Structural Biology and Electron Microscopy of the Autophagy Molecular Machinery
Abstract
:1. Introduction
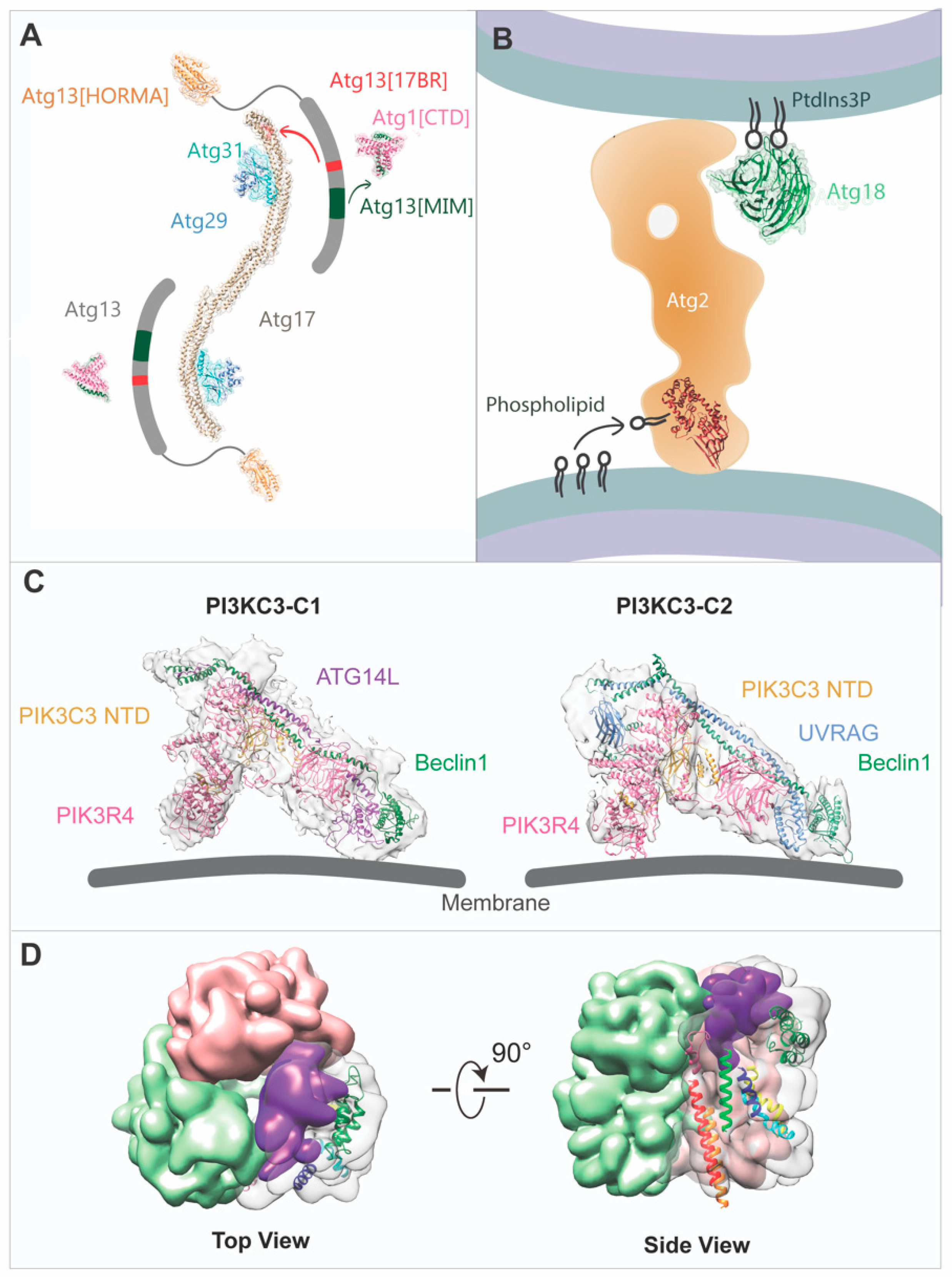
2. The ULK1/ATG1 Complex
3. The PI3KC3 Complex
4. The ATG2-ATG18/WIPI Complex
5. The Transmembrane Protein ATG9
6. Concluding Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of Cells and Tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef] [Green Version]
- Bento, C.F.; Renna, M.; Ghislat, G.; Puri, C.; Ashkenazi, A.; Vicinanza, M.; Menzies, F.M.; Rubinsztein, D.C. Mammalian Autophagy: How Does It Work? Annu. Rev. Biochem. 2016, 85, 685–713. [Google Scholar] [CrossRef]
- Wen, X.; Klionsky, D.J. An overview of macroautophagy in yeast. J. Mol. Biol. 2016, 428, 1681–1699. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Bassham, D.C. Autophagy: Pathways for Self-Eating in Plant Cells. Annu. Rev. Plant. Biol. 2012, 63, 215–237. [Google Scholar] [CrossRef] [Green Version]
- Bozhkov, P.V. Plant autophagy: Mechanisms and functions. J. Exp. Bot. 2018, 69, 1281–1285. [Google Scholar] [CrossRef] [Green Version]
- Janse van Rensburg, H.C.; Van den Ende, W.; Signorelli, S. Autophagy in Plants: Both a Puppet and a Puppet Master of Sugars. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Noda, N.N.; Inagaki, F. Mechanisms of Autophagy. Annu. Rev. Biophys. 2015, 44, 101–122. [Google Scholar] [CrossRef]
- Axe, E.L.; Walker, S.A.; Manifava, M.; Chandra, P.; Roderick, H.L.; Habermann, A.; Griffiths, G.; Ktistakis, N.T. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell. Biol. 2008, 182, 685–701. [Google Scholar] [CrossRef] [Green Version]
- Hayashi-Nishino, M.; Fujita, N.; Noda, T.; Yamaguchi, A.; Yoshimori, T.; Yamamoto, A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 2009, 11, 1433–1437. [Google Scholar] [CrossRef]
- Ravikumar, B.; Moreau, K.; Jahreiss, L.; Puri, C.; Rubinsztein, D.C. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 2010, 12, 747–757. [Google Scholar] [CrossRef] [Green Version]
- Ge, L.; Melville, D.; Zhang, M.; Schekman, R. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife 2013, 2. [Google Scholar] [CrossRef]
- Hamasaki, M.; Furuta, N.; Matsuda, A.; Nezu, A.; Yamamoto, A.; Fujita, N.; Oomori, H.; Noda, T.; Haraguchi, T.; Hiraoka, Y.; et al. Autophagosomes form at ER-mitochondria contact sites. Nature 2013, 495, 389–393. [Google Scholar] [CrossRef]
- Xie, Z.P.; Klionsky, D.J. Autophagosome formation: Core machinery and adaptations. Nat. Cell Biol. 2007, 9, 1102–1109. [Google Scholar] [CrossRef]
- Gomez, R.E.; Joubes, J.; Valentin, N.; Batoko, H.; Satiat-Jeunemaitre, B.; Bernard, A. Lipids in membrane dynamics during autophagy in plants. J. Exp. Bot. 2018, 69, 1287–1299. [Google Scholar] [CrossRef]
- Feng, Y.C.; He, D.; Yao, Z.Y.; Klionsky, D.J. The machinery of macroautophagy. Cell Res. 2014, 24, 24–41. [Google Scholar] [CrossRef] [Green Version]
- Hurley, J.H.; Young, L.N. Mechanisms of Autophagy Initiation. Annu. Rev. Biochem. 2017, 86, 225–244. [Google Scholar] [CrossRef]
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The Role of Atg Proteins in Autophagosome Formation. Annu. Rev. Cell Dev. Biol. 2011, 27, 107–132. [Google Scholar] [CrossRef]
- Soto-Burgos, J.; Zhuang, X.H.; Jiang, L.W.; Bassham, D.C. Dynamics of Autophagosome Formation. Plant. Physiol. 2018, 176, 219–229. [Google Scholar] [CrossRef] [Green Version]
- Suttangkakul, A.; Li, F.Q.; Chung, T.; Vierstra, R.D. The ATG1/ATG13 Protein Kinase Complex Is Both a Regulator and a Target of Autophagic Recycling in Arabidopsis. Plant. Cell 2011, 23, 3761–3779. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, X.H.; Chung, K.P.; Cui, Y.; Lin, W.L.; Gao, C.J.; Kang, B.H.; Jiang, L.W. ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E426–E435. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, X.H.; Chung, K.P.; Luo, M.Q.; Jiang, L.W. Autophagosome Biogenesis and the Endoplasmic Reticulum: A Plant Perspective. Trends Plant. Sci. 2018, 23, 677–692. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Ye, H.; Cui, Y.; Jiang, L. AtSec62 is critical for plant development and is involved in ER-phagy in Arabidopsis thaliana. J. Integr. Plant. Biol. 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Y.; Zhuang, X.H.; Shen, J.B.; Gao, J.; Jiang, L. Organelle biogenesis and function in plants (in Chinese). Sci. Sin. Vitae 2019, 49, 1–16. [Google Scholar]
