Identification and Characterization of Aspergillus nidulans Mutants Impaired in Asexual Development under Phosphate Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Oligonucleotides, Strains, and Culture Conditions
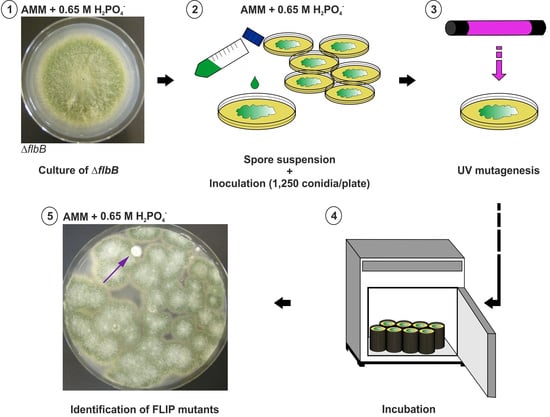
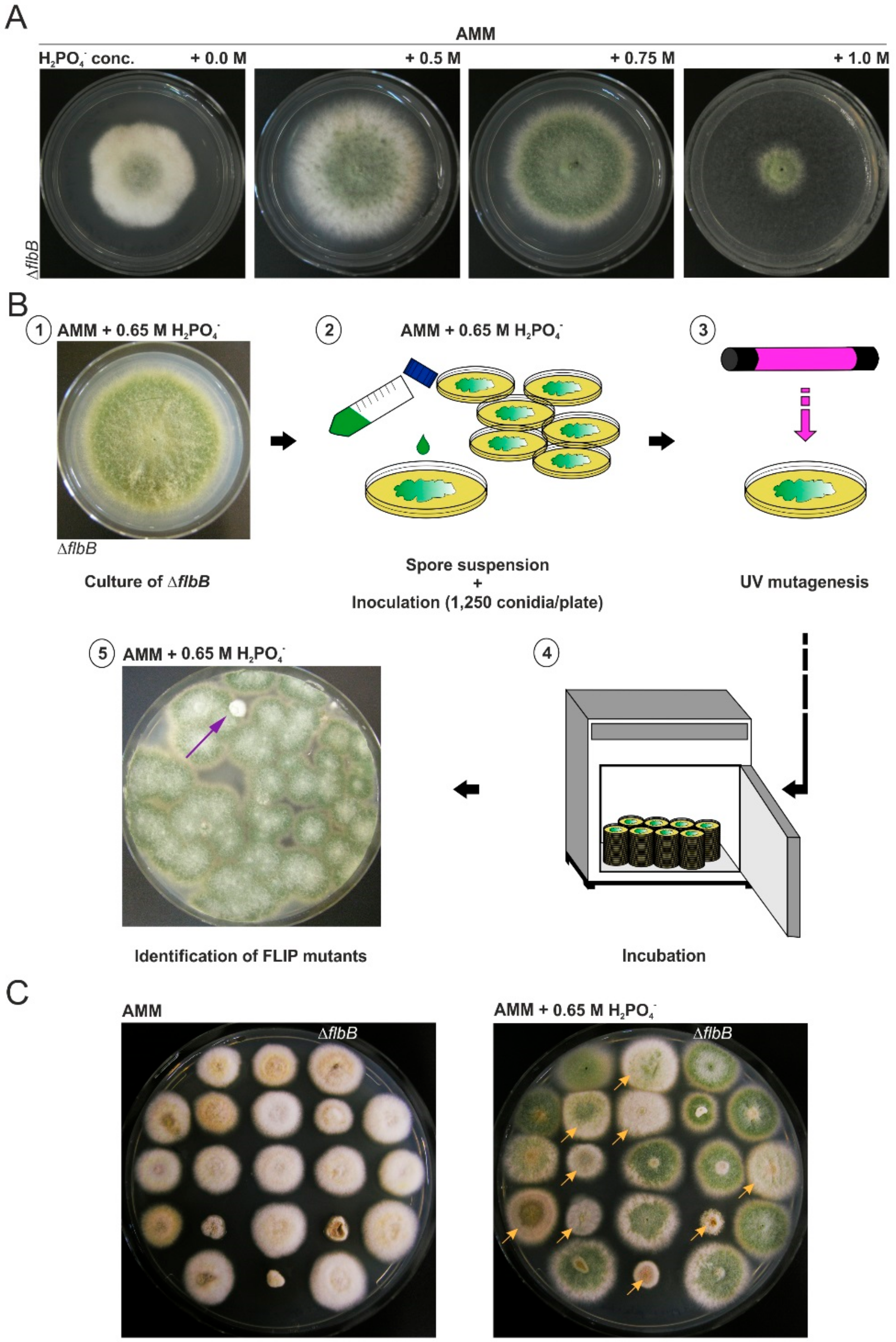
2.2. Mutagenesis Procedure
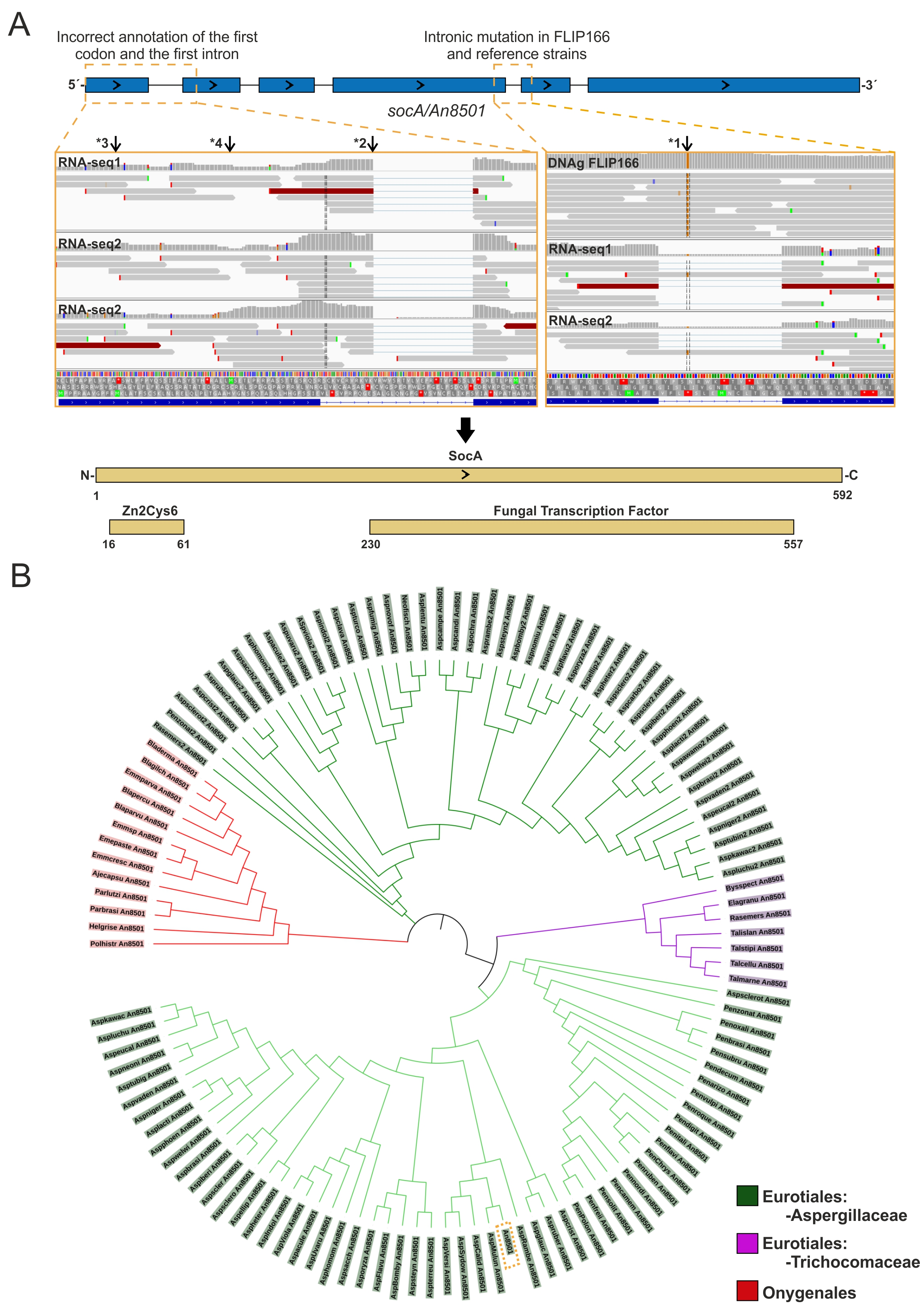
2.3. Cloning of socA
2.4. Bioinformatics
2.5. Fluorescence Microscopy
2.6. Protein Extraction and Immunodetection
3. Results
3.1. Isolation of FLIP (Fluffy in Phosphate) Mutants
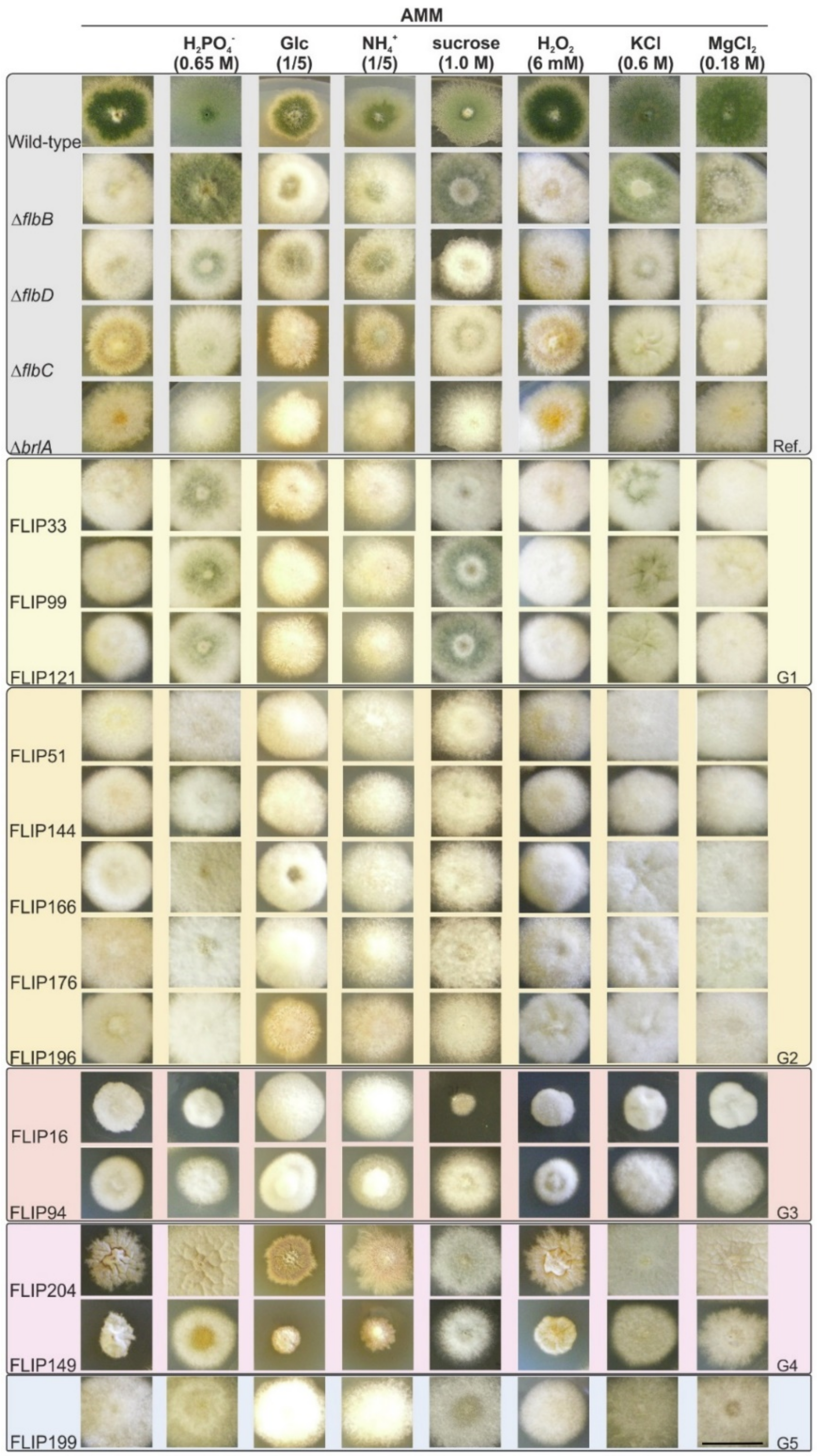
3.2. Phenotypic Characterization of FLIP Mutants
3.3. An8501/socA as a Multicopy Suppressor of the FLIP166 Phenotype: Functional Characterization
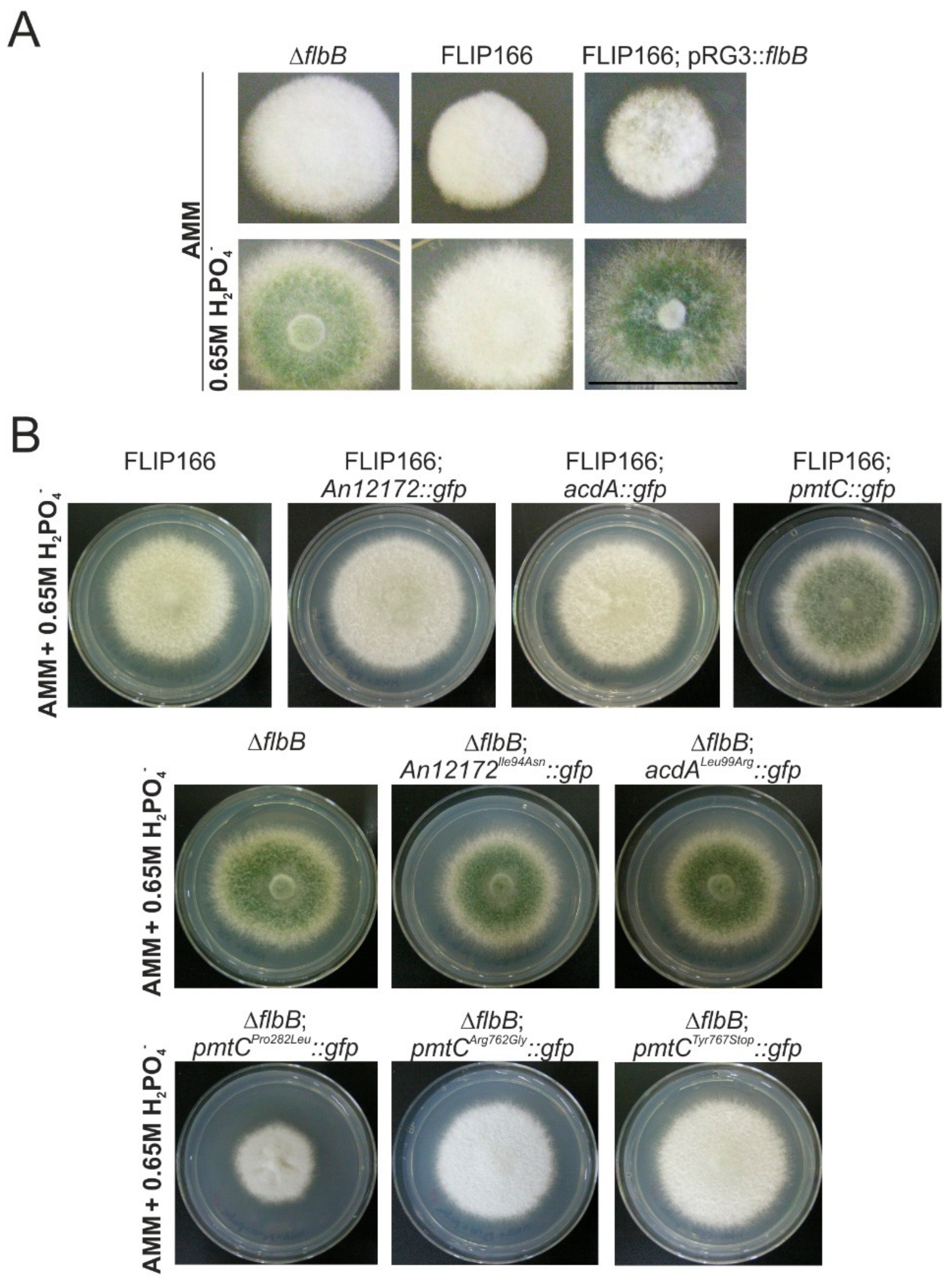
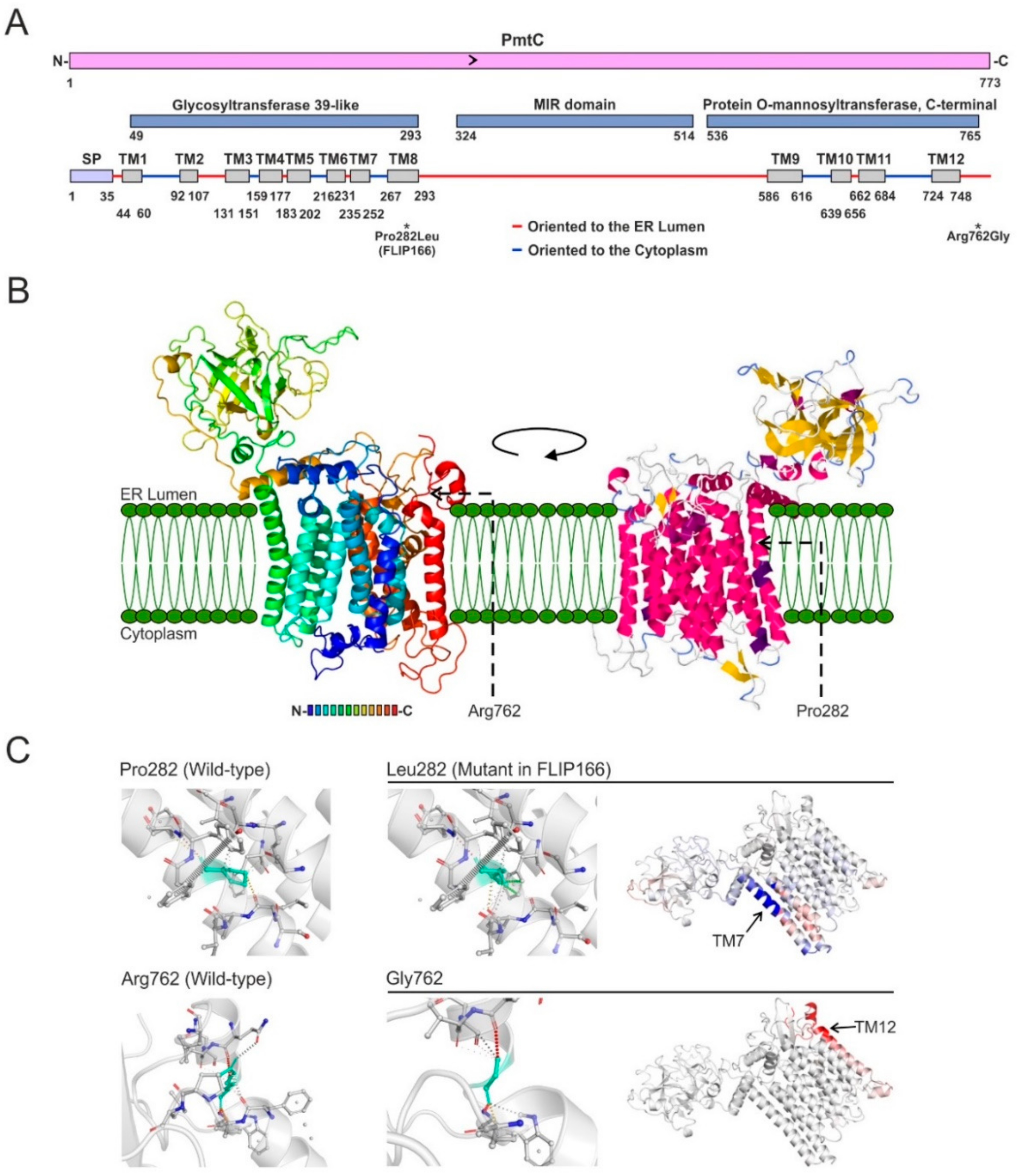
3.4. The Mutant Form Pro282Leu of the Protein O-Mannosyltransferase PmtC Is Responsible for the FLIP166 Phenotype
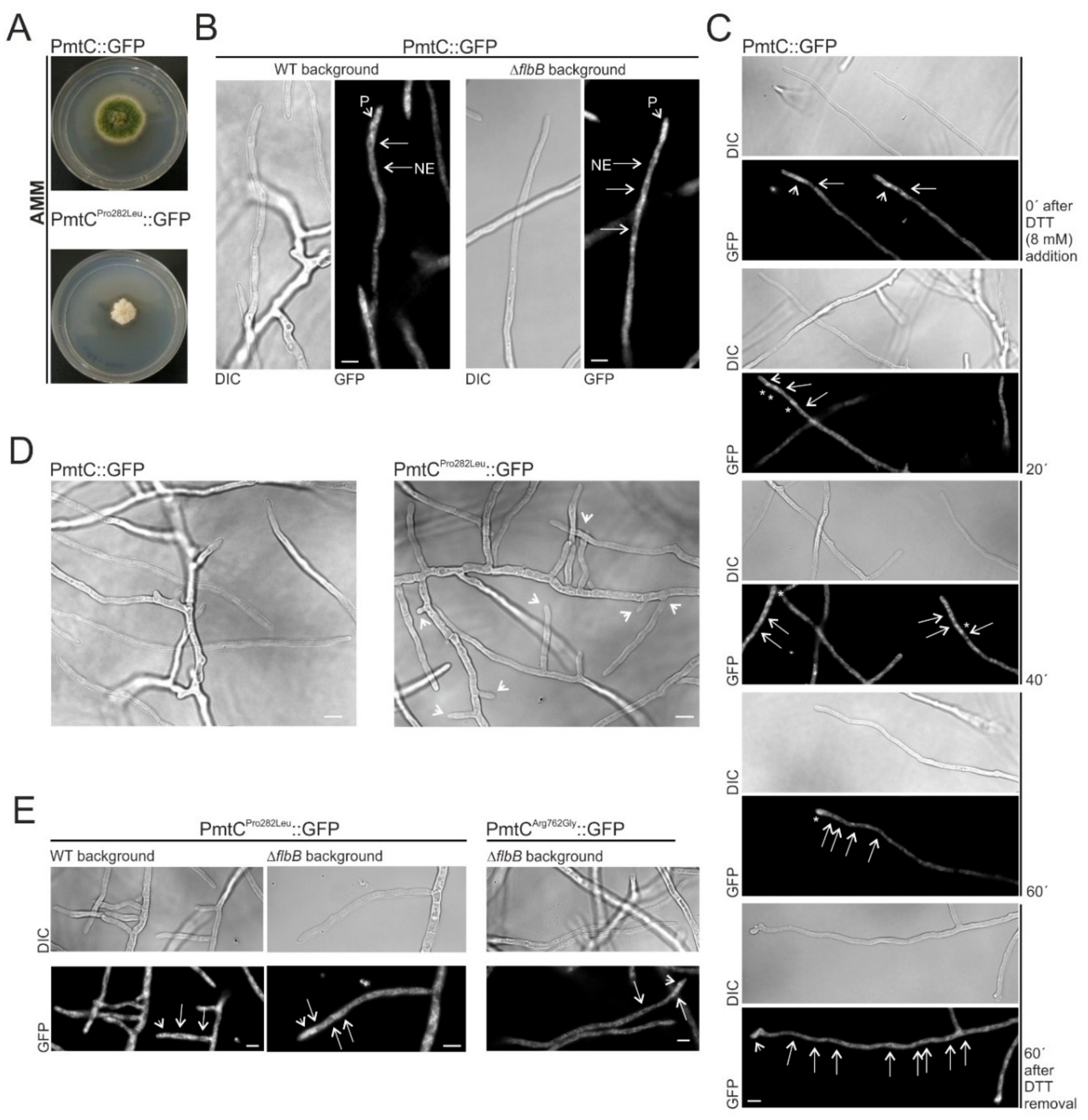
3.5. Substitution of Proline in Position 282 of PmtC Causes an Aconidial Phenotype and an Inhibition of Growth but Not a Delocalization of the Protein
4. Discussion
4.1. A Novel Mutagenesis Strategy as a Tool for the Identification of Hitherto Unknown Genes and Mutations Related to the Control of Conidiation
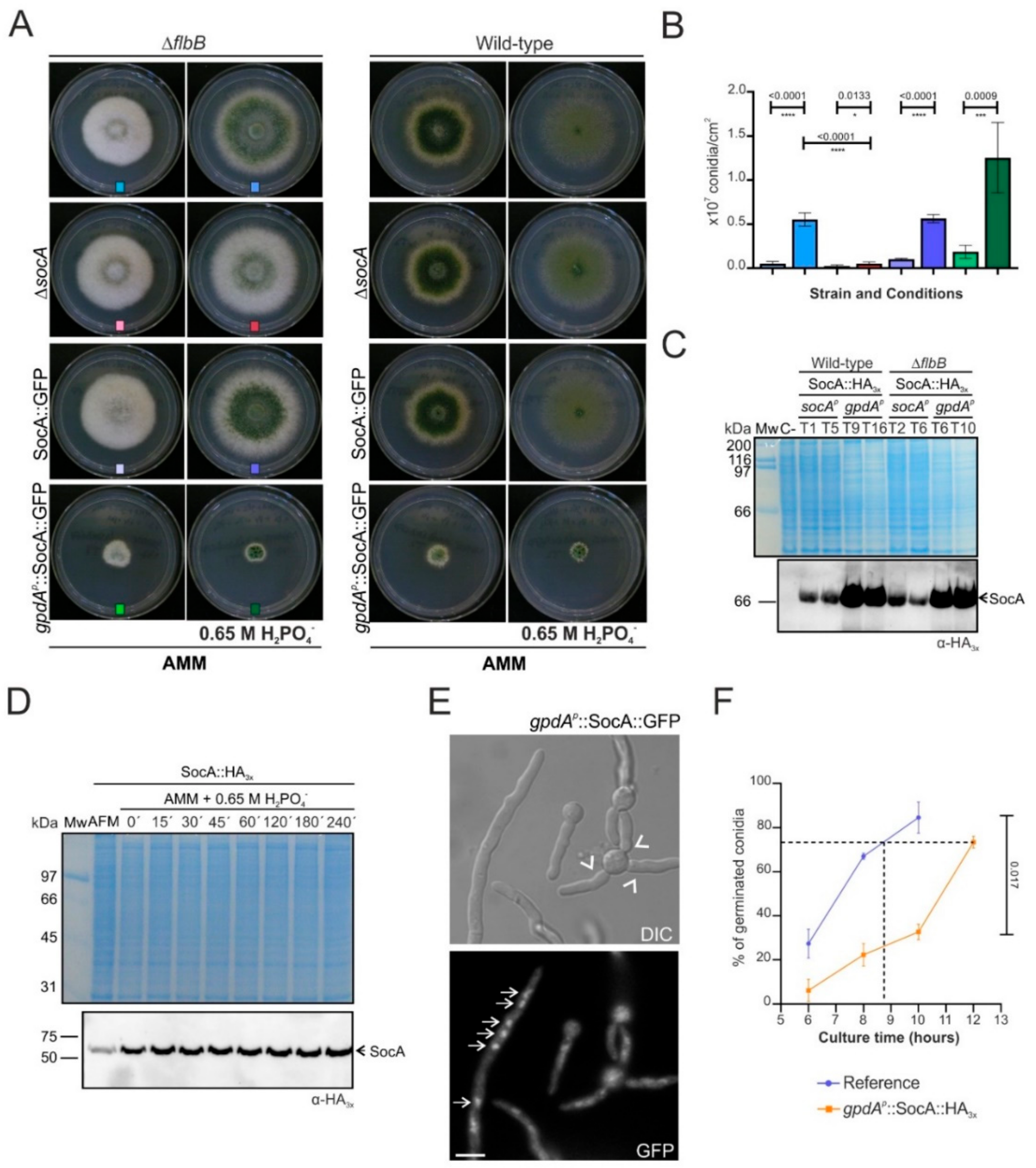
4.2. SocA, a Putative Transcriptional Regulator Probably Involved in the Control of Morphogenesis and Development under Specific Stress Conditions
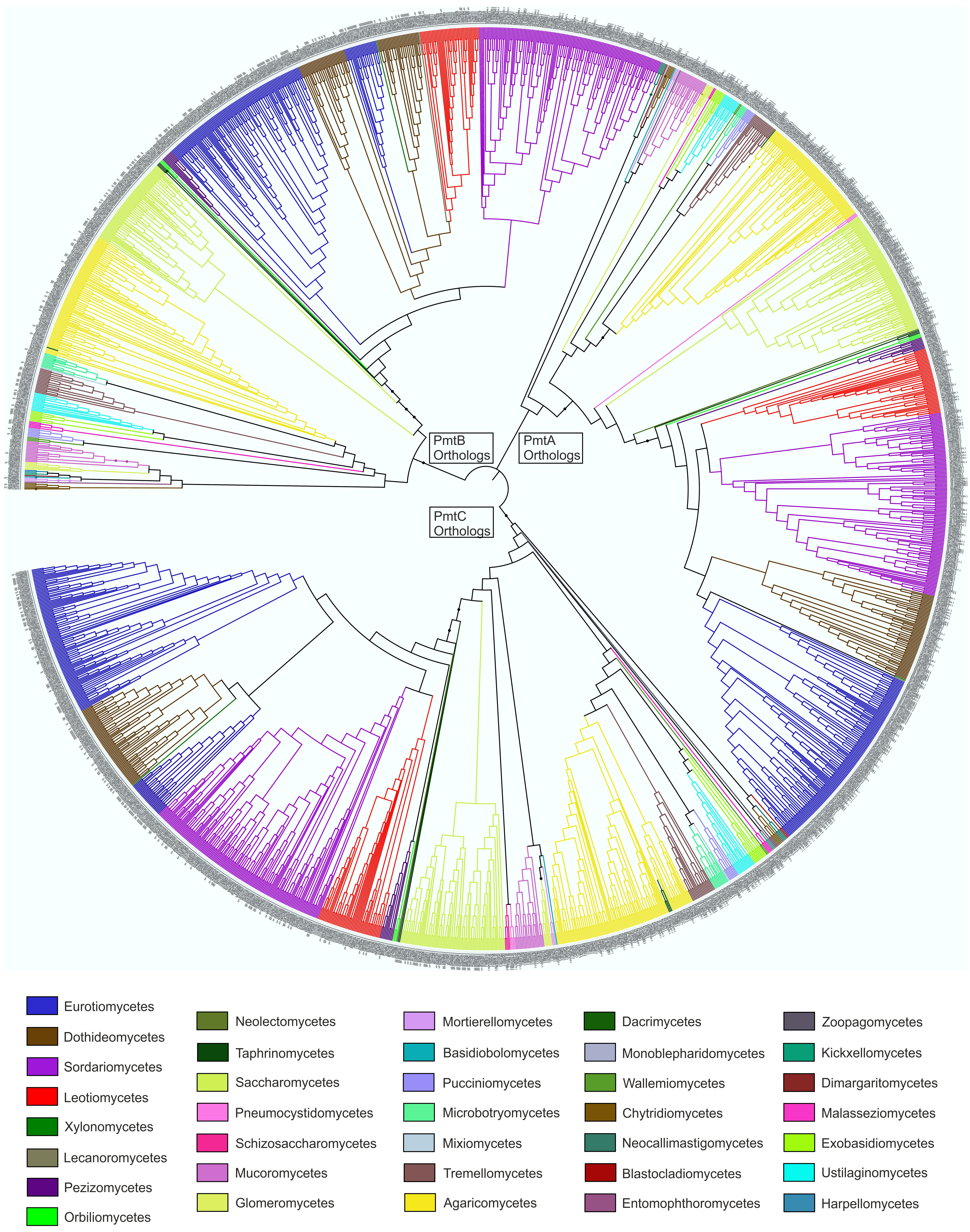
4.3. Revisiting the Function of PmtC
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hawksworth, D.L.; Lücking, R. Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol. Spectr. 2017, 5, 4. [Google Scholar]
- Kirk, P.; Cannon, P.; Minter, D.; Stalpers, J. Dictionary of the Fungi, 10th ed.; CAN International: Wallingford, UK, 2008. [Google Scholar]
