Emerging Roles of Ephexins in Physiology and Disease
Abstract
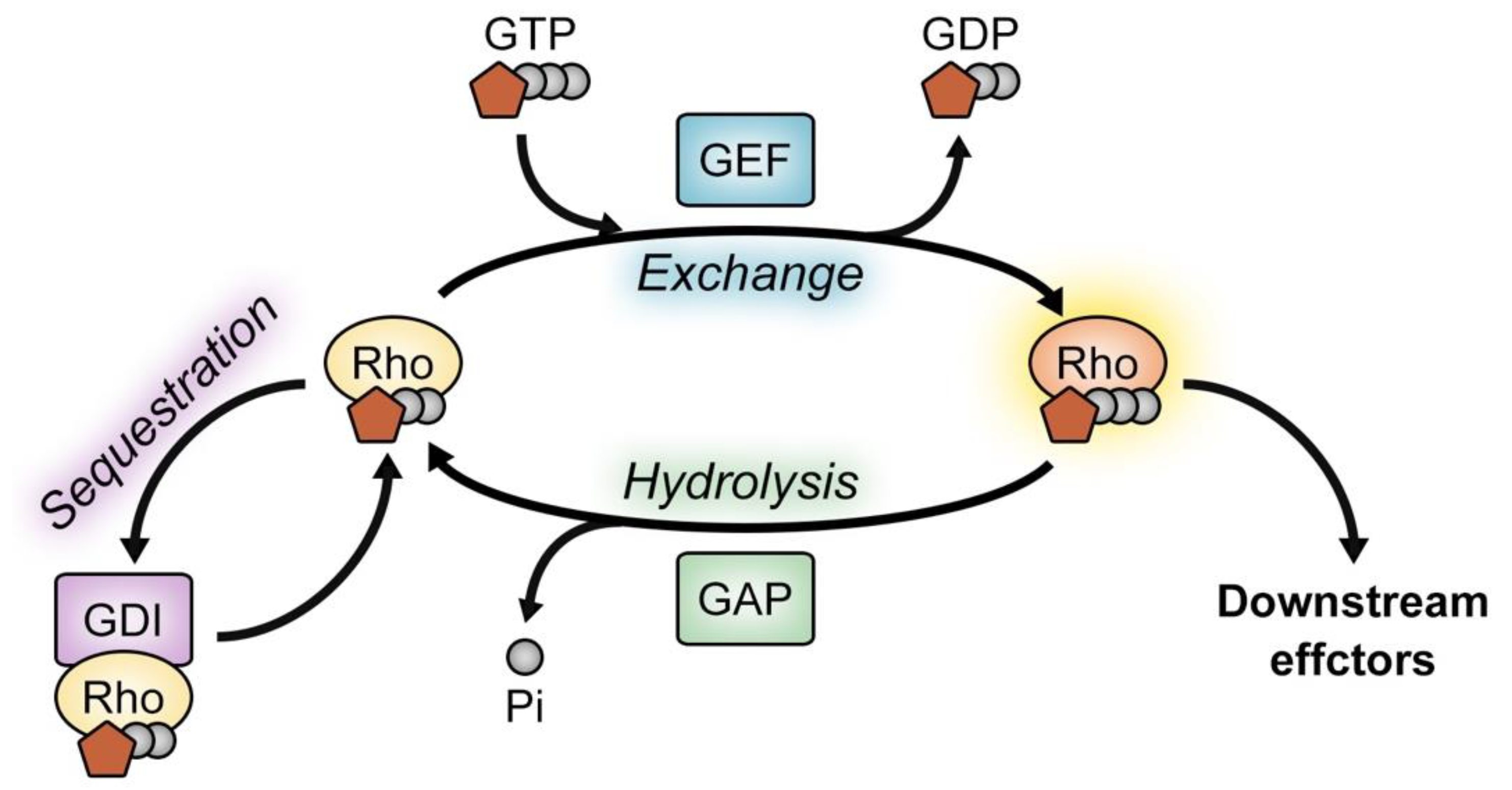
1. Introduction
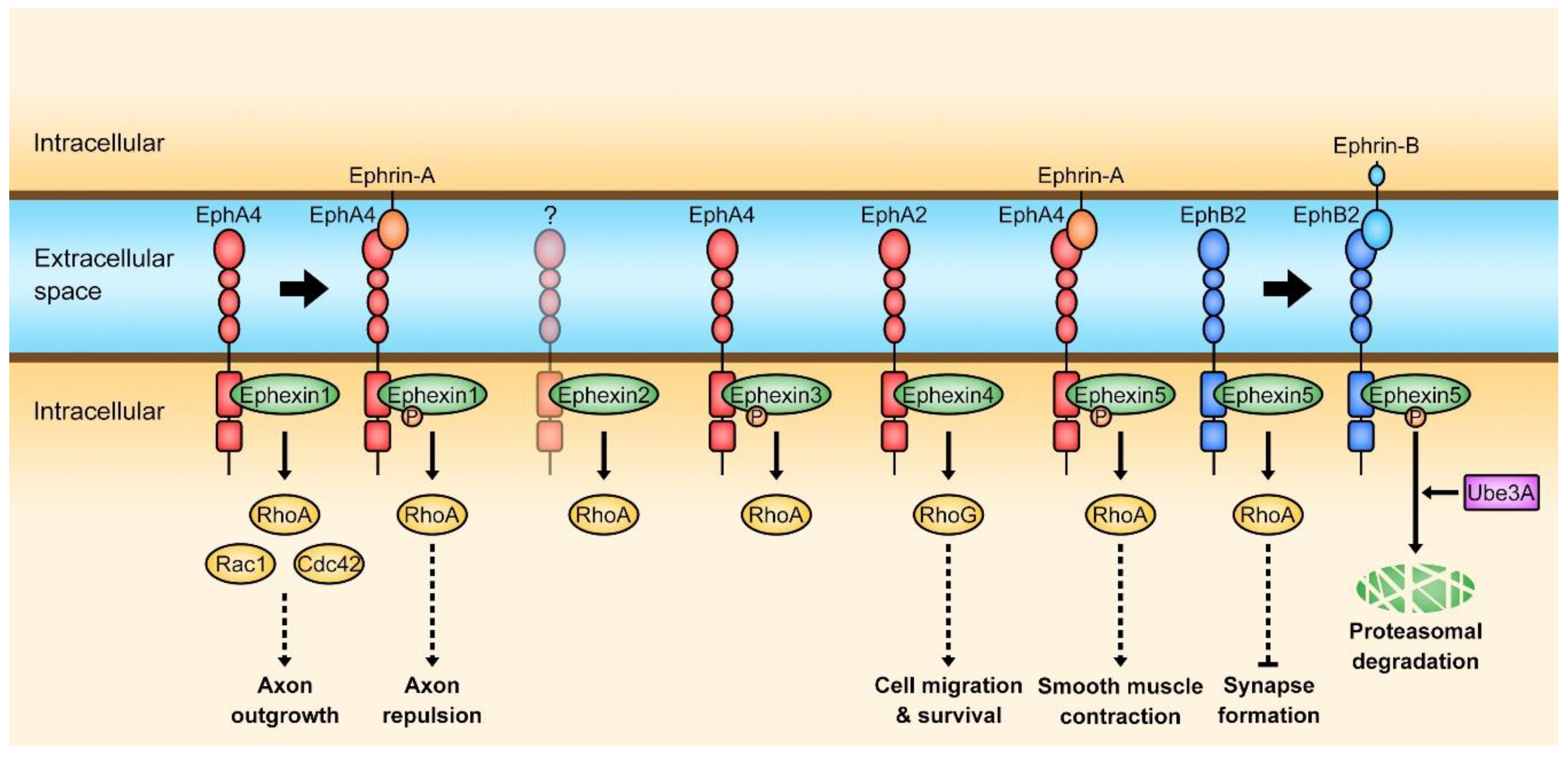
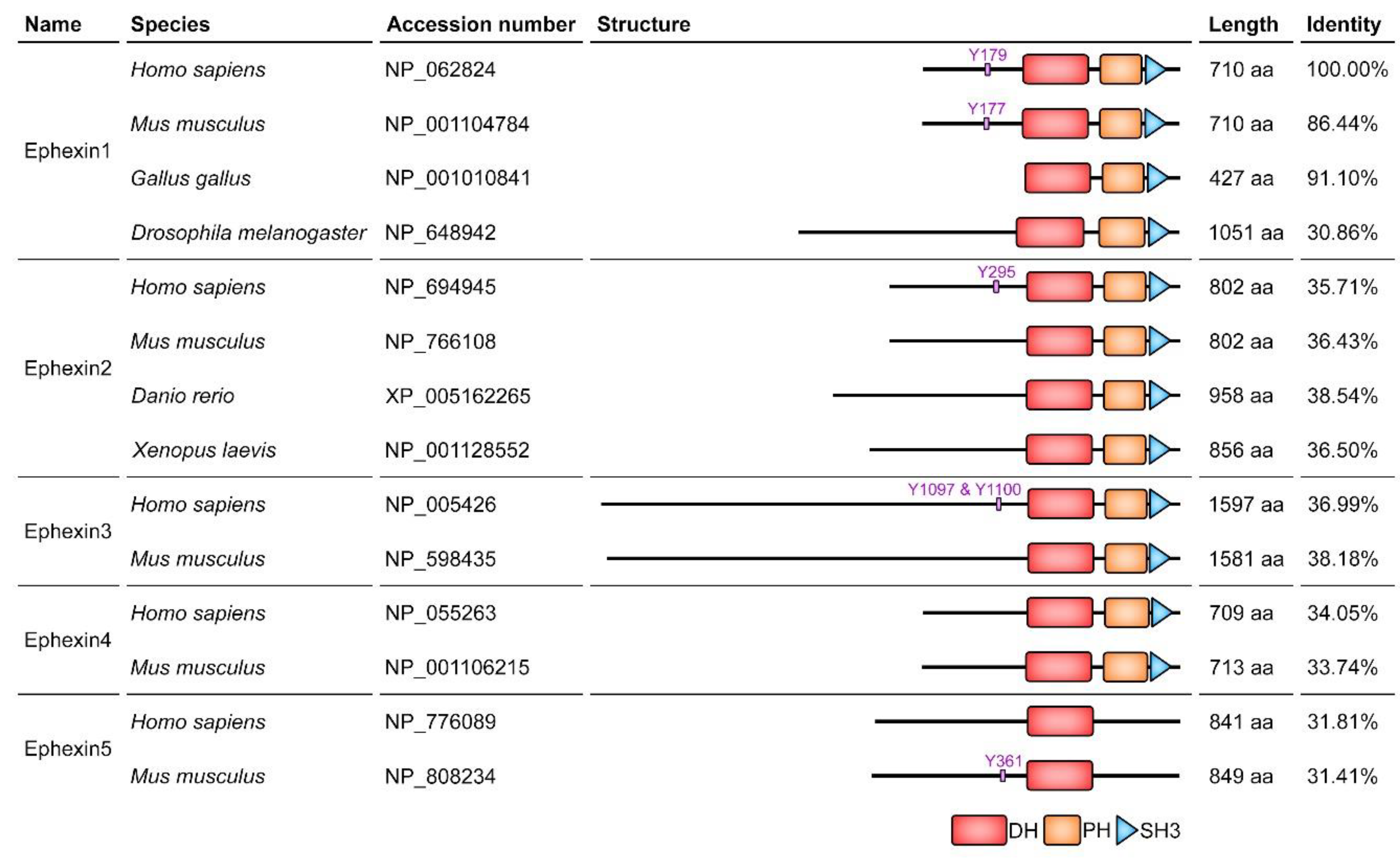
2. Ephexin Family
2.1. Ephexin1
2.2. Ephexin2
2.3. Ephexin3
2.4. Ephexin4
2.5. Ephexin5
3. Targeting Ephexins in Diseases
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van Aelst, L.; D’Souza-Schorey, C. Rho gtpases and signaling networks. Genes. Dev. 1997, 11, 2295–2322. [Google Scholar] [CrossRef] [PubMed]
- Bishop, A.L.; Hall, A. Rho gtpases and their effector proteins. Biochem. J. 2000, 348, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Hall, A. Rho family gtpases. Biochem. Soc. Trans. 2012, 40, 1378–1382. [Google Scholar] [CrossRef] [PubMed]
- Rojas, A.M.; Fuentes, G.; Rausell, A.; Valencia, A. The ras protein superfamily: Evolutionary tree and role of conserved amino acids. J. Cell Biol. 2012, 196, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Etienne-Manneville, S.; Hall, A. Rho gtpases in cell biology. Nature 2002, 420, 629–635. [Google Scholar] [CrossRef]
- Boettner, B.; Van Aelst, L. The role of rho gtpases in disease development. Gene 2002, 286, 155–174. [Google Scholar] [CrossRef]
- Sahai, E.; Marshall, C.J. Rho-gtpases and cancer. Nat. Rev. Cancer 2002, 2, 133–142. [Google Scholar] [CrossRef]
- Hodge, R.G.; Ridley, A.J. Regulating Rho GTPases and their regulators. Nat. Rev. Mol. Cell Biol. 2016, 17, 496–510. [Google Scholar] [CrossRef]
- Rossman, K.L.; Der, C.J.; Sondek, J. Gef means go: Turning on rho gtpases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 2005, 6, 167–180. [Google Scholar] [CrossRef]
- Bos, J.L.; Rehmann, H.; Wittinghofer, A. Gefs and gaps: Critical elements in the control of small g proteins. Cell 2007, 129, 865–877. [Google Scholar] [CrossRef]
- Eva, A.; Vecchio, G.; Rao, C.D.; Tronick, S.R.; Aaronson, S.A. The predicted dbl oncogene product defines a distinct class of transforming proteins. Proc. Natl. Acad. Sci. USA 1988, 85, 2061–2065. [Google Scholar] [CrossRef] [PubMed]
- Hart, M.J.; Eva, A.; Evans, T.; Aaronson, S.A.; Cerione, R.A. Catalysis of guanine nucleotide exchange on the cdc42hs protein by the dbl oncogene product. Nature 1991, 354, 311–314. [Google Scholar] [CrossRef] [PubMed]
- Fort, P.; Blangy, A. The evolutionary landscape of dbl-like rhogef families: Adapting eukaryotic cells to environmental signals. Genome Biol. Evol. 2017, 9, 1471–1486. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, M.; Dvorsky, R.; Ahmadian, M.R. Deciphering the molecular and functional basis of dbl family proteins: A novel systematic approach toward classification of selective activation of the rho family proteins. J. Biol. Chem. 2013, 288, 4486–4500. [Google Scholar] [CrossRef] [PubMed]