- Islam, M.R.; Kwak, J.W.; Lee, J.S.; Hong, S.W.; Khan, M.R.I.; Lee, Y.; Lee, Y.; Lee, S.W.; Hwang, I. Cost-effective production of tag-less recombinant protein in Nicotiana benthamiana. Plant. Biotechnol. J. 2019, 17, 1094–1105. [Google Scholar] [CrossRef] [Green Version]
- Staub, J.M.; Garcia, B.; Graves, J.; Hajdukiewicz, P.T.J.; Hunter, P.; Nehra, N.; Paradkar, V.; Schlittler, M.; Carroll, J.A.; Spatola, L.; et al. High-yield production of a human therapeutic protein in tobacco chloroplasts. Nat. Biotechnol. 2000, 18, 333–338. [Google Scholar] [CrossRef]
- Sohn, E.J.; Lee, Y.; Park, N.; Park, M.; Kim, N.H.; Park, S.; Min, K.; Gu, S.; Park, Y.; Song, J.; et al. Development of Plant-produced E2 Protein for Use as a Green Vaccine Against Classical Swine Fever Virus. J. Plant. Biol. 2018, 61, 241–252. [Google Scholar] [CrossRef]
- Fernandez-Leiro, R.; Scheres, S.H. Unravelling biological macromolecules with cryo-electron microscopy. Nature 2016, 537, 339–346. [Google Scholar] [CrossRef]
- Cheng, Y.F. Single-Particle Cryo-EM at Crystallographic Resolution. Cell 2015, 161, 450–457. [Google Scholar] [CrossRef] [Green Version]
- Merk, A.; Bartesaghi, A.; Banerjee, S.; Falconieri, V.; Rao, P.; Davis, M.I.; Pragani, R.; Boxer, M.B.; Earl, L.A.; Milne, J.L.S.; et al. Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery. Cell 2016, 165, 1698–1707. [Google Scholar] [CrossRef] [Green Version]
- Ragusa, M.J.; Stanley, R.E.; Hurley, J.H. Architecture of the Atg17 Complex as a Scaffold for Autophagosome Biogenesis. Cell 2012, 151, 1501–1512. [Google Scholar] [CrossRef] [Green Version]
- Jao, C.C.; Ragusa, M.J.; Stanley, R.E.; Hurley, J.H. A HORMA domain in Atg13 mediates PI 3-kinase recruitment in autophagy. Proc. Natl. Acad. Sci. USA 2013, 110, 5486–5491. [Google Scholar] [CrossRef] [Green Version]
- Chew, L.H.; Setiaputra, D.; Klionsky, D.J.; Yip, C.K. Structural characterization of the Saccharomyces cerevisiae autophagy regulatory complex Atg17-Atg31-Atg29. Autophagy 2013, 9, 1467–1474. [Google Scholar] [CrossRef] [Green Version]
- Fujioka, Y.; Suzuki, S.W.; Yamamoto, H.; Kondo-Kakuta, C.; Kimura, Y.; Hirano, H.; Akada, R.; Inagaki, F.; Ohsumi, Y.; Noda, N.N. Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat. Struct. Mol. Biol. 2014, 21, 513–521. [Google Scholar] [CrossRef]
- Suzuki, H.; Kaizuka, T.; Mizushima, N.; Noda, N.N. Structure of the Atg101-Atg13 complex reveals essential roles of Atg101 in autophagy initiation. Nat. Struct. Mo.L Biol. 2015, 22, 572–580. [Google Scholar] [CrossRef]
- Chew, L.H.; Lu, S.; Liu, X.; Li, F.K.; Yu, A.Y.; Klionsky, D.J.; Dong, M.Q.; Yip, C.K. Molecular interactions of the Saccharomyces cerevisiae Atg1 complex provide insights into assembly and regulatory mechanisms. Autophagy 2015, 11, 891–905. [Google Scholar] [CrossRef] [Green Version]
- Kofinger, J.; Ragusa, M.J.; Lee, I.H.; Hummer, G.; Hurley, J.H. Solution Structure of the Atg1 Complex: Implications for the Architecture of the Phagophore Assembly Site. Structure 2015, 23, 809–818. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, H.; Fujioka, Y.; Suzuki, S.W.; Noshiro, D.; Suzuki, H.; Kondo-Kakuta, C.; Kimura, Y.; Hirano, H.; Ando, T.; Noda, N.N.; et al. The Intrinsically Disordered Protein Atg13 Mediates Supramolecular Assembly of Autophagy Initiation Complexes. Dev. Cell 2016, 38, 86–99. [Google Scholar] [CrossRef] [Green Version]
- Nanji, T.; Liu, X.; Chew, L.H.; Li, F.K.; Biswas, M.; Yu, Z.Q.; Lu, S.; Dong, M.Q.; Du, L.L.; Klionsky, D.J.; et al. Conserved and unique features of the fission yeast core Atg1 complex. Autophagy 2017, 13, 2018–2027. [Google Scholar] [CrossRef]
- Lazarus, M.B.; Novotny, C.J.; Shokat, K.M. Structure of the Human Autophagy Initiating Kinase ULK1 in Complex with Potent Inhibitors. ACS Chem. Biol. 2015, 10, 257–261. [Google Scholar] [CrossRef] [Green Version]
- Lazarus, M.B.; Shokat, K.M. Discovery and structure of a new inhibitor scaffold of the autophagy initiating kinase ULK1. Bioorgan. Med. Chem. 2015, 23, 5483–5488. [Google Scholar] [CrossRef] [Green Version]
- Qi, S.Q.; Kim, D.J.; Stjepanovic, G.; Hurley, J.H. Structure of the Human Atg13-Atg101 HORMA Heterodimer: An Interaction Hub within the ULK1 Complex. Structure 2015, 23, 1848–1857. [Google Scholar] [CrossRef] [Green Version]
- Michel, M.; Schwarten, M.; Decker, C.; Nagel-Steger, L.; Willbold, D.; Weiergraber, O.H. The mammalian autophagy initiator complex contains 2 HORMA domain proteins. Autophagy 2015, 11, 2300–2308. [Google Scholar] [CrossRef] [Green Version]
