Fibronectin and Its Role in Human Infective Diseases
Abstract
1. Introduction
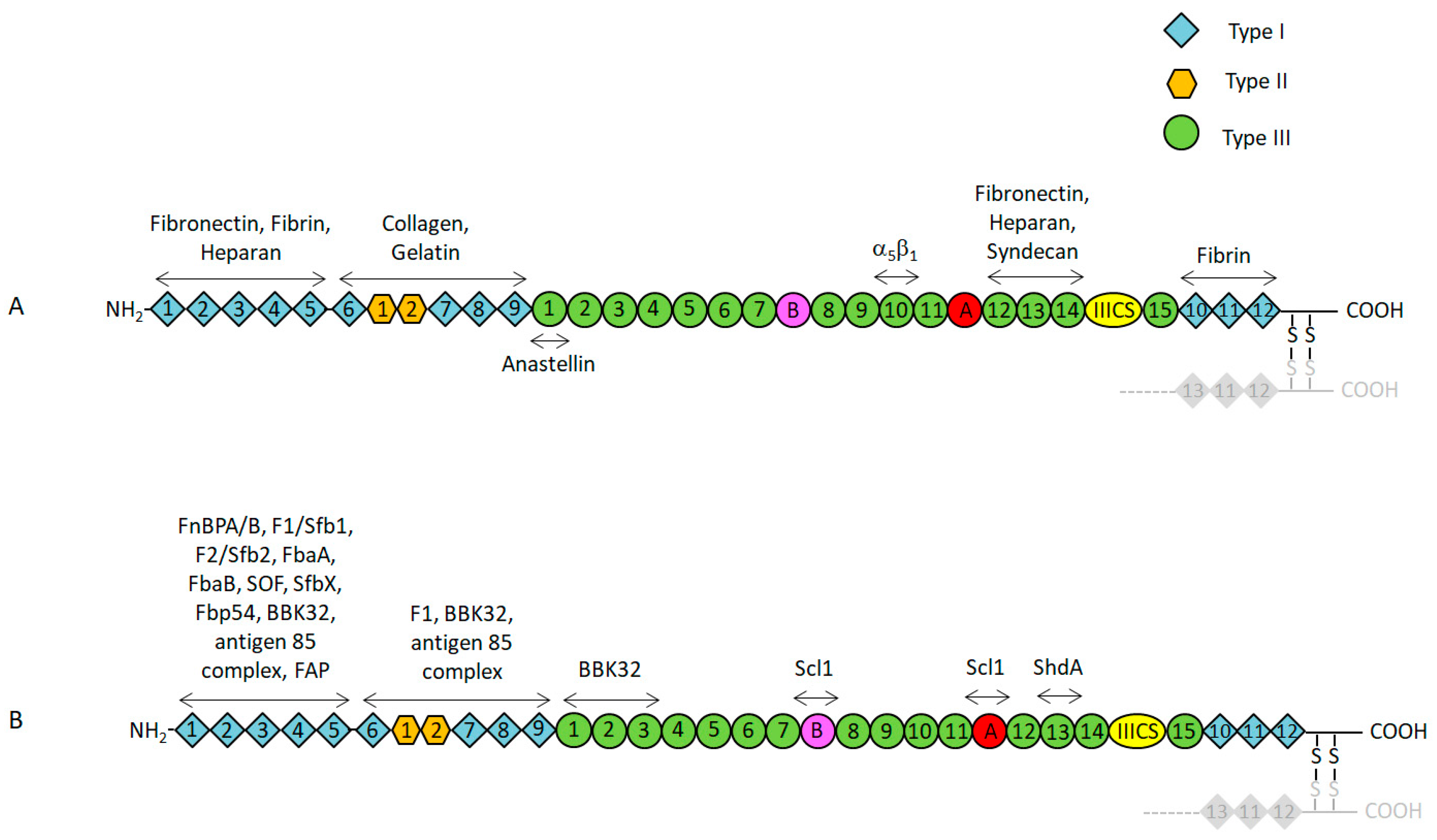
2. Functional and Structural Properties of Fn
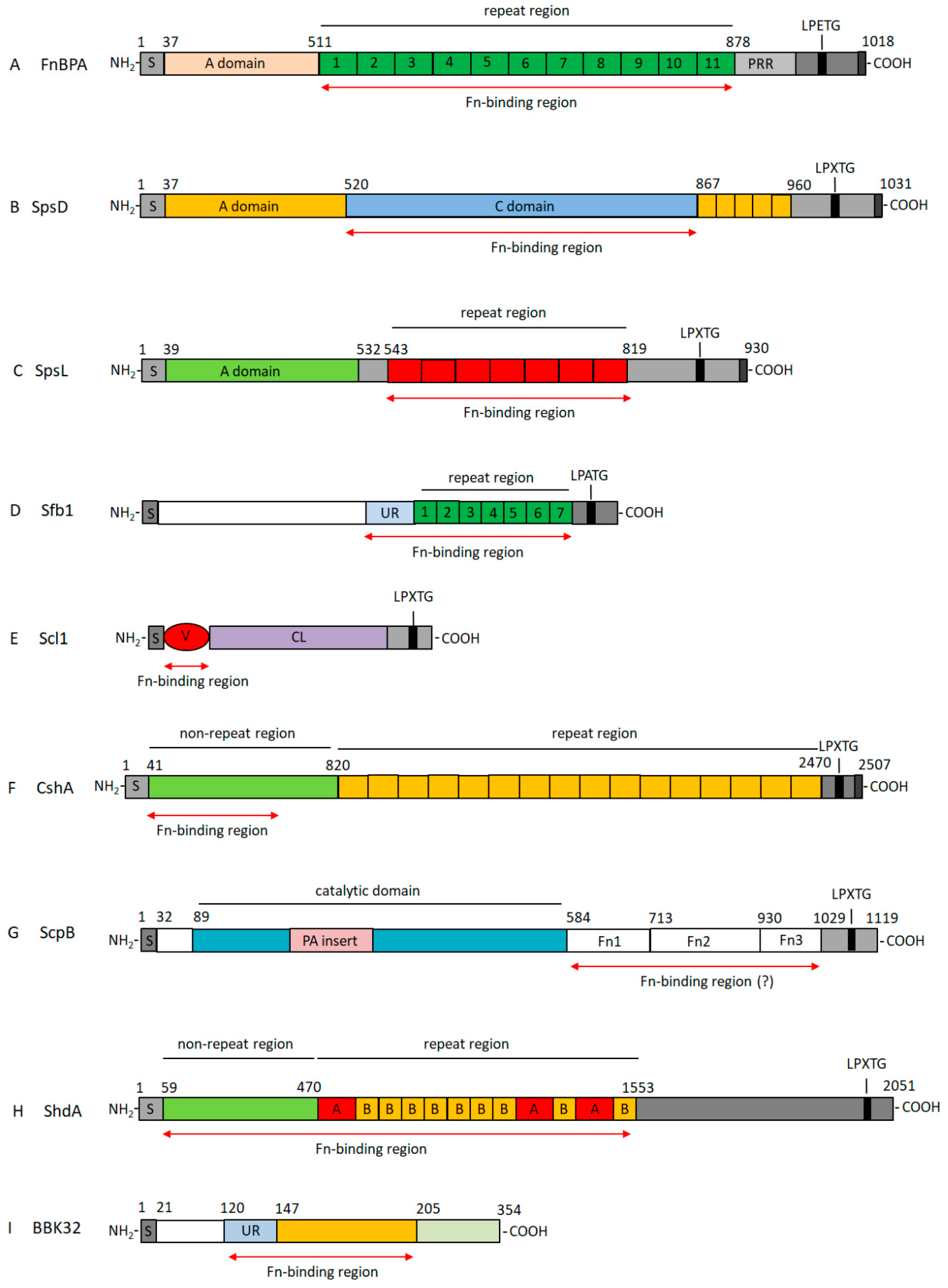
3. Properties of Bacterial Fn-Binding Proteins
3.1. Fn-Binding Proteins from Staphylococcus aureus as a Prototype of a Wide Class of Bacterial Adhesins
3.2. Fibronectin-Binding Proteins from Streptococci
3.2.1. Fn-Binding Proteins from Streptococcus pyogenes
3.2.2. Fn-Binding Proteins from Other Streptococcal Species
3.3. Fn-Binding Proteins from Other Gram-Positive Bacteria
3.3.1. Enterococci
3.3.2. Mycobacteria
3.3.3. Listeria Monocytogenes
3.3.4. Clostridia
3.4. Fn-Binding Proteins from Other Gram Negative-Bacteria
3.4.1. Escherichia coli
3.4.2. Campilobacter Jejuni
3.4.3. Salmonella enterica Serotype Typhimurium
3.4.4. Borrelia burgdorferi
4. Fn-Binding Proteins as Virulence Factors
5. Targeting Fn-Binding Proteins as a Means of Infection Control
6. Concluding Remarks and Outlook
Author Contributions
Funding
Conflicts of Interest
References
- Naba, A.; Clauser, K.R.; Ding, H.; Whittaker, C.A.; Carr, S.A.; Hynes, R.O. The extracellular matrix: Tools and insights for the “omics” era. Matrix Biol. 2016, 49, 10–24. [Google Scholar] [CrossRef]
- Ricard-Blum, S. The collagen family. Cold Spring Harb. Perspect. Biol. 2011, 3, a004978. [Google Scholar] [CrossRef] [PubMed]
- Iozzo, R.V.; Murdoch, A.D. Proteoglycans of the extracellular environment: clues from the gene and protein side offer novel perspectives in molecular diversity and function. FASEB J. 1996, 10, 598–614. [Google Scholar] [CrossRef] [PubMed]
- Hynes, R.O.; Naba, A. Overview of the matrisome--an inventory of extracellular matrix constituents and functions. Cold Spring Harb. Perspect. Biol. 2012, 4, a004903. [Google Scholar] [CrossRef] [PubMed]
- Aumailley, M.; Bruckner-Tuderman, L.; Carter, W.G.; Deutzmann, R.; Edgar, D.; Ekblom, P.; Engel, J.; Engvall, E.; Hohenester, E.; Jones, J.C.; et al. A simplified laminin nomenclature. Matrix Biol. 2005, 24, 326–332. [Google Scholar] [CrossRef] [PubMed]
- Yurchenco, P.D. Basement membranes: cell scaffoldings and signaling platforms. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef]
- Schwarzbauer, J.E.; DeSimone, D.W. Fibronectins, their fibrillogenesis, and in vivo functions. Cold Spring Harb. Perspect. Biol. 2011, 3, a005041. [Google Scholar] [CrossRef]