- Goicoechea, S.M.; Awadia, S.; Garcia-Mata, R. I’m coming to GEF you: Regulation of rhogefs during cell migration. Cell Adh. Migr. 2014, 8, 535–549. [Google Scholar] [CrossRef] [PubMed]
- Gadea, G.; Blangy, A. Dock-family exchange factors in cell migration and disease. Eur. J. Cell Biol. 2014, 93, 466–477. [Google Scholar] [CrossRef] [PubMed]
- Sahin, M.; Greer, P.L.; Lin, M.Z.; Poucher, H.; Eberhart, J.; Schmidt, S.; Wright, T.M.; Shamah, S.M.; O’Connell, S.; Cowan, C.W.; et al. Eph-dependent tyrosine phosphorylation of ephexin1 modulates growth cone collapse. Neuron 2005, 46, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Ogita, H.; Kunimoto, S.; Kamioka, Y.; Sawa, H.; Masuda, M.; Mochizuki, N. Epha4-mediated rho activation via vsm-rhogef expressed specifically in vascular smooth muscle cells. Circ. Res. 2003, 93, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Shamah, S.M.; Lin, M.Z.; Goldberg, J.L.; Estrach, S.; Sahin, M.; Hu, L.; Bazalakova, M.; Neve, R.L.; Corfas, G.; Debant, A.; et al. Epha receptors regulate growth cone dynamics through the novel guanine nucleotide exchange factor ephexin. Cell 2001, 105, 233–244. [Google Scholar] [CrossRef]
- Yohe, M.E.; Rossman, K.L.; Gardner, O.S.; Karnoub, A.E.; Snyder, J.T.; Gershburg, S.; Graves, L.M.; Der, C.J.; Sondek, J. Auto-inhibition of the dbl family protein tim by an n-terminal helical motif. J. Biol. Chem. 2007, 282, 13813–13823. [Google Scholar] [CrossRef]
- Hiramoto-Yamaki, N.; Takeuchi, S.; Ueda, S.; Harada, K.; Fujimoto, S.; Negishi, M.; Katoh, H. Ephexin4 and epha2 mediate cell migration through a rhog-dependent mechanism. J. Cell Biol. 2010, 190, 461–477. [Google Scholar] [CrossRef] [PubMed]
- Margolis, S.S.; Salogiannis, J.; Lipton, D.M.; Mandel-Brehm, C.; Wills, Z.P.; Mardinly, A.R.; Hu, L.; Greer, P.L.; Bikoff, J.B.; Ho, H.Y.; et al. Ephb-mediated degradation of the rhoa gef ephexin5 relieves a developmental brake on excitatory synapse formation. Cell 2010, 143, 442–455. [Google Scholar] [CrossRef]
- Kullander, K.; Klein, R. Mechanisms and functions of eph and ephrin signalling. Nat. Rev. Mol. Cell Biol. 2002, 3, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Egea, J.; Nissen, U.V.; Dufour, A.; Sahin, M.; Greer, P.; Kullander, K.; Mrsic-Flogel, T.D.; Greenberg, M.E.; Kiehn, O.; Vanderhaeghen, P.; et al. Regulation of epha 4 kinase activity is required for a subset of axon guidance decisions suggesting a key role for receptor clustering in eph function. Neuron 2005, 47, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.Y.; Chen, Y.; Sahin, M.; Zhao, X.S.; Shi, L.; Bikoff, J.B.; Lai, K.O.; Yung, W.H.; Fu, A.K.; Greenberg, M.E.; et al. Cdk5 regulates epha4-mediated dendritic spine retraction through an ephexin1-dependent mechanism. Nat. Neurosci. 