- Kim, B.W.; Jin, Y.; Kim, J.; Kim, J.H.; Jung, J.; Kang, S.; Kim, I.Y.; Kim, J.; Cheong, H.; Song, H.K. The C-terminal region of ATG101 bridges ULK1 and PtdIns3K complex in autophagy initiation. Autophagy 2018, 14, 2104–2116. [Google Scholar] [CrossRef] [Green Version]
- Turco, E.; Witt, M.; Abert, C.; Bock-Bierbaum, T.; Su, M.Y.; Trapannone, R.; Sztacho, M.; Danieli, A.; Shi, X.; Zaffagnini, G.; et al. FIP200 Claw Domain Binding to p62 Promotes Autophagosome Formation at Ubiquitin Condensates. Mol. Cell 2019, 74, 330–346 e11. [Google Scholar] [CrossRef] [Green Version]
- Chaikuad, A.; Koschade, S.E.; Stolz, A.; Zivkovic, K.; Pohl, C.; Shaid, S.; Ren, H.; Lambert, L.J.; Cosford, N.D.P.; Brandts, C.H.; et al. Conservation of structure, function and inhibitor binding in UNC-51-like kinase 1 and 2 (ULK1/2). Biochem. J. 2019, 476, 875–887. [Google Scholar] [CrossRef]
- Shi, X.Y.; Adam, L.; Wang, C.; Young, L.N.; Youle, R.J. ULK complex organization in autophagy by a C-shaped FIP200 N-terminal domain dimer. BioRxiv 2019. [Google Scholar]
- Watanabe, Y.; Kobayashi, T.; Yamamoto, H.; Hoshida, H.; Akada, R.; Inagaki, F.; Ohsumi, Y.; Noda, N.N. Structure-based Analyses Reveal Distinct Binding Sites for Atg2 and Phosphoinositides in Atg18. J. Biol. Chem. 2012, 287, 31681–31690. [Google Scholar] [CrossRef] [Green Version]
- Baskaran, S.; Ragusa, M.J.; Boura, E.; Hurley, J.H. Two-Site Recognition of Phosphatidylinositol 3-Phosphate by PROPPINs in Autophagy. Mol. Cell 2012, 47, 339–348. [Google Scholar] [CrossRef] [Green Version]
- Krick, R.; Busse, R.A.; Scacioc, A.; Stephan, M.; Janshoff, A.; Thumm, M.; Kuhnel, K. Structural and functional characterization of the two phosphoinositide binding sites of PROPPINs, a beta-propeller protein family. Proc. Natl. Acad. Sci. USA 2012, 109, E2042–E2049. [Google Scholar] [CrossRef] [Green Version]
- Osawa, T.; Kotani, T.; Kawaoka, T.; Hirata, E.; Suzuki, K.; Nakatogawa, H.; Ohsumi, Y.; Noda, N.N. Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat. Struct. Mol. Biol. 2019, 26, 281–288. [Google Scholar] [CrossRef]
- Zheng, J.X.; Li, Y.; Ding, Y.H.; Liu, J.J.; Zhang, M.J.; Dong, M.Q.; Wang, H.W.; Yu, L. Architecture of the ATG2B-WDR45 complex and an aromatic Y/HF motif crucial for complex formation. Autophagy 2017, 13, 1870–1883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chowdhury, S.; Otomo, C.; Leitner, A.; Ohashi, K.; Aebersold, R.; Lander, G.C.; Otomo, T. Insights into autophagosome biogenesis from structural and biochemical analyses of the ATG2A-WIPI4 complex. Proc. Natl. Acad. Sci. USA 2018, 115, E9792–E9801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valverde, D.P.; Yu, S.; Boggavarapu, V.; Kumar, N.; Lees, J.A.; Walz, T.; Reinisch, K.M.; Melia, T.J. ATG2 transports lipids to promote autophagosome biogenesis. J. Cell Biol. 2019, 218, 1787–1798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heenan, E.J.; Vanhooke, J.L.; Temple, B.R.; Betts, L.; Sondek, J.E.; Dohlman, H.G. Structure and Function of Vps15 in the Endosomal G Protein Signaling Pathway. Biochemistry 2009, 48, 6390–6401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noda, N.N.; Kobayashi, T.; Adachi, W.; Fujioka, Y.; Ohsumi, Y.; Inagaki, F. Structure of the Novel C-terminal Domain of Vacuolar Protein Sorting 30/Autophagy-related Protein 6 and Its Specific Role in Autophagy. J. Biol. Chem. 2012, 287, 16256–16266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rostislavleva, K.; Soler, N.; Ohashi, Y.; Zhang, L.; Pardon, E.; Burke, J.E.; Masson, G.R.; Johnson, C.; Steyaert, J.; Ktistakis, N.T.; et al. Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science 2015, 350, aac7365. [Google Scholar] [CrossRef] [Green Version]
- Ohashi, Y.; Soler, N.; Garcia Ortegon, M.; Zhang, L.; Kirsten, M.L.; Perisic, O.; Masson, G.R.; Burke, J.E.; Jakobi, A.J.; Apostolakis, A.A.; et al. Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex. Autophagy 2016, 12, 2129–2144. [Google Scholar] [CrossRef]
- Miller, S.; Tavshanjian, B.; Oleksy, A.; Perisic, O.; Houseman, B.T.; Shokat, K.M.; Williams, R.L. Shaping Development of Autophagy Inhibitors with the Structure of the Lipid Kinase Vps34. Science 2010, 327, 1638–1642. [Google Scholar] [CrossRef]