- Bentley, A.A.; Adams, J.C. The evolution of thrombospondins and their ligand-binding activities. Mol. Biol. Evol. 2010, 27, 2187–2197. [Google Scholar] [CrossRef]
- Bergmeier, W.; Hynes, R.O. Extracellular matrix proteins in hemostasis and thrombosis. Cold Spring Harb. Perspect. Biol. 2012, 4, a005132. [Google Scholar] [CrossRef]
- Hynes, R.O. Integrins: bidirectional, allosteric signaling machines. Cell 2002, 110, 673–687. [Google Scholar] [CrossRef]
- Campbell, I.D.; Humphries, M.J. Integrin structure, activation, and interactions. Cold Spring Harb. Perspect. Biol. 2011, 3, a004994. [Google Scholar] [CrossRef] [PubMed]
- Foster, T.J.; Geoghegan, J.A.; Ganesh, V.K.; Höök, M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 2014, 12, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Foster, T.J. The MSCRAMM Family of Cell-Wall-Anchored Surface Proteins of Gram-Positive Cocci. Trends Microbiol. 2019, 27, 927–941. [Google Scholar] [CrossRef] [PubMed]
- Burke, F.M.; Di Poto, A.; Speziale, P.; Foster, T.J. The A domain of fibronectin-binding protein B of Staphylococcus aureus contains a novel fibronectin binding site. FEBS J. 2011, 278, 2359–2371. [Google Scholar] [CrossRef] [PubMed]
- Roche, F.M.; Downer, R.; Keane, F.; Speziale, P.; Park, P.W.; Foster, T.J. The N-terminal A domain of fibronectin-binding proteins A and B promotes adhesion of Staphylococcus aureus to elastin. J. Biol. Chem. 2004, 279, 38433–38440. [Google Scholar] [CrossRef]
- Pietrocola, G.; Nobile, G.; Gianotti, V.; Zapotoczna, M.; Foster, T.J.; Geoghegan, J.A.; Speziale, P. Molecular Interactions of Human Plasminogen with Fibronectin-binding Protein B (FnBPB), a Fibrinogen/Fibronectin-binding Protein from Staphylococcus aureus. J. Biol. Chem. 2016, 291, 18148–18162. [Google Scholar] [CrossRef]
- Jönsson, K.; Signäs, C.; Müller, H.P.; Lindberg, M. Two different genes encode fibronectin binding proteins in Staphylococcus aureus. The complete nucleotide sequence and characterization of the second gene. Eur J. Biochem. 1991, 202, 1041–1048. [Google Scholar] [CrossRef]
- Hynes, R.O. Structure of Fibronectins. In Fibronectins; Springer: New York, NY, USA, 1990; pp. 113–175. [Google Scholar] [CrossRef]
- Skorstengaard, K.; Jensen, M.S.; Petersen, T.E.; Magnusson, S. Purification and complete primary structures of the heparin-, cell-, and DNA-binding domains of bovine plasma fibronectin. Eur. J. Biochem. 1986, 154, 15–29. [Google Scholar] [CrossRef]