2007, 10, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Defourny, J.; Poirrier, A.L.; Lallemend, F.; Mateo Sanchez, S.; Neef, J.; Vanderhaeghen, P.; Soriano, E.; Peuckert, C.; Kullander, K.; Fritzsch, B.; et al. Ephrin-a5/epha4 signalling controls specific afferent targeting to cochlear hair cells. Nat. Commun. 2013, 4, 1438. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.J.; Chang, M.Y.; Chou, S.Y.; Huang, C.C.; Chuang, J.Y.; Hsu, T.I.; Chang, H.F.; Wu, Y.H.; Wu, C.C.; Morales, D.; et al. Ephexin1 is required for eph-mediated limb trajectory of spinal motor axons. J. Neurosci. 2018, 38, 2043–2056. [Google Scholar] [CrossRef] [PubMed]
- Fiore, L.; Medori, M.; Spelzini, G.; Carreno, C.O.; Carri, N.G.; Sanchez, V.; Scicolone, G. Regulation of axonal epha4 forward signaling is involved in the effect of epha3 on chicken retinal ganglion cell axon growth during retinotectal mapping. Exp. Eye Res. 2018, 178, 46–60. [Google Scholar] [CrossRef]
- Sardana, J.; Organisti, C.; Grunwald Kadow, I.C. Eph receptor effector ephexin mediates olfactory dendrite targeting in drosophila. Dev. Neurobiol. 2018, 78, 873–888. [Google Scholar] [CrossRef]
- Shah, K.; Rossie, S. Tale of the good and the bad cdk5: Remodeling of the actin cytoskeleton in the brain. Mol. Neurobiol. 2018, 55, 3426–3438. [Google Scholar] [CrossRef]
- Frank, C.A.; Pielage, J.; Davis, G.W. A presynaptic homeostatic signaling system composed of the eph receptor, ephexin, cdc42, and cav2.1 calcium channels. Neuron 2009, 61, 556–569. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Butt, B.; Ip, F.C.; Dai, Y.; Jiang, L.; Yung, W.H.; Greenberg, M.E.; Fu, A.K.; Ip, N.Y. Ephexin1 is required for structural maturation and neurotransmission at the neuromuscular junction. Neuron 2010, 65, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Fu, A.K.; Ip, N.Y. Multiple roles of the rho gef ephexin1 in synapse remodeling. Commun. Integr. Biol. 2010, 3, 622–624. [Google Scholar] [CrossRef] [PubMed]