- Oberstein, A.; Jeffrey, P.D.; Shi, Y.G. Crystal structure of the Bcl-X-L-beclin 1 peptide complex-Beclin 1 is a novel BH3-only protein. J. Biol. Chem. 2007, 282, 13123–13132. [Google Scholar] [CrossRef] [Green Version]
- Feng, W.; Huang, S.Y.; Wu, H.; Zhang, M.J. Molecular basis of Bcl-xL’s target recognition versatility revealed by the structure of Bcl-xL in complex with the BH3 domain of beclin-1. J. Mol. Biol. 2007, 372, 223–235. [Google Scholar] [CrossRef]
- Ku, B.; Woo, J.S.; Liang, C.; Lee, K.H.; Hong, H.S.; E, X.; Kim, K.S.; Jung, J.U.; Oh, B.H. Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine gamma-herpesvirus 68. PLoS Pathog. 2008, 4. [Google Scholar]
- Sinha, S.; Colbert, C.L.; Becker, N.; Wei, Y.; Levine, B. Molecular basis of the regulation of Beclin 1-dependent autophagy by the gamma-herpesvirus 68 Bcl-2 homolog M11. Autophagy 2008, 4, 989–997. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; He, L.; Che, K.H.; Funderburk, S.F.; Pan, L.; Pan, N.; Zhang, M.; Yue, Z.; Zhao, Y. Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat. Commun. 2012, 3, 662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, W.; Choi, W.; Hu, W.; Mi, N.; Guo, Q.; Ma, M.; Liu, M.; Tian, Y.; Lu, P.; Wang, F.L.; et al. Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein. Cell Res. 2012, 22, 473–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dowdle, W.E.; Nyfeler, B.; Murphy, L. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Mol. Biol. Cell 2014, 25. [Google Scholar] [CrossRef] [PubMed]
- Ronan, B.; Flamand, O.; Vescovi, L.; Dureuil, C.; Durand, L.; Fassy, F.; Bachelot, M.F.; Lamberton, A.; Mathieu, M.; Bertrand, T.; et al. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat. Chem. Biol. 2014, 10, 1013. [Google Scholar] [CrossRef] [PubMed]
- Baskaran, S.; Carlson, L.A.; Stjepanovic, G.; Young, L.N.; Kim, D.J.; Grob, P.; Stanley, R.E.; Nogales, E.; Hurley, J.H. Architecture and Dynamics of the Autophagic Phosphatidylinositol 3-Kinase Complex. Elife 2014, 3. [Google Scholar] [CrossRef]
- Mei, Y.; Glover, K.; Su, M.F.; Sinha, S.C. Conformational flexibility of BECN1: Essential to its key role in autophagy and beyond. Protein Sci. 2016, 25, 1767–1785. [Google Scholar] [CrossRef] [Green Version]
- Mei, Y.; Su, M.; Sanishvili, R.; Chakravarthy, S.; Colbert, C.L.; Sinha, S.C. Identification of BECN1 and ATG14 Coiled-Coil Interface Residues That Are Important for Starvation-Induced Autophagy. Biochemistry 2016, 55, 4239–4253. [Google Scholar] [CrossRef] [Green Version]
- Young, L.N.; Cho, K.; Lawrence, R.; Zoncu, R.; Hurley, J.H. Dynamics and architecture of the NRBF2-containing phosphatidylinositol 3-kinase complex I of autophagy. Proc. Natl. Acad. Sci. USA 2016, 113, 8224–8229. [Google Scholar] [CrossRef] [Green Version]
- Ma, M.; Liu, J.J.; Li, Y.; Huang, Y.; Ta, N.; Chen, Y.; Fu, H.; Ye, M.D.; Ding, Y.; Huang, W.; et al. Cryo-EM structure and biochemical analysis reveal the basis of the functional difference between human PI3KC3-C1 and -C2. Cell Res. 2017, 27, 989–1001. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; He, Y.; Qiu, X.; Yang, W.; Liu, W.; Li, X.; Li, Y.; Shen, H.M.; Wang, R.; Yue, Z.; et al. Targeting the potent Beclin 1-UVRAG coiled-coil interaction with designed peptides enhances autophagy and endolysosomal trafficking. Proc. Natl. Acad. Sci. USA 2018, 115, E5669–E5678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, L.N.; Goerdeler, F.; Hurley, J.H. Structural pathway for allosteric activation of the autophagic PI 3-kinase complex I. Proc. Natl. Acad. Sci. USA 2019, 116, 21508–21513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, L.T.F.; Yu, C.; Wong, J.S.K.; Lo, H.S.; Benlekbir, S.; Jiang, L.; Lau, W.C.Y. Subnanometer resolution cryo-EM structure of Arabidopsis thaliana ATG9. Autophagy 2019, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Scheres, S.H.W. Processing of Structurally Heterogeneous Cryo-EM Data in RELION. Method Enzymol. 2016, 579, 125–157. [Google Scholar]
- Hurley, J.H.; Nogales, E. Next-generation electron microscopy in autophagy research. Curr. Opin. Struc. Biol. 2016, 41, 211–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, H.; Osawa, T.; Fujioka, Y.; Noda, N.N. Structural biology of the core autophagy machinery. Curr. Opin. Struc. Biol. 2017, 43, 10–17. [Google Scholar] [CrossRef] [Green Version]
- Stjepanovic, G.; Baskaran, S.; Lin, M.G.; Hurley, J.H. Vps34 Kinase Domain Dynamics Regulate the Autophagic PI 3-Kinase Complex. Mol. Cell 2017, 67, 528–534. [Google Scholar] [CrossRef]