- Pankov, R.; Yamada, K.M. Fibronectin at a glance. J. Cell Sci. 2002, 115, 3861–3863. [Google Scholar] [CrossRef]
- Zardi, L.; Carnemolla, B.; Siri, A.; Petersen, T.E.; Paolella, G.; Sebastio, G.; Baralle, F.E. Transformed human cells produce a new fibronectin isoform by preferential alternative splicing of a previously unobserved exon. EMBO J. 1987, 6, 2337–2342. [Google Scholar] [CrossRef]
- Ffrench-Constant, C.; Van De Water, L.; Dvorak, H.F.; Hynes, R.O. Reappearance of an embryonic pattern of fibronectin splicing during wound healing in the adult rat. J. Cell Biol. 1989, 109, 903–914. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, T.; Erickson, H.P. Domain unfolding plays a role in superfibronectin formation. J. Biol. Chem. 2005, 280, 39143–39151. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, T.; Erickson, H.P. Fibronectin aggregation and assembly: The unfolding of the second fibronectin type III domain. J. Biol Chem. 2011, 286, 39188–39199. [Google Scholar] [CrossRef] [PubMed]
- Lowy, F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef]
- Chambers, H.F. Community-associated MRSA resistance and virulence converge. N. Engl. J. Med. 2005, 1485–1487. [Google Scholar] [CrossRef]
- Stemberk, V.; Jones, R.P.; Moroz, O.; Atkin, K.E.; Edwards, A.M.; Turkenburg, J.P.; Leech, A.P.; Massey, R.C.; Potts, J.R. Evidence for steric regulation of fibrinogen binding to Staphylococcus aureus fibronectin-binding protein A (FnBPA). J. Biol. Chem. 2014, 289, 12842–12851. [Google Scholar] [CrossRef]
- Schwarz-Linek, U.; Werner, J.M.; Pickford, A.R.; Gurusiddappa, S.; Kim, J.H.; Pilka, E.S.; Briggs, J.A.; Gough, T.S.; Höök, M.; Campbell, I.D.; et al. Pathogenic bacteria attach to human fibronectin through a tandem beta-zipper. Nature 2003, 423, 177–181. [Google Scholar] [CrossRef]
- Raibaud, S.; Schwarz-Linek, U.; Kim, J.H.; Jenkins, H.T.; Baines, E.R.; Gurusiddappa, S.; Höök, M.; Potts, J.R. Borrelia burgdorferi binds fibronectin through a tandem beta-zipper, a common mechanism of fibronectin binding in staphylococci, streptococci, and spirochetes. J. Biol. Chem. 2005, 280, 18803–18809. [Google Scholar] [CrossRef]
- Fröman, G.; Switalski, L.M.; Speziale, P.; Höök, M. Isolation and characterization of a fibronectin receptor from Staphylococcus aureus. J. Biol. Chem. 1987, 262, 6564–6571. [Google Scholar]