- Kusuhara, S.; Fukushima, Y.; Fukuhara, S.; Jakt, L.M.; Okada, M.; Shimizu, Y.; Hata, M.; Nishida, K.; Negi, A.; Hirashima, M.; et al. Arhgef15 promotes retinal angiogenesis by mediating vegf-induced cdc42 activation and potentiating rhoj inactivation in endothelial cells. PLoS ONE 2012, 7, e45858. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Park, B.; Kim, G.; Kim, K.; Pak, J.; Kim, K.; Ye, M.B.; Park, S.G.; Park, D. Arhgef16, a novel elmo1 binding partner, promotes clearance of apoptotic cells via rhog-dependent rac1 activation. Biochim. Biophys. Acta 2014, 1843, 2438–2447. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Lee, J.; Lee, S.A.; Moon, H.; Park, B.; Kim, D.; Joo, Y.E.; Park, D. Intermolecular steric inhibition of ephexin4 is relieved by elmo1. Sci. Rep. 2017, 7, 4404. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Lee, J.; Moon, H.; Lee, S.A.; Kim, D.; Yang, S.; Lee, D.H.; Lee, G.; Park, D. The intermolecular interaction of ephexin4 leads to autoinhibition by impeding binding of rhog. Cells 2018, 7, 211. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, H.; Yasumoto, M.; Ogasawara, S.; Akiba, J.; Kitasato, Y.; Nakayama, M.; Naito, Y.; Ishida, Y.; Okabe, Y.; Yasunaga, M.; et al. Arhgef15 overexpression worsens the prognosis in patients with pancreatic ductal adenocarcinoma through enhancing the motility and proliferative activity of the cancer cells. Mol. Cancer 2016, 15, 32. [Google Scholar] [CrossRef]
- Zhou, J.; Tao, Y.; Peng, C.; Gu, P.; Wang, W. Mir-503 regulates metastatic function through rho guanine nucleotide exchanger factor 19 in hepatocellular carcinoma. J. Surg. Res. 2014, 188, 129–136. [Google Scholar] [CrossRef]
- Li, Y.; Ye, Z.; Chen, S.; Pan, Z.; Zhou, Q.; Li, Y.Z.; Shuai, W.D.; Kuang, C.M.; Peng, Q.H.; Shi, W.; et al. Arhgef19 interacts with braf to activate mapk signaling during the tumorigenesis of non-small cell lung cancer. Int. J. Cancer 2018, 142, 1379–1391. [Google Scholar] [CrossRef]
- Xie, X.; Chang, S.W.; Tatsumoto, T.; Chan, A.M.; Miki, T. TIM, a dbl-related protein, regulates cell shape and cytoskeletal organization in a rho-dependent manner. Cell. Signal. 2005, 17, 461–471. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Wu, W.; Wang, H.; Liao, K.; Zhang, W.; Xiong, G.; Wu, F.; Meng, G.; Yang, K. Co-expression of rho guanine nucleotide exchange factor 5 and src associates with poor prognosis of patients with resected non-small cell lung cancer. Oncol. Rep. 2013, 30, 2864–2870. [Google Scholar] [CrossRef] [PubMed]
- Komiya, Y.; Onodera, Y.; Kuroiwa, M.; Nomimura, S.; Kubo, Y.; Nam, J.M.; Kajiwara, K.; Nada, S.; Oneyama, C.; Sabe, H.; et al. The rho guanine nucleotide exchange factor arhgef5 promotes tumor malignancy via epithelial-mesenchymal transition. Oncogenesis 2016, 5, e258. [Google Scholar] [CrossRef] [PubMed]