- Chang, C.M.; Young, L.N.; Morris, K.L.; von Bulow, S.; Schoneberg, J.; Yamamoto-Imoto, H.; Oe, Y.; Yamamoto, K.; Nakamura, S.; Stjepanovic, G.; et al. Bidirectional Control of Autophagy by BECN1 BARA Domain Dynamics. Mol. Cell 2019, 73, 339–356. [Google Scholar] [CrossRef] [Green Version]
- Kamada, Y.; Funakoshi, T.; Shintani, T.; Nagano, K.; Ohsumi, M.; Ohsumi, Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 2000, 150, 1507–1513. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egan, D.F.; Shackelford, D.B.; Mihaylova, M.M.; Gelino, S.; Kohnz, R.A.; Mair, W.; Vasquez, D.S.; Joshi, A.; Gwinn, D.M.; Taylor, R.; et al. Phosphorylation of ULK1 (hATG1) by AMP-Activated Protein Kinase Connects Energy Sensing to Mitophagy. Science 2011, 331, 456–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganley, I.G.; Lam, D.H.; Wang, J.R.; Ding, X.J.; Chen, S.; Jiang, X.J. ULK1 center dot ATG13 center dot FIP200 Complex Mediates mTOR Signaling and Is Essential for Autophagy. J. Biol. Chem. 2009, 284, 12297–12305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosokawa, N.; Hara, T.; Kaizuka, T.; Kishi, C.; Takamura, A.; Miura, Y.; Iemura, S.; Natsume, T.; Takehana, K.; Yamada, N.; et al. Nutrient-dependent mTORC1 Association with the ULK1-Atg13-FIP200 Complex Required for Autophagy. Mol. Biol. Cell 2009, 20, 1981–1991. [Google Scholar] [CrossRef] [Green Version]
- Kabeya, Y.; Noda, N.N.; Fujioka, Y.; Suzuki, K.; Inagaki, F.; Ohsumi, Y. Characterization of the Atg17-Atg29-Atg31 complex specifically required for starvation-induced autophagy in Saccharomyces cerevisiae. Biochem. Bioph. Res. Commun. 2009, 389, 612–615. [Google Scholar] [CrossRef]
- Li, F.; Vierstra, R.D. Arabidopsis ATG11, a scaffold that links the ATG1-ATG13 kinase complex to general autophagy and selective mitophagy. Autophagy 2014, 10, 1466–1467. [Google Scholar] [CrossRef] [Green Version]
- Russell, R.C.; Tian, Y.; Yuan, H.; Park, H.W.; Chang, Y.Y.; Kim, J.; Kim, H.; Neufeld, T.P.; Dillin, A.; Guan, K.L. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell. Biol. 2013, 15, 741–750. [Google Scholar] [CrossRef] [Green Version]
- Morris, D.H.; Yip, C.K.; Shi, Y.; Chait, B.T.; Wang, Q.J. Beclin 1-Vps34 Complex Architecture: Understanding the Nuts and Bolts of Therapeutic Targets. Front. Biol. (Beijing) 2015, 10, 398–426. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, S.W.; Yamamoto, H.; Oikawa, Y.; Kondo-Kakuta, C.; Kimura, Y.; Hirano, H.; Ohsumi, Y. Atg13 HORMA domain recruits Atg9 vesicles during autophagosome formation. Proc. Natl. Acad. Sci. USA 2015, 112, 3350–3355. [Google Scholar] [CrossRef] [Green Version]
- Reggiori, F.; Tucker, K.A.; Stromhaug, P.E.; Klionsky, D.J. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev. Cell 2004, 6, 79–90. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Contento, A.L.; Bassham, D.C. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant. J. 2005, 42, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Kotani, T.; Kirisako, H.; Koizumi, M.; Ohsumi, Y.; Nakatogawa, H. The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc. Natl. Acad. Sci. USA 2018, 115, 10363–10368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez-Sanchez, R.; Rose, J.; Guimaraes, R.; Mari, M.; Papinski, D.; Rieter, E.; Geerts, W.J.; Hardenberg, R.; Kraft, C.; Ungermann, C.; et al. Atg9 establishes Atg2-dependent contact sites between the endoplasmic reticulum and phagophores. J. Cell Biol. 2018, 217, 2743–2763. [Google Scholar] [CrossRef] [PubMed]
- Shin, K.D.; Lee, H.N.; Chung, T. A Revised Assay for Monitoring Autophagic Flux in Arabidopsis thaliana Reveals Involvement of AUTOPHAGY-RELATED9 in Autophagy. Mol. Cells 2014, 37, 399–405. [Google Scholar] [CrossRef] [Green Version]
- Hara, T.; Mizushima, N. Role of ULK-FIP200 complex in mammalian autophagy FIP200, a counterpart of yeast Atg 17? Autophagy 2009, 5, 85–87. [Google Scholar] [CrossRef] [Green Version]
- Hosokawa, N.; Sasaki, T.; Iemura, S.; Natsume, T.; Hara, T.; Mizushima, N. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 2009, 5, 973–979. [Google Scholar] [CrossRef] [Green Version]
- Noda, N.N.; Mizushima, N. Atg101: Not Just an Accessory Subunit in the Autophagy-initiation Complex. Cell Struct. Funct. 2016, 41, 13–20. [Google Scholar] [CrossRef] [Green Version]
