Chronological Aging Standard Curves of Telomere Length and Mitochondrial DNA Copy Number in Twelve Tissues of C57BL/6 Male Mouse
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
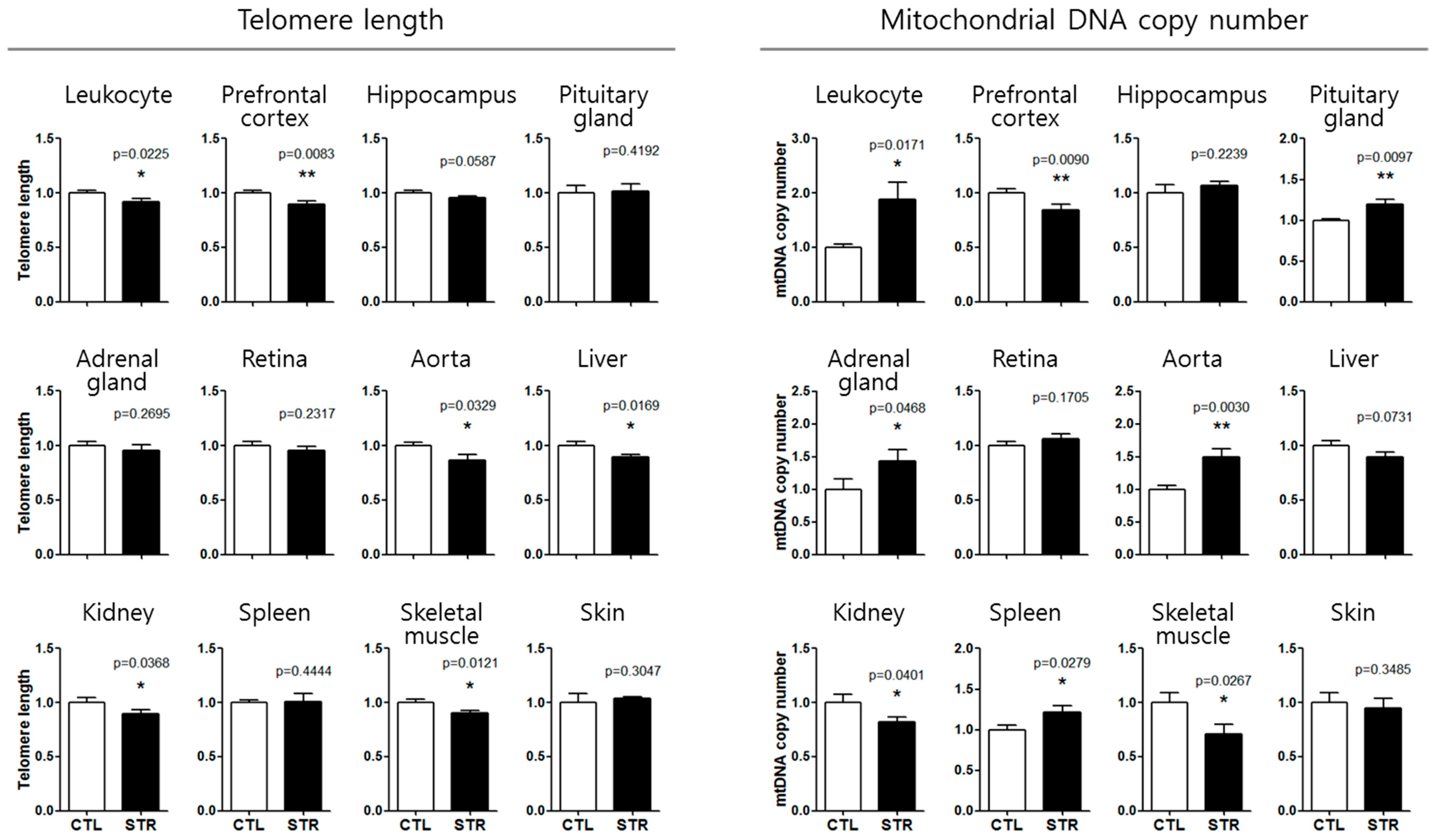
2.2. Chronic Immobilization Stress
2.3. Telomere Length and mtDNAcn Analysis by Quantitative Real-Time PCR (qPCR)
2.4. Statistical Analysis
3. Results
3.1. qPCR Condition Optimization
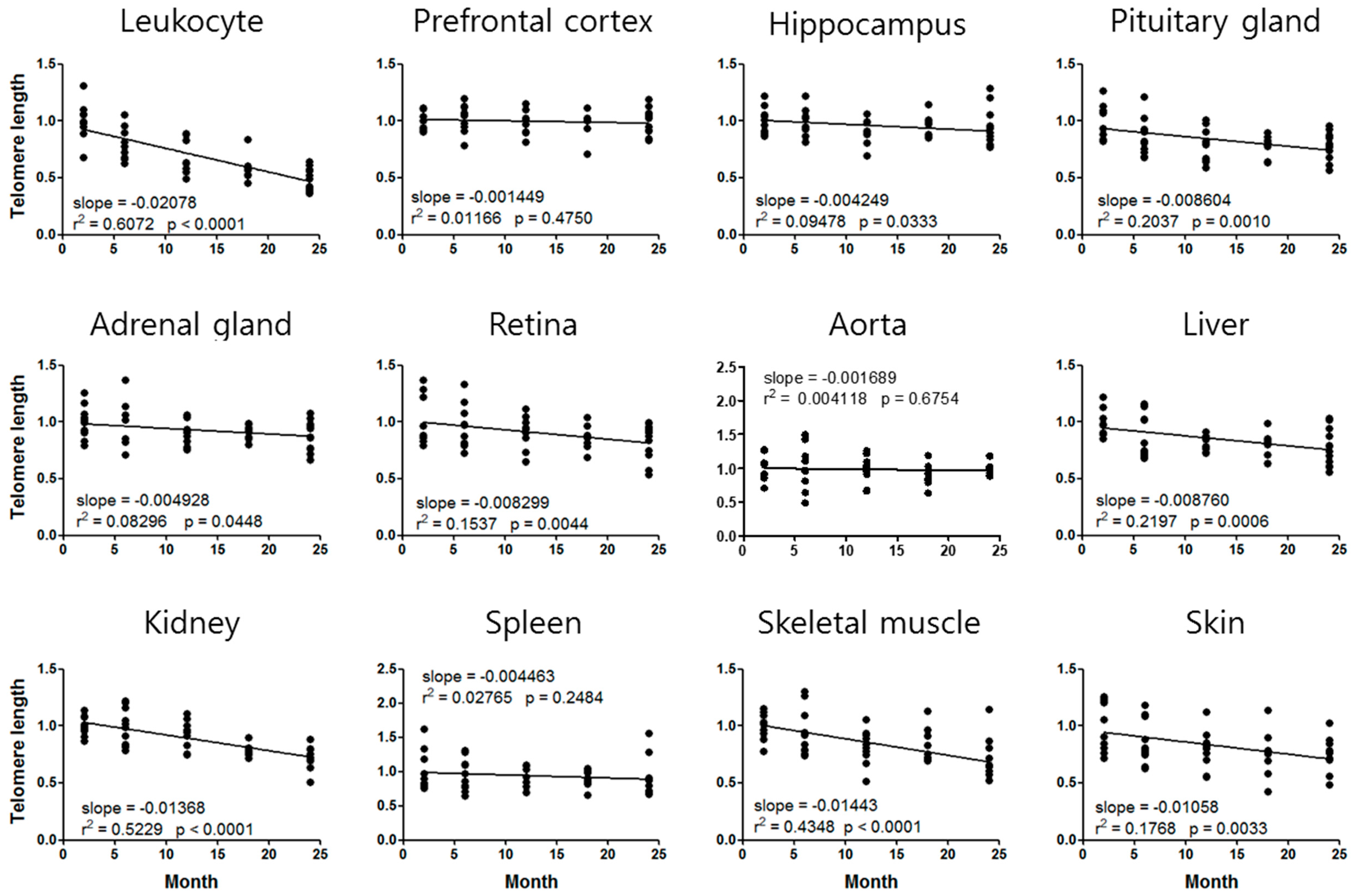
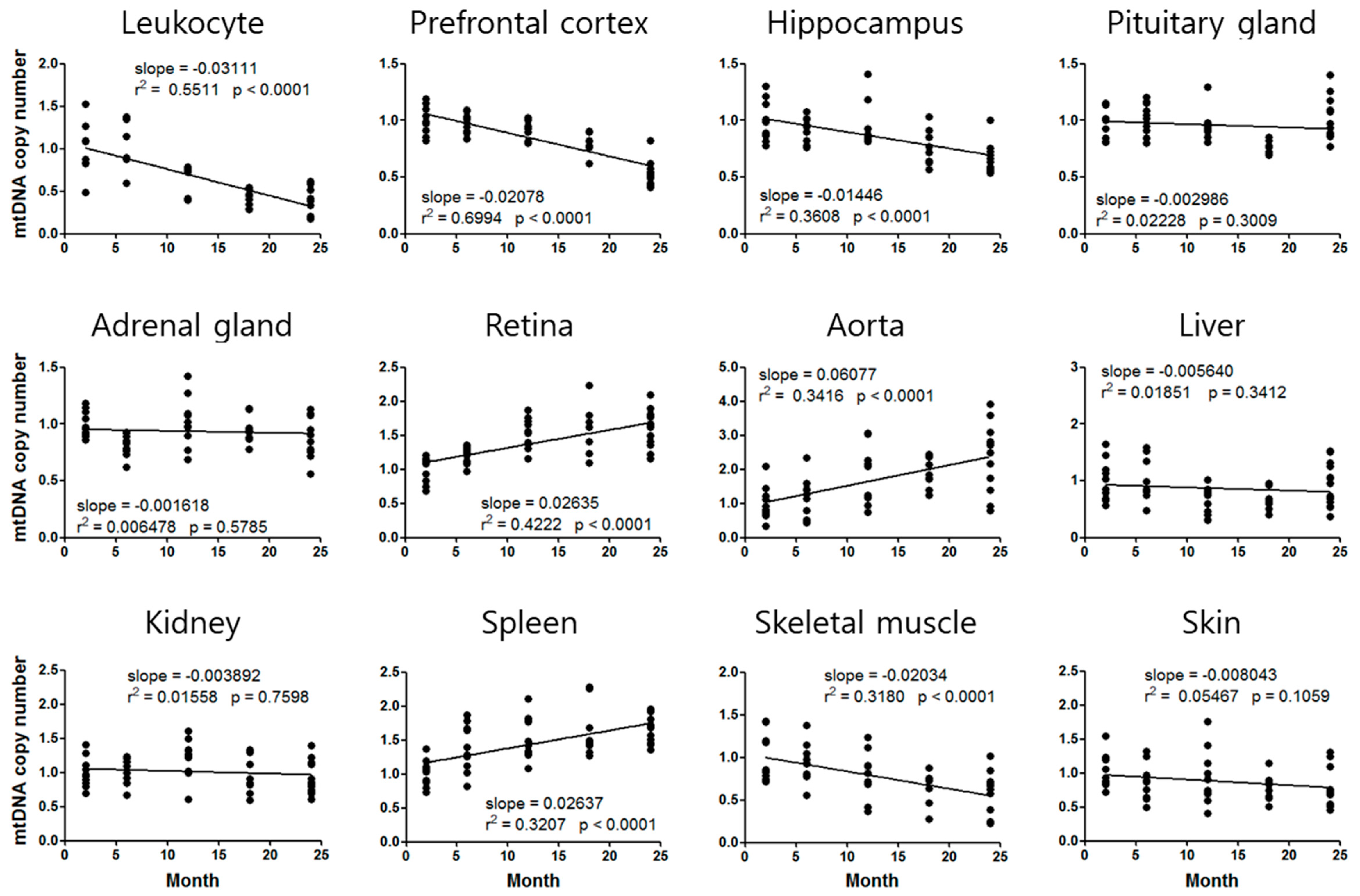
3.2. Mouse Age-Dependent Telomere Length and mtDNAcn Standard Curves
3.3. Application of the Aging Standard Curves for Chronological Age Estimation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bratic, A.; Larsson, N.G. The role of mitochondria in aging. J. Clin. Investig. 2013, 123, 951–957. [Google Scholar] [CrossRef] [PubMed]
- Calado, R.T.; Dumitriu, B. Telomere dynamics in mice and humans. Semin. Hematol. 2013, 50, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Chang, S. Role of telomeres and telomerase in genomic instability, senescence and cancer. Lab. Investig. 2007, 87, 1071. [Google Scholar] [CrossRef] [PubMed]
- Kazachkova, N.; Ramos, A.; Santos, C.; Lima, M. Mitochondrial DNA damage patterns and aging: Revising the evidences for humans and mice. Aging Dis. 2013, 4, 337–350. [Google Scholar] [CrossRef] [PubMed]
- Lagouge, M.; Larsson, N.G. The role of mitochondrial DNA mutations and free radicals in disease and ageing. J. Intern. Med. 2013, 273, 529–543. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, S.; Raza, S.T.; Mahdi, F. Telomere length variations in aging and age-related diseases. Current Aging Sci. 2014, 7, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Arai, Y.; Martin-Ruiz, C.M.; Takayama, M.; Abe, Y.; Takebayashi, T.; Koyasu, S.; Suematsu, M.; Hirose, N.; von Zglinicki, T. Inflammation, but not Telomere Length, Predicts Successful Ageing at Extreme Old Age: A Longitudinal Study of Semi-supercentenarians. EBioMedicine 2015, 2, 1549–1558. [Google Scholar] [CrossRef] [PubMed]