- Debily, M.A.; Camarca, A.; Ciullo, M.; Mayer, C.; El Marhomy, S.; Ba, I.; Jalil, A.; Anzisi, A.; Guardiola, J.; Piatier-Tonneau, D. Expression and molecular characterization of alternative transcripts of the arhgef5/tim oncogene specific for human breast cancer. Hum. Mol. Genet. 2004, 13, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Kuroiwa, M.; Oneyama, C.; Nada, S.; Okada, M. The guanine nucleotide exchange factor arhgef5 plays crucial roles in src-induced podosome formation. J. Cell Sci. 2011, 124, 1726–1738. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Wu, W.; Yang, K.; Tan, D.; Tang, M.; Liu, H.; Wu, T.; Zhang, S.; Wang, H. Rho guanine nucleotide exchange factor 5 increases lung cancer cell tumorigenesis via mmp-2 and cyclin d1 upregulation. Mol. Cancer Ther. 2015, 14, 1671–1679. [Google Scholar] [CrossRef]
- Mihira, H.; Suzuki, H.I.; Akatsu, Y.; Yoshimatsu, Y.; Igarashi, T.; Miyazono, K.; Watabe, T. Tgf-beta-induced mesenchymal transition of ms-1 endothelial cells requires smad-dependent cooperative activation of rho signals and mrtf-a. J. Biochem. 2012, 151, 145–156. [Google Scholar] [CrossRef]
- Cheng, S.L.; Liu, R.H.; Sheu, J.N.; Chen, S.T.; Sinchaikul, S.; Tsay, G.J. Toxicogenomics of kojic acid on gene expression profiling of a375 human malignant melanoma cells. Biol. Pharm. Bull. 2006, 29, 655–669. [Google Scholar] [CrossRef]
- Bralten, L.B.; Nouwens, S.; Kockx, C.; Erdem, L.; Hoogenraad, C.C.; Kros, J.M.; Moorhouse, M.J.; Sillevis Smitt, P.A.; van der Spek, P.; van Ijcken, W.; et al. Absence of common somatic alterations in genes on 1p and 19q in oligodendrogliomas. PLoS ONE 2011, 6, e22000. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Hiramoto-Yamaki, N.; Negishi, M.; Katoh, H. Ephexin4 and epha2 mediate resistance to anoikis through rhog and phosphatidylinositol 3-kinase. Exp. Cell Res. 2011, 317, 1701–1713. [Google Scholar] [CrossRef]
- Kawai, H.; Kobayashi, M.; Hiramoto-Yamaki, N.; Harada, K.; Negishi, M.; Katoh, H. Ephexin4-mediated promotion of cell migration and anoikis resistance is regulated by serine 897 phosphorylation of epha2. FEBS Open Bio 2013, 3, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Akada, M.; Harada, K.; Negishi, M.; Katoh, H. Ephb6 promotes anoikis by modulating epha2 signaling. Cell. Signal. 2014, 26, 2879–2884. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Wang, Y.; Xu, L.; Chen, L.; Cheng, M.; Shi, W.; Xiong, H.; Zalli, D.; Luo, S. Gli2 promotes cell proliferation and migration through transcriptional activation of arhgef16 in human glioma cells. J. Exp. Clin. Cancer Res. 2018, 37, 247. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, N.R.; Theodosiou, A.M.; Nesbit, M.A.; Campbell, L.; Tandle, A.T.; Saranath, D.; Davies, K.E. Characterization of ngef, a novel member of the dbl family of genes expressed predominantly in the caudate nucleus. Genomics 2000, 65, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Suzuki, H.; Yokoo, T.; Tada-Iida, K.; Kihara, R.; Miura, M.; Watanabe, K.; Sone, H.; Shimano, H.; Toyoshima, H.; et al. Wgef is a novel rhogef expressed in intestine, liver, heart, and kidney. Biochem. Biophys. Res. Commun. 2004, 324, 1053–1058. [Google Scholar] [CrossRef]
- Hampson, L.; He, X.T.; Oliver, A.W.; Hadfield, J.A.; Kemp, T.; Butler, J.; McGown, A.; Kitchener, H.C.; Hampson, I.N. Analogues of y27632 increase gap junction communication and suppress the formation of transformed nih3t3 colonies. Br. J. Cancer 2009, 101, 829–839. [Google Scholar] [CrossRef] [PubMed]
- Oliver, A.W.; He, X.; Borthwick, K.; Donne, A.J.; Hampson, L.; Hampson, I.N. The hpv16 e6 binding protein tip-1 interacts with arhgef16, which activates cdc42. Br. J. Cancer 2011, 104, 324–331. [Google Scholar] [CrossRef]
- Egea, J.; Klein, R. Bidirectional eph-ephrin signaling during axon guidance. Trends Cell Biol. 2007, 17, 230–238. [Google Scholar] [CrossRef]
- Harboe, M.; Torvund-Jensen, J.; Kjaer-Sorensen, K.; Laursen, L.S. Ephrin-a1-epha4 signaling negatively regulates myelination in the central nervous system. Glia 2018, 66, 934–950. [Google Scholar] [CrossRef]
- Yohe, M.E.; Rossman, K.; Sondek, J. Role of the c-terminal sh3 domain and n-terminal tyrosine phosphorylation in regulation of tim and related dbl-family proteins. Biochemistry 2008, 47, 6827–6839. [Google Scholar] [CrossRef]
- Zhang, J.C.; Yao, W.; Qu, Y.; Nakamura, M.; Dong, C.; Yang, C.; Ren, Q.; Ma, M.; Han, M.; Shirayama, Y.; et al. Increased epha4-ephexin1 signaling in the medial prefrontal cortex plays a role in depression-like phenotype. Sci. Rep. 2017, 7, 7133. [Google Scholar] [CrossRef] [PubMed]
- Rosas, O.R.; Figueroa, J.D.; Torrado, A.I.; Rivera, M.; Santiago, J.M.; Konig-Toro, F.; Miranda, J.D. Expression and activation of ephexin is altered after spinal cord injury. Dev. Neurobiol. 2011, 71, 595–607. [Google Scholar] [CrossRef] [PubMed]
- Rosas, O.R.; Torrado, A.I.; Santiago, J.M.; Rodriguez, A.E.; Salgado, I.K.; Miranda, J.D. Long-term treatment with pp2 after spinal cord injury resulted in functional locomotor recovery and increased spared tissue. Neural. Regen. Res. 2014, 9, 2164–2173. [Google Scholar] [PubMed]