- Young, P.G.; Passalacqua, M.J.; Chappell, K.; Llinas, R.J.; Bartel, B. A facile forward-genetic screen for Arabidopsis autophagy mutants reveals twenty-one loss-of-function mutations disrupting six ATG genes. Autophagy 2019, 15, 941–959. [Google Scholar] [CrossRef] [Green Version]
- Volinia, S.; Dhand, R.; Vanhaesebroeck, B.; Macdougall, L.; Stein, R.; Zvelebil, M.J.; Domin, J.; Panaretou, C.; Waterfield, M.D. Human Phosphatidylinositol 3-Kinase Complex Related to the Yeast Vps34p-Vps15p Protein Sorting System. Embo, J. 1995, 14, 3339–3348. [Google Scholar] [CrossRef]
- Stephens, L.; Smrcka, A.; Cooke, F.T.; Jackson, T.R.; Sternweis, P.C.; Hawkins, P.T. A Novel Phosphoinositide-3 Kinase-Activity in Myeloid-Derived Cells Is Activated by G-Protein Beta-Gamma-Subunits. Cell 1994, 77, 83–93. [Google Scholar] [CrossRef]
- Kihara, A.; Noda, T.; Ishihara, N.; Ohsumi, Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 2001, 152, 519–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Q.M.; Fan, W.L.; Chen, K.L.; Ding, X.J.; Chen, S.; Zhong, Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. USA 2008, 105, 19211–19216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsunaga, K.; Saitoh, T.; Tabata, K.; Omori, H.; Satoh, T.; Kurotori, N.; Maejima, I.; Shirahama-Noda, K.; Ichimura, T.; Isobe, T.; et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell. Biol. 2009, 11, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.Y.; Lee, J.S.; Inn, K.S.; Gack, M.U.; Li, Q.L.; Roberts, E.A.; Vergne, I.; Deretic, V.; Feng, P.H.; Akazawa, C.; et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 2008, 10, 776–787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, S.S.; Ni, D.J.; Ma, B.Y.; Lee, J.H.; Zhang, T.; Ghozalli, I.; Pirooz, S.D.; Zhao, Z.; Bharatham, N.; Li, B.H.; et al. PtdIns(3)P-bound UVRAG coordinates Golgi-ER retrograde and Atg9 transport by differential interactions with the ER tether and the beclin 1 complex. Nat. Cell. Biol. 2013, 15, 1206–1219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.H.; Kwon, C.; Lee, J.H.; Chung, T. Genes for plant Autophagy: Functions and interactions. Mol. Cells 2012, 34, 413–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slessareva, J.E.; Routt, S.M.; Temple, B.; Bankaitis, V.A.; Dohlman, H.G. Activation of the phosphatidylinositol 3-kinase Vps34 by a G protein alpha subunit at the endosome. Cell 2006, 126, 191–203. [Google Scholar] [CrossRef] [Green Version]
- Mei, Y.; Su, M.F.; Soni, G.; Salem, S.; Colbert, C.L.; Sinha, S.C. Intrinsically disordered regions in autophagy proteins. Proteins 2014, 82, 565–578. [Google Scholar] [CrossRef] [Green Version]
- Glover, K.; Li, Y.; Mukhopadhyay, S.; Leuthner, Z.; Chakravarthy, S.; Colbert, C.L.; Sinha, S.C. Structural transitions in conserved, ordered Beclin 1 domains essential to regulating autophagy. J. Biol. Chem. 2017, 292, 16235–16248. [Google Scholar] [CrossRef] [Green Version]
- Nakatogawa, H.; Ichimura, Y.; Ohsumi, Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 2007, 130, 165–178. [Google Scholar] [CrossRef] [Green Version]
- Geng, J.F.; Klionsky, D.J. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. Embo Rep. 2008, 9, 859–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaufmann, A.; Beier, V.; Franquelim, H.G.; Wollert, T. Molecular Mechanism of Autophagic Membrane-Scaffold Assembly and Disassembly. Cell 2014, 156, 469–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klionsky, D.J.; Schulman, B.A. Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nat. Struct. Mol. Biol. 2014, 21, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Velikkakath, A.K.G.; Nishimura, T.; Oita, E.; Ishihara, N.; Mizushima, N. Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol. Biol. Cell 2012, 23, 896–909. [Google Scholar] [CrossRef] [PubMed]
- Proikas-Cezanne, T.; Waddell, S.; Gaugel, A.; Frickey, T.; Lupas, A.; Nordheim, A. WIPI-1 alpha (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy. Oncogene 2004, 23, 9314–9325. [Google Scholar] [CrossRef] [Green Version]
- Krick, R.; Henke, S.; Tolstrup, J.; Thumm, M. Dissecting the localization and function of Atg18, Atg21 and Ygr223c. Autophagy 2008, 4, 896–910. [Google Scholar] [CrossRef] [Green Version]
- Graef, M.; Friedman, J.R.; Graham, C.; Babu, M.; Nunnari, J. ER exit sites are physical and functional core autophagosome biogenesis components. Mol. Biol. Cell 2013, 24, 2918–2931. [Google Scholar] [CrossRef]