- Etxebeste, O.; Otamendi, A.; Garzia, A.; Espeso, E.A.; Cortese, M.S. Rewiring of transcriptional networks as a major event leading to the diversity of asexual multicellularity in fungi. Crit. Rev. Microbiol. 2019, 7, 1–16. [Google Scholar] [CrossRef]
- Ojeda-López, M.; Chen, W.; Eagle, C.E.; Gutiérrez, G.; Jia, W.L.; Swilaiman, S.S.; Huang, Z.; Park, H.-S.; Yu, J.-H.; Cánovas, D.; et al. Evolution of asexual and sexual reproduction in the aspergilli. Stud. Mycol. 2018, 91, 37–59. [Google Scholar] [CrossRef]
- De Vries, R.P.; Riley, R.; Wiebenga, A.; Aguilar-Osorio, G.; Amillis, S.; Uchima, C.A.; Anderluh, G.; Asadollahi, M.; Askin, M.; Barry, K.; et al. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 2017, 18, 28. [Google Scholar] [CrossRef]
- Park, H.-S.; Yu, J.-H. Genetic control of asexual sporulation in filamentous fungi. Curr. Opin. Microbiol. 2012, 15, 669–677. [Google Scholar] [CrossRef]
- Fischer, R.; Kües, U. Asexual sporulation in mycelial fungi. In Growth, Differentiation and Sexuality; Kües, U., Fischer, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 263–292. [Google Scholar]
- Kües, U.; Badalyan, S.M.; Gießler, A.; Dörnte, B. Asexual sporulation in Agaricomycetes. In Growth, Differentiation and Sexuality; Wendland, J., Ed.; Springer International Publishing: Basel, Switzerland, 2016; pp. 269–328. [Google Scholar]
- Mims, C.W.; Richardson, E.A.; Timberlake, W.E. Ultrastructural analysis of conidiophore development in the fungus Aspergillus nidulans using freeze-substitution. Protoplasma 1988, 144, 132–141. [Google Scholar] [CrossRef]
- Adams, T.H.; Wieser, J.K.; Yu, J.-H. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 1998, 62, 35–54. [Google Scholar]
- Etxebeste, O.; Espeso, E.A. Aspergillus nidulans in the post-genomic era: A top-model filamentous fungus for the study of signaling and homeostasis mechanisms. Int. Microbiol. 2019, 2, 1–18. [Google Scholar] [CrossRef]