- Okuda, K.; Bardeguez, A.; Gardner, J.P.; Rodriguez, P.; Ganesh, V.; Kimura, M.; Skurnick, J.; Awad, G.; Aviv, A. Telomere Length in the Newborn. Pediatr. Res. 2002, 52, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Henle, E.S.; Han, Z.; Tang, N.; Rai, P.; Luo, Y.; Linn, S. Sequence-specific DNA cleavage by Fe2+-mediated fenton reactions has possible biological implications. J. Biol. Chem. 1999, 274, 962–971. [Google Scholar] [CrossRef] [PubMed]
- Petersen, S.; Saretzki, G.; von Zglinicki, T. Preferential accumulation of single-stranded regions in telomeres of human fibroblasts. Exp. Cell Res. 1998, 239, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, S.; Tada-Oikawa, S.; Kawanishi, S. Site-specific DNA damage at the GGG sequence by UVA involves acceleration of telomere shortening. Biochemistry 2001, 40, 4763–4768. [Google Scholar] [CrossRef] [PubMed]
- Wolkowitz, O.M.; Epel, E.S.; Reus, V.I.; Mellon, S.H. Depression gets old fast: Do stress and depression accelerate cell aging? Depress. Anxiety 2010, 27, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Bernal, A.; Tusell, L. Telomeres: Implications for Cancer Development. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Gilley, D.; Tanaka, H.; Herbert, B.S. Telomere dysfunction in aging and cancer. Int. J. Biochem. Cell Biol. 2005, 37, 1000–1013. [Google Scholar] [CrossRef] [PubMed]
- Picard, M.; McEwen, B.S. Psychological Stress and Mitochondria: A Systematic Review. Psychosom. Med. 2018, 80, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Yakes, F.M.; Van Houten, B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc. Natl. Acad. Sci. USA 1997, 94, 514–519. [Google Scholar] [CrossRef] [PubMed]
- Ylikallio, E.; Tyynismaa, H.; Tsutsui, H.; Ide, T.; Suomalainen, A. High mitochondrial DNA copy number has detrimental effects in mice. Hum. Mol. Genet. 2010, 19, 2695–2705. [Google Scholar] [CrossRef] [PubMed]