- Meenan, N.A.; Visai, L.; Valtulina, V.; Schwarz-Linek, U.; Norris, N.C.; Gurusiddappa, S.; Höök, M.; Speziale, P.; Potts, J.R. The tandem beta-zipper model defines high affinity fibronectin-binding repeats within Staphylococcus aureus FnBPA. J. Biol. Chem. 2007, 282, 25893–25902. [Google Scholar] [CrossRef]
- Casillas-Ituarte, N.N.; DiBartola, A.C.; Broughton, M.J.; Pérez-Guzmán, L.; Wheeler, R.M.; Ibaraki, M.; Lower, B.A.; Dunn, J.A.; Lower, B.H.; Fowler, V.G., Jr.; et al. Fibrinogen binding is affected by amino acid substitutions in C-terminal repeat region of fibronectin binding protein A. Sci. Rep. 2019, 9, 11619. [Google Scholar] [CrossRef] [PubMed]
- Sinha, B.; François, P.P.; Nüsse, O.; Foti, M.; Hartford, O.M.; Vaudaux, P.; Foster, T.J.; Lew, D.P.; Herrmann, M.; Krause, K.H. Fibronectin-binding protein acts as Staphylococcus aureus invasin via fibronectin bridging to integrin alpha5beta1. Cell Microbiol. 1999, 1, 101–117. [Google Scholar] [CrossRef] [PubMed]
- Edwards, A.M.; Potts, J.R.; Josefsson, E.; Massey, R.C. Staphylococcus aureus host cell invasion and virulence in sepsis is facilitated by the multiple repeats within FnBPA. PLoS Pathog. 2010, 6, e1000964. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Garcia, B.L.; Visai, L.; Prabhakaran, S.; Meenan, N.A.; Potts, J.R.; Humphries, M.J.; Höök, M. Allosteric Regulation of Fibronectin/α5β1 Interaction by Fibronectin-Binding MSCRAMMs. PLoS ONE 2016, 11, e0159118. [Google Scholar] [CrossRef]
- Prystopiuk, V.; Feuillie, C.; Herman-Bausier, P.; Viela, F.; Alsteens, D.; Pietrocola, G.; Speziale, P.; Dufrêne, Y.F. Mechanical Forces Guiding Staphylococcus aureus Cellular Invasion. ACS Nano 2018, 12, 3609–3622. [Google Scholar] [CrossRef]
- Bingham, R.J.; Rudiño-Piñera, E.; Meenan, N.A.; Schwarz-Linek, U.; Turkenburg, J.P.; Höök, M.; Garman, E.F.; Potts, J.R. Crystal structures of fibronectin-binding sites from Staphylococcus aureus FnBPA in complex with fibronectin domains. Proc. Natl. Acad. Sci. USA 2008, 105, 12254–12258. [Google Scholar] [CrossRef]
- Fowler, T.; Johansson, S.; Wary, K.K.; Höök, M. Src kinase has a central role in in vitro cellular internalization of Staphylococcus aureus. Cell Microbiol. 2003, 5, 417–426. [Google Scholar] [CrossRef]
- Agerer, F.; Lux, S.; Michel, A.; Rohde, M.; Ohlsen, K.; Hauck, C.R. Cellular invasion by Staphylococcus aureus reveals a functional link between focal adhesion kinase and cortactin in integrin-mediated internalisation. J. Cell Sci. 2005, 118, 2189–2200. [Google Scholar] [CrossRef]