- Oiartzabal-Arano, E.; Perez-de-Nanclares-Arregi, E.; Espeso, E.A.; Etxebeste, O. Apical control of conidiation in Aspergillus nidulans. Curr. Genet. 2016, 62, 371–377. [Google Scholar] [CrossRef]
- Park, H.-S.; Lee, M.-K.; Han, K.-H.; Kim, M.-J.; Yu, J.-H. Developmental Decisions in Aspergillus nidulans. In Biology of the Fungal Cell; Hoffmeister, D., Gressler, M., Eds.; Springer International Publishing: Basel, Switzerland, 2019; pp. 63–80. [Google Scholar]
- Otamendi, A.; Perez-de-Nanclares-Arregi, E.; Oiartzabal-Arano, E.; Cortese, M.S.; Espeso, E.A.; Etxebeste, O. Developmental regulators FlbE/D orchestrate the polarity site-to-nucleus dynamics of the fungal bZIP transcription factor FlbB. Cell. Mol. Life Sci. 2019, 5, 1–22. [Google Scholar] [CrossRef]
- Herrero-Garcia, E.; Perez-de-Nanclares-Arregi, E.; Cortese, M.S.; Markina-Iñarrairaegui, A.; Oiartzabal-Arano, E.; Etxebeste, O.; Ugalde, U.; Espeso, E.A. Tip-to-nucleus migration dynamics of the asexual development regulator FlbB in vegetative cells. Mol. Microbiol. 2015, 98, 607–624. [Google Scholar] [CrossRef]
- Garzia, A.; Etxebeste, O.; Herrero-García, E.; Ugalde, U.; Espeso, E.A. The concerted action of bZip and cMyb transcription factors FlbB and FlbD induces brlA expression and asexual development in Aspergillus nidulans. Mol. Microbiol. 2010, 75, 1314–1324. [Google Scholar] [CrossRef]
- Etxebeste, O.; Ni, M.; Garzia, A.; Kwon, N.J.; Fischer, R.; Yu, J.H.; Espeso, E.A.; Ugalde, U. Basic-zipper-type transcription factor FlbB controls asexual development in Aspergillus nidulans. Eukaryot. Cell 2008, 7, 38–48. [Google Scholar] [CrossRef]
- Rodríguez-Urra, A.B.; Jiménez, C.; Nieto, M.I.; Rodríguez, J.; Hayashi, H.; Ugalde, U. Signaling the Induction of Sporulation Involves the Interaction of Two Secondary Metabolites in Aspergillus nidulans. ACS Chem. Biol. 2012, 7, 599–606. [Google Scholar] [CrossRef]