- Fuke, S.; Kubota-Sakashita, M.; Kasahara, T.; Shigeyoshi, Y.; Kato, T. Regional variation in mitochondrial DNA copy number in mouse brain. Biochim. Biophys. Acta 2011, 1807, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Veltri, K.L.; Espiritu, M.; Singh, G. Distinct genomic copy number in mitochondria of different mammalian organs. J. Cell. Physiol. 1990, 143, 160–164. [Google Scholar] [CrossRef] [PubMed]
- Gilley, D.; Herbert, B.S.; Huda, N.; Tanaka, H.; Reed, T. Factors impacting human telomere homeostasis and age-related disease. Mech. Ageing Dev. 2008, 129, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Yao, X.; Shen, Y. Altered mitochondrial DNA copy number contributes to human cancer risk: Evidence from an updated meta-analysis. Sci. Rep. 2016, 6, 35859. [Google Scholar] [CrossRef] [PubMed]
- Monroy-Jaramillo, N.; Dyukova, E.; Walss-Bass, C. Telomere length in psychiatric disorders: Is it more than an ageing marker? World J. Biol. Psychiatry 2017. [Google Scholar] [CrossRef] [PubMed]
- Zgheib, N.K.; Sleiman, F.; Nasreddine, L.; Nasrallah, M.; Nakhoul, N.; Isma’eel, H.; Tamim, H. Short Telomere Length is Associated with Aging, Central Obesity, Poor Sleep and Hypertension in Lebanese Individuals. Aging Dis. 2018, 9, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Joo, Y.; Choi, K.M.; Lee, Y.H.; Kim, G.; Lee, D.H.; Roh, G.S.; Kang, S.S.; Cho, G.J.; Choi, W.S.; Kim, H.J. Chronic immobilization stress induces anxiety- and depression-like behaviors and decreases transthyretin in the mouse cortex. Neurosci. Lett. 2009, 461, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Son, H.; Baek, J.H.; Go, B.S.; Jung, D.H.; Sontakke, S.B.; Chung, H.J.; Lee, D.H.; Roh, G.S.; Kang, S.S.; Cho, G.J.; et al. Glutamine has antidepressive effects through increments of glutamate and glutamine levels and glutamatergic activity in the medial prefrontal cortex. Neuropharmacology 