- Fan, R.; Enkhjargal, B.; Camara, R.; Yan, F.; Gong, L.; Yao, S.; Tang, J.; Chen, Y.; Zhang, J.H. Critical role of epha4 in early brain injury after subarachnoid hemorrhage in rat. Exp. Neurol. 2017, 296, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Tanegashima, K.; Zhao, H.; Dawid, I.B. Wgef activates rho in the wnt-pcp pathway and controls convergent extension in xenopus gastrulation. EMBO J. 2008, 27, 606–617. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.K.; Canny, S.G.; Jang, C.W.; Cho, K.; Ji, H.; Wagner, D.S.; Jones, E.A.; Habas, R.; McCrea, P.D. Pronephric tubulogenesis requires daam1-mediated planar cell polarity signaling. J. Am. Soc. Nephrol. 2011, 22, 1654–1664. [Google Scholar] [CrossRef]
- Wang, Z.; Kumamoto, Y.; Wang, P.; Gan, X.; Lehmann, D.; Smrcka, A.V.; Cohn, L.; Iwasaki, A.; Li, L.; Wu, D. Regulation of immature dendritic cell migration by rhoa guanine nucleotide exchange factor arhgef5. J. Biol. Chem. 2009, 284, 28599–28606. [Google Scholar] [CrossRef] [PubMed]
- Kanwal, M.; Ding, X.J.; Ma, Z.H.; Li, L.W.; Wang, P.; Chen, Y.; Huang, Y.C.; Cao, Y. Characterization of germline mutations in familial lung cancer from the chinese population. Gene 2018, 641, 94–104. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Tan, D.L.; Liu, H.X.; Lv, F.L.; Wu, W. The auto-inhibitory state of rho guanine nucleotide exchange factor arhgef5/tim can be relieved by targeting its sh3 domain with rationally designed peptide aptamers. Biochimie 2015, 111, 10–18. [Google Scholar] [CrossRef]
- Huang, O.; Wu, D.; Xie, F.; Lin, L.; Wang, X.; Jiang, M.; Li, Y.; Chen, W.; Shen, K.; Hu, X. Targeting rho guanine nucleotide exchange factor arhgef5/tim with auto-inhibitory peptides in human breast cancer. Amino Acids 2015, 47, 1239–1246. [Google Scholar] [CrossRef] [PubMed]
- Matsukawa, T.; Morita, K.; Omizu, S.; Kato, S.; Koriyama, Y. Mechanisms of rhoa inactivation and cdc42 and rac1 activation during zebrafish optic nerve regeneration. Neurochem. Int. 2018, 112, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, K.S.; Lorenz, U. Engulfment of apoptotic cells: Signals for a good meal. Nat. Rev. Immunol. 2007, 7, 964–974. [Google Scholar] [CrossRef] [PubMed]
- Dalva, M.B. Ephecting excitatory synapse development. Cell 2010, 143, 341–342. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, A.M.; Lambert, J.T.; Parajuli, L.K.; Vivas, O.; Park, D.K.; Stein, I.S.; Jahncke, J.N.; Greenberg, M.E.; Margolis, S.S.; Zito, K. A dual role for the rhogef ephexin5 in regulation of dendritic spine outgrowth. Mol. Cell Neurosci. 2017, 80, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Risau, W. Mechanisms of angiogenesis. Nature 1997, 386, 671–674. [Google Scholar] [CrossRef] [PubMed]