- Skromne, I.; Sanchez, O.; Aguirre, J. Starvation stress modulates the expression of the Aspergillus nidulans brlA regulatory gene. Microbiology 1995, 141, 21–28. [Google Scholar] [CrossRef]
- Etxebeste, O.; Herrero-García, E.; Cortese, M.S.; Garzia, A.; Oiartzabal-Arano, E.; de los Ríos, V.; Ugalde, U.; Espeso, E.A. GmcA is a putative glucose-methanol-choline oxidoreductase required for the induction of asexual development in Aspergillus nidulans. PLoS ONE 2012, 7, e40292. [Google Scholar] [CrossRef]
- Etxebeste, O.; Herrero-García, E.; Araújo-Bazán, L.; Rodríguez-Urra, A.B.; Garzia, A.; Ugalde, U.; Espeso, E.A. The bZIP-type transcription factor FIbB regulates distinct morphogenetic stages of colony formation in Aspergillus nidulans. Mol. Microbiol. 2009, 73, 775–789. [Google Scholar] [CrossRef]
- Osherov, N.; Mathew, J.; May, G.S. Polarity-defective mutants of Aspergillus nidulans. Fungal Genet. Biol. 2000, 31, 181–188. [Google Scholar] [CrossRef][Green Version]
- Cove, D.J. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta Enzymol. Biol. Oxid. 1966, 113, 51–56. [Google Scholar] [CrossRef]
- Käfer, E. Origins of translocations in Aspergillus nidulans. Genetics 1965, 52, 217–232. [Google Scholar]
- Galindo, A.; Calcagno-Pizarelli, A.M.; Arst, H.N.; Peñalva, M.Á. An ordered pathway for the assembly of fungal ESCRT-containing ambient pH signalling complexes at the plasma membrane. J. Cell Sci. 2012, 125, 1784–1795. [Google Scholar] [CrossRef]
- Garzia, A.; Etxebeste, O.; Herrero-Garcia, E.; Fischer, R.; Espeso, E.A.; Ugalde, U. Aspergillus nidulans FlbE is an upstream developmental activator of conidiation functionally associated with the putative transcription factor FlbB. Mol. Microbiol. 2009, 71, 172–184. [Google Scholar] [CrossRef]
- Peñalva, M.A. Tracing the endocytic pathway of Aspergillus nidulans with FM4-64. Fungal Genet. Biol. 2005, 42, 963–975. [Google Scholar] [CrossRef]