- Suzuki, K.; Akioka, M.; Kondo-Kakuta, C.; Yamamoto, H.; Ohsumi, Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J. Cell Sci. 2013, 126, 2534–2544. [Google Scholar] [CrossRef] [Green Version]
- Obara, K.; Sekito, T.; Niimi, K.; Ohsumi, Y. The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J. Biol. Chem. 2008, 283, 23972–23980. [Google Scholar] [CrossRef] [Green Version]
- Baskaran, S.; Ragusa, M.J.; Hurley, J.H. How Atg18 and the WIPIs sense phosphatidylinositol 3-phosphate. Autophagy 2012, 8, 1851–1852. [Google Scholar] [CrossRef] [Green Version]
- Young, A.R.J.; Chan, E.Y.W.; Hu, X.W.; Koch, R.; Crawshaw, S.G.; High, S.; Hailey, D.W.; Lippincott-Schwartz, J.; Tooze, S.A. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 2006, 119, 3888–3900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mari, M.; Griffith, J.; Rieter, E.; Krishnappa, L.; Klionsky, D.J.; Reggiori, F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J. Cell. Biol. 2010, 190, 1005–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orsi, A.; Razi, M.; Dooley, H.C.; Robinson, D.; Weston, A.E.; Collinson, L.M.; Tooze, S.A. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol. Biol. Cell 2012, 23, 1860–1873. [Google Scholar] [CrossRef]
- Yamamoto, H.; Kakuta, S.; Watanabe, T.M.; Kitamura, A.; Sekito, T.; Kondo-Kakuta, C.; Ichikawa, R.; Kinjo, M.; Ohsumi, Y. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J. Cell Biol. 2012, 198, 219–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noda, T. Autophagy in the context of the cellular membrane-trafficking system: The enigma of Atg9 vesicles. Biochem. Soc. T. 2017, 45, 1323–1331. [Google Scholar] [CrossRef] [Green Version]
- Karanasios, E.; Walker, S.A.; Okkenhaug, H.; Manifava, M.; Hummel, E.; Zimmermann, H.; Ahmed, Q.; Domart, M.C.; Collinson, L.; Ktistakis, N.T. Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef]
- Rao, Y.J.; Perna, M.G.; Hofmann, B.; Beier, V.; Wollert, T. The Atg1-kinase complex tethers Atg9-vesicles to initiate autophagy. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef]
- He, C.C.; Baba, M.; Cao, Y.; Klionsky, D.J. Self-Interaction Is Critical for Atg9 Transport and Function at the Phagophore Assembly Site during Autophagy. Mol. Biol. Cell 2008, 19, 5506–5516. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.Q.; Ma, K.L.; Gao, R.Z.; Mu, C.L.; Chen, L.B.; Liu, Q.Q.; Luo, Q.; Feng, D.; Zhu, Y.S.; Chen, Q. Regulation of mATG9 trafficking by Src- and ULK1-mediated phosphorylation in basal and starvation-induced autophagy. Cell Res. 2017, 27, 184–201. [Google Scholar] [CrossRef] [Green Version]
- Papinski, D.; Schuschnig, M.; Reiter, W.; Wilhelm, L.; Barnes, C.A.; Maiolica, A.; Hansmann, I.; Pfaffenwimmer, T.; Kijanska, M.; Stoffel, I.; et al. Early Steps in Autophagy Depend on Direct Phosphorylation of Atg9 by the Atg1 Kinase. Mol. Cell 2014, 53, 471–483. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Cao, W.H.; He, Y.L.; Zhao, Q.; Wakazaki, M.; Zhuang, X.H.; Gao, J.Y.; Zeng, Y.L.; Gao, C.J.; Ding, Y.; et al. A whole-cell electron tomography model of vacuole biogenesis in Arabidopsis root cells. Nat. Plants 2019, 5, 95–105. [Google Scholar] [CrossRef] [PubMed]
| Complex | Component | Origin | Method | Resolution (Å) | Year | References |
|---|---|---|---|---|---|---|
| ATG1 complex | Atg17-Atg31-Atg29 | Yeast | X-ray | 3.05 | 2012 | [30] |
| Atg13 HORMA | Yeast (Lt) | X-ray | 2.3 | 2013 | [31] | |
| Atg17-Atg31-Atg29 | Yeast | Negative stain | 37 | 2013 | [32] | |
| C-terminal region of Atg1(MIT)-ATG13MIM | Yeast (Km) | X-ray | 2.2 | 2014 | [33] | |
| Atg17-Atg29-Atg31-Atg13(17BR) | Yeast (Lt) | X-ray | 3.2 | 2014 | [33] | |
| Atg101-Atg13 | Yeast (Sp) | X-ray | 3 | 2015 | [34] | |
| Atg1 complex (Atg17-Atg31-Atg29 and Atg17-Atg31-Atg29-Atg1[CTD]-Atg13[CTD]) | Yeast | Negative stain | / | 2015 | [35] | |
| Atg1-Atg13 and Atg17-Atg31-Atg29 subcomplexes and the Atg1 complex | Yeast (Kl) | SAXS | / | 2015 | [36] | |
| Atg17–Atg29–Atg31-Atg13(17BR)-Atg13(17LR) | Yeast (Lt) | X-ray | 3.2 | 2016 | [37] | |
| Atg17 | Yeast (Sp) | Negative stain | / | 2017 | [38] | |
| Kinase domain of ULK1 with inhibitor | Mammal | X-ray | 1.88 | 2015 | [39] | |
| Kinase domain of ULK1 with inhibitor | Mammal | X-ray | 1.74 | 2015 | [40] | |
| ATG13 HORMA-ATG101 HORMA | Mammal | X-ray | 2.2 | 2015 | [41] | |
| ATG101 | Mammal | X-ray | 1.9 | 2015 | [42] | |
| ATG101-ATG13HORMA | Mammal | X-ray | 2.5 | 2018 | [43] | |