- Lamalice, L.; Houle, F.; Jourdan, G.; Huot, J. Phosphorylation of tyrosine 1214 on vegfr2 is required for vegf-induced activation of cdc42 upstream of sapk2/p38. Oncogene 2004, 23, 434–445. [Google Scholar] [CrossRef]
- Fukushima, Y.; Okada, M.; Kataoka, H.; Hirashima, M.; Yoshida, Y.; Mann, F.; Gomi, F.; Nishida, K.; Nishikawa, S.; Uemura, A. Sema3e-plexind1 signaling selectively suppresses disoriented angiogenesis in ischemic retinopathy in mice. J. Clin. Invest. 2011, 121, 1974–1985. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Garcia, R.; Iruela-Arispe, M.L.; Reyes-Cruz, G.; Vazquez-Prado, J. Endothelial rhogefs: A systematic analysis of their expression profiles in vegf-stimulated and tumor endothelial cells. Vascul. Pharmacol. 2015, 74, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Veeramah, K.R.; Johnstone, L.; Karafet, T.M.; Wolf, D.; Sprissler, R.; Salogiannis, J.; Barth-Maron, A.; Greenberg, M.E.; Stuhlmann, T.; Weinert, S.; et al. Exome sequencing reveals new causal mutations in children with epileptic encephalopathies. Epilepsia 2013, 54, 1270–1281. [Google Scholar] [CrossRef]
- Sell, G.L.; Schaffer, T.B.; Margolis, S.S. Reducing expression of synapse-restricting protein ephexin5 ameliorates alzheimer’s-like impairment in mice. J. Clin. Invest. 2017, 127, 1646–1650. [Google Scholar] [CrossRef]
| Member | Aliases | Expression | GEF Specificity | Interacting Receptors | References |
|---|---|---|---|---|---|
| Ephexin1 | Arhgef27, Ngef, Ephexin | Brain, spinal cord | RhoA, Rac1, Cdc42 | EphA4 | [19,54] |
| Ephexin2 | Arhgef19, Wgef | Liver, kidney, heart, intestine | RhoA | – 1 | [55] |
| Ephexin3 | Arhgef5, Tim | Liver, kidney, colon, trachea, prostate, pancreas | RhoA, Rac1, Cdc42 2 | EphA4 | [20,41,44] |
| Ephexin4 | Arhgef16 | – 3 | RhoG, Cdc42 | EphA2 | [21,56,57] |
| Ephexin5 | Arhgef15, Vsm-RhoGEF | Brain, vascular smooth muscles (liver, kidney, heart, spleen) | RhoA, Rac1, Cdc42 4 | EphA4, EphB2 | [18,22,34] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.; Lee, S.-A.; Park, D. Emerging Roles of Ephexins in Physiology and Disease. Cells 2019, 8, 87. https://doi.org/10.3390/cells8020087
Kim K, Lee S-A, Park D. Emerging Roles of Ephexins in Physiology and Disease. Cells. 2019; 8(2):87. https://doi.org/10.3390/cells8020087
Chicago/Turabian StyleKim, Kwanhyeong, Sang-Ah Lee, and Daeho Park. 2019. "Emerging Roles of Ephexins in Physiology and Disease" Cells 8, no. 2: 87. https://doi.org/10.3390/cells8020087
APA StyleKim, K., Lee, S.-A., & Park, D. (2019). Emerging Roles of Ephexins in Physiology and Disease. Cells, 8(2), 87. https://doi.org/10.3390/cells8020087