- Pantazopoulou, A.; Peñalva, M.A. Organization and dynamics of the Aspergillus nidulans Golgi during apical extension and mitosis. Mol. Biol. Cell 2009, 20, 4335–4347. [Google Scholar] [CrossRef]
- Yang, L.; Ukil, L.; Osmani, A.; Nahm, F.; Davies, J.; De Souza, C.P.C.; Dou, X.; Perez-Balaguer, A.; Osmani, S.A. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot. Cell 2004, 3, 1359–1362. [Google Scholar] [CrossRef]
- Tilburn, J.; Scazzocchio, C.; Taylor, G.G.; Zabicky-Zissman, J.H.; Lockington, R.A.; Davies, R.W. Transformation by integration in Aspergillus nidulans. Gene 1983, 26, 205–221. [Google Scholar] [CrossRef]
- Szewczyk, E.; Nayak, T.; Oakley, C.E.; Edgerton, H.; Xiong, Y.; Taheri-Talesh, N.; Osmani, S.A.; Oakley, B.R. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 2007, 1, 3111–3120. [Google Scholar] [CrossRef]
- Todd, R.B.; Davis, M.A.; Hynes, M.J. Genetic manipulation of Aspergillus nidulans: Meiotic progeny for genetic analysis and strain construction. Nat. Protoc. 2007, 2, 811–821. [Google Scholar] [CrossRef]
- Todd, R.B.; Davis, M.A.; Hynes, M.J. Genetic manipulation of Aspergillus nidulans: Heterokaryons and diploids for dominance, complementation and haploidization analyses. Nat. Protoc. 2007, 2, 822–830. [Google Scholar] [CrossRef]
- Upshall, A.; Giddings, B.; Mortimore, I.D. The use of benlate for distinguishing between haploid and diploid strains of Aspergillus nidulans and Aspergillus terreus. Microbiology 1977, 100, 413–418. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Mitchell, A.L.; Attwood, T.K.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.-Y.; El-Gebali, S.; Fraser, M.I.; et al. InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019, 47, 351–360. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, 256–259. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, 296–303. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Rodrigues, C.H.M.; Pires, D.E.V.; Ascher, D.B. DynaMut: Predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018, 46, 350–355. [Google Scholar] [CrossRef]