2018, 143, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Son, H.; Jung, S.; Shin, J.H.; Kang, M.J.; Kim, H.J. Anti-Stress and Anti-Depressive Effects of Spinach Extracts on a Chronic Stress-Induced Depression Mouse Model through Lowering Blood Corticosterone and Increasing Brain Glutamate and Glutamine Levels. J. Clin. Med. 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Cawthon, R.M. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009, 37, e21. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.P.; Price, N.L.; Ling, A.J.; Moslehi, J.J.; Montgomery, M.K.; Rajman, L.; White, J.P.; Teodoro, J.S.; Wrann, C.D.; Hubbard, B.P.; et al. Declining NAD+ induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell 2013, 155, 1624–1638. [Google Scholar] [CrossRef] [PubMed]
- Callicott, R.J.; Womack, J.E. Real-time PCR assay for measurement of mouse telomeres. Comp. Med. 2006, 56, 17–22. [Google Scholar] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Cawthon, R.M. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002, 30, e47. [Google Scholar] [CrossRef] [PubMed]
- Aubert, G.; Hills, M.; Lansdorp, P.M. Telomere length measurement-caveats and a critical assessment of the available technologies and tools. Mutat. Res. 2012, 730, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Martin-Ruiz, C.M.; Baird, D.; Roger, L.; Boukamp, P.; Krunic, D.; Cawthon, R.; Dokter, M.M.; van der Harst, P.; Bekaert, S.; de Meyer, T.; et al. Reproducibility of telomere length assessment: An international collaborative study. Int. J. Epidemiol. 2015, 44, 1673–1683. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Ju, Z.; Rudolph, K.L. Telomere shortening and ageing. Z. Gerontol. Geriatr. 2007, 40, 314–324. [Google Scholar] [CrossRef] [PubMed]