| FIP200 CTR | Mammal | X-ray | 3.2 | 2019 | [44] | |
| Kinase domain of ULK2 with inhibitor | Mammal | X-ray | 2.5 | 2019 | [45] | |
| ULK1 complex | Mammal | Cryo-EM | 12-15 | 2019 | [46] | |
| ATG2-18 | Hsv2 (ATG18 homolog) | Yeast (Km) | X-ray | 2.6 | 2012 | [47] |
| Hsv2 (ATG18 homolog) | Yeast | X-ray | 3 | 2012 | [48] | |
| Hsv2 (ATG18 homolog) | Yeast (Kl) | X-ray | 3 | 2012 | [49] | |
| N-terminal domain of Atg2 | Yeast (Sp) | X-ray | 3.2 | 2019 | [50] | |
| ATG2B(human)-WDR45(rat) | Mammal | Negative stain | / | 2017 | [51] | |
| ATG2A-WIPI4 | Mammal | Negative stain | / | 2018 | [52] | |
| ATG2A | Mammal | Cryo-EM | 15 | 2019 | [53] | |
| PI3K complex | VPS15 WD repeat domain | Yeast | X-ray | 1.8 | 2009 | [54] |
| VPS30 BARA domain | Yeast | X-ray | 2.3 | 2012 | [55] | |
| PI3KC3-C2 | Yeast | X-ray | 4.4 | 2015 | [56] | |
| ATG38 C-terminal domain | Yeast | X-ray | 2.2 | 2016 | [57] | |
| VPS15-VPS34 | Yeast | Negative stain | 28 | 2016 | [57] | |
| VPS34 with inhibitors | Drosophila | X-ray | 2.9–3.5 | 2010 | [58] | |
| Bcl-XL-Beclin 1 BH3 | Mammal | X-ray | 2.5 | 2007 | [59] | |
| Bcl-XL-Beclin 1 BH3 | Mammal | NMR | / | 2007 | [60] | |
| M11-Beclin1 BH3 | Mammal | X-ray | 2.3 | 2008 | [61] | |
| M11-Beclin 1 BH3 | Mammal | X-ray, NMR | 2008 | [62] | ||
| Beclin 1 CC domain | Mammal | X-ray | 1.9 | 2012 | [63] | |
| Beclin 1 ECD domain | Mammal | X-ray | 1.55 | 2012 | [64] | |
| VPS34 with PIK-III | Mammal | X-ray | 2.8 | 2014 | [65] | |
| VPS34 with SAR405 | Mammal | X-ray | 2.9 | 2014 | [66] | |
| PI3KC3-C1 | Mammal | Negative stain | 27.5 | 2014 | [67] | |
| Beclin 1 FHD domain | Mammal | X-ray | 1.95 | 2016 | [68] | |
| Beclin 1 CC domain | Mammal | X-ray | 1.46 | 2016 | [69] | |
| ATG14 CC domain with/without Beclin 1 CC domain | Mammal | SAXS | / | 2016 | [69] | |
| PI3KC3-C1 with NRBF2 | Mammal | Negative stain | / | 2016 | [57] | |
| PI3KC3-C1 with NRBF2 | Mammal | Negative stain | / | 2017 | [70] | |
| PI3KC3-C1 and PI3KC3-C2 | Mammal | Cryo-EM | 8.5 (C1) and 8.6 (C2) | 2017 | [71] | |
| Beclin 1-UVRAG CC domain | Mammal | X-ray | 1.9 | 2018 | [72] | |
| PI3KC3-C1 with NRBF2 dimer | Mammal | Cryo-EM | 6.6 | 2019 | [73] | |
| ATG9 | ATG9 | Plant | Cryo-EM | 7.8 | 2019 | [74] |
| Yeast (Saccharomyces cerevisiae) | Mammal (Homo sapiens) | Plant (Arabidopsis thaliana) | Function in Autophagy | Protein Interactions* | Reference | |
|---|---|---|---|---|---|---|
| ATG1 complex | Atg1 | ULK1 | ATG1a ATG1b ATG1c ATG1t | S/T kinase | S. cerevisiae  | [17,32,77,85,86] |
| Atg13 | ATG13 | ATG13a ATG13b | Regulatory subunit | H. sapiens  | ||
| Atg11 Atg17 Atg29 Atg31 | FIP200 ATG101 | ATG11 ATG101 | Scaffold and regulatory | A. thaliana  | ||
| Class III PI3K complex I | Vps34 | VPS34 | VPS34 | PI kinase | S. cerevisiae 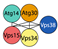 | [56,86,87,88] |
| Vps15 | VPS15 | VPS15 | Scaffold | |||
| Vps30/Atg6 | BECN1 | ATG6 | Regulatory subunit | H. Sapiens 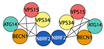 | ||
| Atg14 | ATG14 | ATG14a ATG14b | PAS targeting (N.C. in plant) | |||
| Atg38 | NBRF2 | - | Activator | |||
| ATG9 vesicle | Atg9 | ATG9A/B | ATG9 | Phagophore formation and expansion (Autophagosome progression and closure in plant) | S. cerevisiae 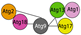 | [20,73,90,91,92] |
H. Sapiens  | ||||||
A. thaliana  | ||||||
| ATG2-ATG18 complex | Atg2 | ATG2A/B | ATG2 | PAS targeting and lipid binding | S. cerevisiae  | [47,92,93,94,95] |
| Atg18 | WIPI1/2 | ATG18a-h |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, L.T.F.; Ye, H.; Zhang, W.; Jiang, L.; Lau, W.C.Y. Structural Biology and Electron Microscopy of the Autophagy Molecular Machinery. Cells 2019, 8, 1627. https://doi.org/10.3390/cells8121627
Lai LTF, Ye H, Zhang W, Jiang L, Lau WCY. Structural Biology and Electron Microscopy of the Autophagy Molecular Machinery. Cells. 2019; 8(12):1627. https://doi.org/10.3390/cells8121627
Chicago/Turabian StyleLai, Louis Tung Faat, Hao Ye, Wenxin Zhang, Liwen Jiang, and Wilson Chun Yu Lau. 2019. "Structural Biology and Electron Microscopy of the Autophagy Molecular Machinery" Cells 8, no. 12: 1627. https://doi.org/10.3390/cells8121627
APA StyleLai, L. T. F., Ye, H., Zhang, W., Jiang, L., & Lau, W. C. Y. (2019). Structural Biology and Electron Microscopy of the Autophagy Molecular Machinery. Cells, 8(12), 1627. https://doi.org/10.3390/cells8121627