- Hervás-Aguilar, A.; Peñalva, M.A. Endocytic machinery protein SlaB is dispensable for polarity establishment but necessary for polarity maintenance in hyphal tip cells of Aspergillus nidulans. Eukaryot. Cell 2010, 9, 1504–1518. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Oiartzabal-Arano, E.; Garzia, A.; Gorostidi, A.; Ugalde, U.; Espeso, E.A.; Etxebeste, O. Beyond asexual development: Modifications in the gene expression profile caused by the absence of the Aspergillus nidulans transcription factor FlbB. Genetics 2015, 199, 1127–1142. [Google Scholar] [CrossRef]
- Garzia, A.; Etxebeste, O.; Rodríguez-Romero, J.; Fischer, R.; Espeso, E.A.; Ugalde, U. Transcriptional changes in the transition from vegetative cells to asexual development in the model fungus Aspergillus nidulans. Eukaryot. Cell 2013, 12, 311–321. [Google Scholar] [CrossRef]
- Reiser, K.; Davis, M.A.; Hynes, M.J. AoxA is a major peroxisomal long chain fatty acyl-CoA oxidase required for β-oxidation in A. nidulans. Curr. Genet. 2010, 56, 139–150. [Google Scholar] [CrossRef]
- Goto, M.; Harada, Y.; Oka, T.; Matsumoto, S.; Takegawa, K.; Furukawa, K. Protein O-Mannosyltransferases B and C support hyphal development and differentiation in Aspergillus nidulans. Eukaryot. Cell 2009, 8, 1465–1474. [Google Scholar] [CrossRef]
- Kriangkripipat, T.; Momany, M. Aspergillus nidulans Protein O-Mannosyltransferases play roles in cell wall integrity and developmental patterning. Eukaryot. Cell 2009, 8, 1475–1485. [Google Scholar] [CrossRef]
- Le, T.H.T.; Oki, A.; Goto, M.; Shimizu, K. Protein O-mannosyltransferases are required for sterigmatocystin production and developmental processes in Aspergillus nidulans. Curr. Genet. 2018, 64, 1043–1056. [Google Scholar] [CrossRef]
- Willer, T.; Valero, M.C.; Tanner, W.; Cruces, J.; Strahl, S. O-mannosyl glycans: From yeast to novel associations with human disease. Curr. Opin. Struct. Biol. 2003, 13, 621–630. [Google Scholar] [CrossRef]