- Bogenhagen, D.F. Does mtDNA nucleoid organization impact aging? Exp. Gerontol. 2010, 45, 473–477. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.W.; Turnbull, D.M. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 2005, 6, 389–402. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Wen, S.; Sun, X.; Fang, Q.; Huang, L.; Liu, S.; Li, W.; Qiu, M. Elevated Mitochondrial DNA Copy Number in Peripheral Blood and Tissue Predict the Opposite Outcome of Cancer: A Meta-Analysis. Sci. Rep. 2016, 6, 37404. [Google Scholar] [CrossRef] [PubMed]
- Lan, Q.; Lim, U.; Liu, C.S.; Weinstein, S.J.; Chanock, S.; Bonner, M.R.; Virtamo, J.; Albanes, D.; Rothman, N. A prospective study of mitochondrial DNA copy number and risk of non-Hodgkin lymphoma. Blood 2008, 112, 4247–4249. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.Y.; Liu, Y.; Yang, H.R.; Yang, H.Y.; Liu, J.X.; Ma, Y.N.; Qi, Y. Reference Intervals of Mitochondrial DNA Copy Number in Peripheral Blood for Chinese Minors and Adults. Chin. Med. J. (Engl.) 2017, 130, 2435–2440. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, I.; Izumi, T.; Boku, S.; Kimura, A.; Zhang, Y.; Mouri, K.; Okazaki, S.; Shiroiwa, K.; Takahashi, M.; Ueno, Y.; et al. Aberrant telomere length and mitochondrial DNA copy number in suicide completers. Sci. Rep. 2017, 7, 3176. [Google Scholar] [CrossRef] [PubMed]
- Cai, N.; Chang, S.; Li, Y.; Li, Q.; Hu, J.; Liang, J.; Song, L.; Kretzschmar, W.; Gan, X.; Nicod, J.; et al. Molecular Signatures of Major Depression. Curr. Biol. 2015, 25, 1146–1156. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.Y.; Huang, Y.C.; Hung, C.F. Shortened telomere length in patients with depression: A meta-analytic study. J. Psychiatr. Res. 2016, 76, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Prowse, K.R.; Greider, C.W. Developmental and tissue-specific regulation of mouse telomerase and telomere length. Proc. Natl. Acad. Sci. USA 1995, 92, 4818–4822. [Google Scholar] [CrossRef] [PubMed]
| Target | Name | Sequence | Ref. |
|---|---|---|---|
| Telomere | telg | 5′-ACACTAAGGTTTGGGTTTGGGTTTGGGTTTGGGTTAGTGT-3′ | [27] |
