubiquitin Ligase Mutated in Limb Girdle Muscular Dystrophy 2H. Cells 2019, 8, 254. https://doi.org/10.3390/cells8030254
Lazzari E, El-Halawany MS, De March M, Valentino F, Cantatore F, Migliore C, Onesti S, Meroni G. Analysis of the Zn-Binding Domains of TRIM32, the E3 Ubiquitin Ligase Mutated in Limb Girdle Muscular Dystrophy 2H. Cells. 2019; 8(3):254. https://doi.org/10.3390/cells8030254
Chicago/Turabian StyleLazzari, Elisa, Medhat S. El-Halawany, Matteo De March, Floriana Valentino, Francesco Cantatore, Chiara Migliore, Silvia Onesti, and Germana Meroni. 2019. "Analysis of the Zn-Binding Domains of TRIM32, the E3 Ubiquitin Ligase Mutated in Limb Girdle Muscular Dystrophy 2H" Cells 8, no. 3: 254. https://doi.org/10.3390/cells8030254
APA StyleLazzari, E., El-Halawany, M. S., De March, M., Valentino, F., Cantatore, F., Migliore, C., Onesti, S., & Meroni, G. (2019). Analysis of the Zn-Binding Domains of TRIM32, the E3 Ubiquitin Ligase Mutated in Limb Girdle Muscular Dystrophy 2H. Cells, 8(3), 254. https://doi.org/10.3390/cells8030254