- Loibl, M.; Strahl, S. Protein O-mannosylation: What we have learned from baker’s yeast. Biochim. Biophys. Acta Mol. Cell Res. 2013, 1833, 2438–2446. [Google Scholar] [CrossRef]
- Markina-Iñarrairaegui, A.; Pantazopoulou, A.; Espeso, E.A.; Peñalva, M.A. The Aspergillus nidulans peripheral ER: Disorganization by ER stress and persistence during mitosis. PLoS ONE 2013, 8, e67154. [Google Scholar] [CrossRef]
- Bravo-Plaza, I.; Hernández-González, M.; Pinar, M.; Díaz, J.F.; Peñalva, M.A. Identification of the guanine nucleotide exchange factor for SAR1 in the filamentous fungal model Aspergillus nidulans. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 118551. [Google Scholar] [CrossRef]
- Timberlake, W.E. Developmental gene regulation in Aspergillus nidulans. Dev. Biol. 1980, 78, 497–510. [Google Scholar] [CrossRef]
- Martinelli, S.D.; Clutterbuck, A.J. A quantitative survey of conidiation mutants in Aspergillus nidulans. J. Gen. Microbiol. 1971, 69, 261–268. [Google Scholar] [CrossRef][Green Version]
- Wortman, J.R.; Gilsenan, J.M.; Joardar, V.; Deegan, J.; Clutterbuck, J.; Andersen, M.R.; Archer, D.; Bencina, M.; Braus, G.; Coutinho, P.; et al. The 2008 update of the Aspergillus nidulans genome annotation: A community effort. Fungal Genet Biol 2009, 46, 2–13. [Google Scholar] [CrossRef]
- Sorrells, T.R.; Johnson, A.D. Making Sense of Transcription Networks. Cell 2015, 161, 714–723. [Google Scholar] [CrossRef]
- Voordeckers, K.; Pougach, K.; Verstrepen, K.J. How do regulatory networks evolve and expand throughout evolution? Curr. Opin. Biotechnol. 2015, 34, 180–188. [Google Scholar] [CrossRef]
- Dai, Z. Transcription Factors indirectly regulate genes through nuclear colocalization. Cells 2019, 8, 754. [Google Scholar] [CrossRef]