| (constant length) | telc | 5′-TGTTAGGTATCCCTATCCCTATCCCTATCCCTATCCCTAACA-3′ | |
| Cox1 | Cox1-F | 5′-CCCAGATATAGCATTCCCACG-3′ | [28] |
| Cox1-R | 5′-ACTGTTCATCCTGTTCCTGC-3′ | ||
| Cox2 | Cox2-F | 5′-ATAACCGAGTCGTTCTGCCAAT-3′ | [28] |
| Cox2-R | 5′-TTTCAGAGCATTGGCCATAGAA-3′ | ||
| 36b4 | 36b4d | 5′-ACTGGTCTAGGACCCGAGAAG-3′ | [29] |
| 36b4d | 5′-TCAATGGTGCCTCTGGAGATT-3′ |
| Telomere Length | STR (CTL = 1) | Slope (/Month) | Telomere Age (Actual Age = 2.5 months) |
| Leukocyte | 0.9204 | −0.02078 | 6.3 |
| Liver | 0.8968 | −0.00876 | 14.3 |
| Kidney | 0.8953 | −0.01368 | 10.2 |
| Skeletal muscle | 0.9044 | −0.01443 | 9.1 |
| mtDNAcn | STR (CTL = 1) | Slope (/Month) | mtDNA Age (Actual Age = 2.5 months) |
| Prefrontal cortex | 0.8460 | −0.02780 | 8.0 |
| Aorta | 1.4990 | 0.06077 | 10.7 |
| Spleen | 1.2170 | 0.02637 | 10.7 |
| Skeletal muscle | 0.7116 | −0.02034 | 16.7 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baek, J.H.; Son, H.; Jeong, Y.-H.; Park, S.W.; Kim, H.J. Chronological Aging Standard Curves of Telomere Length and Mitochondrial DNA Copy Number in Twelve Tissues of C57BL/6 Male Mouse. Cells 2019, 8, 247. https://doi.org/10.3390/cells8030247
Baek JH, Son H, Jeong Y-H, Park SW, Kim HJ. Chronological Aging Standard Curves of Telomere Length and Mitochondrial DNA Copy Number in Twelve Tissues of C57BL/6 Male Mouse. Cells. 2019; 8(3):247. https://doi.org/10.3390/cells8030247
Chicago/Turabian StyleBaek, Ji Hyeong, Hyeonwi Son, Young-Hoon Jeong, Sang Won Park, and Hyun Joon Kim. 2019. "Chronological Aging Standard Curves of Telomere Length and Mitochondrial DNA Copy Number in Twelve Tissues of C57BL/6 Male Mouse" Cells 8, no. 3: 247. https://doi.org/10.3390/cells8030247
APA StyleBaek, J. H., Son, H., Jeong, Y.-H., Park, S. W., & Kim, H. J. (2019). Chronological Aging Standard Curves of Telomere Length and Mitochondrial DNA Copy Number in Twelve Tissues of C57BL/6 Male Mouse. Cells, 8(3), 247. https://doi.org/10.3390/cells8030247