- Riquelme, M.; Aguirre, J.; Bartnicki-García, S.; Braus, G.H.; Feldbrügge, M.; Fleig, U.; Hansberg, W.; Herrera-Estrella, A.; Kämper, J.; Kück, U.; et al. Fungal Morphogenesis, from the Polarized Growth of Hyphae to Complex Reproduction and Infection Structures. Microbiol. Mol. Biol. Rev. 2018, 82, e00068-17. [Google Scholar] [CrossRef]
- Futagami, T.; Nakao, S.; Kido, Y.; Oka, T.; Kajiwara, Y.; Takashita, H.; Omori, T.; Furukawa, K.; Goto, M. Putative stress sensors WscA and WscB are involved in hypo-osmotic and acidic pH stress tolerance in Aspergillus nidulans. Eukaryot. Cell 2011, 10, 1504–1515. [Google Scholar] [CrossRef]
- Virag, A.; Lee, M.P.; Si, H.; Harris, S.D. Regulation of hyphal morphogenesis by cdc42 and rac1 homologues in Aspergillus nidulans. Mol. Microbiol. 2007, 66, 1579–1596. [Google Scholar] [CrossRef]
- Kriangkripipat, T.; Momany, M. Aspergillus nidulans Pmts form heterodimers in all pairwise combinations. FEBS Open Bio. 2014, 4, 335–341. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Otamendi, A.; Espeso, E.A.; Etxebeste, O. Identification and Characterization of Aspergillus nidulans Mutants Impaired in Asexual Development under Phosphate Stress. Cells 2019, 8, 1520. https://doi.org/10.3390/cells8121520
Otamendi A, Espeso EA, Etxebeste O. Identification and Characterization of Aspergillus nidulans Mutants Impaired in Asexual Development under Phosphate Stress. Cells. 2019; 8(12):1520. https://doi.org/10.3390/cells8121520
Chicago/Turabian StyleOtamendi, Ainara, Eduardo A. Espeso, and Oier Etxebeste. 2019. "Identification and Characterization of Aspergillus nidulans Mutants Impaired in Asexual Development under Phosphate Stress" Cells 8, no. 12: 1520. https://doi.org/10.3390/cells8121520
APA StyleOtamendi, A., Espeso, E. A., & Etxebeste, O. (2019). Identification and Characterization of Aspergillus nidulans Mutants Impaired in Asexual Development under Phosphate Stress. Cells, 8(12), 1520. https://doi.org/10.3390/cells8121520