- Cheng, A.G.; Missiakas, D.; Schneewind, O. The giant protein Ebh is a determinant of Staphylococcus aureus cell size and complement resistance. J. Bacteriol. 2014, 196, 971–981. [Google Scholar] [CrossRef]
- Kuroda, M.; Tanaka, Y.; Aoki, R.; Shu, D.; Tsumoto, K.; Ohta, T. Staphylococcus aureus giant protein Ebh is involved in tolerance to transient hyperosmotic pressure. Biochem. Biophys. Res. Commun. 2008, 374, 237–241. [Google Scholar] [CrossRef]
- Clarke, S.R.; Harris, L.G.; Richards, R.G.; Foster, S.J. Analysis of Ebh, a 1.1-megadalton cell wall-associated fibronectin-binding protein of Staphylococcus aureus. Infect. Immun. 2002, 70, 6680–6687. [Google Scholar] [CrossRef] [PubMed]
- Jönsson, K.; McDevitt, D.; McGavin, M.H.; Patti, J.M.; Höök, M. Staphylococcus aureus expresses a major histocompatibility complex class II analog. J. Biol Chem. 1995, 270, 21457–21460. [Google Scholar] [CrossRef] [PubMed]
- Flock, M.; Flock, J.I. Rebinding of extracellular adherence protein Eap to Staphylococcus aureus can occur through a surface-bound neutral phosphatase. J. Bacteriol. 2001, 183, 3999–4003. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.; Haggar, A.; Heilmann, C.; Peters, G.; Flock, J.I.; Herrmann, M. Insertional inactivation of Eap in Staphylococcus aureus strain Newman confers reduced staphylococcal binding to fibroblasts. Infect. Immun. 2002, 70, 2933–2940. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hussain, M.; Becker, K.; von Eiff, C.; Schrenzel, J.; Peters, G.; Herrmann, M. Identification and characterization of a novel 38.5-kilodalton cell surface protein of Staphylococcus aureus with extended-spectrum binding activity for extracellular matrix and plasma proteins. J. Bacteriol. 2001, 183, 6778–6786. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Geraci, J.; Neubauer, S.; Pöllath, C.; Hansen, U.; Rizzo, F.; Krafft, C.; Westermann, M.; Hussain, M.; Peters, G.; Pletz, M.W.; et al. The Staphylococcus aureus extracellular matrix protein (Emp) has a fibrous structure and binds to different extracellular matrices. Sci. Rep. 2017, 7, 13665. [Google Scholar] [CrossRef]
- Kneidl, J.; Löffler, B.; Erat, M.C.; Kalinka, J.; Peters, G.; Roth, J.; Barczyk, K. Soluble CD163 promotes recognition, phagocytosis and killing of Staphylococcus aureus via binding of specific fibronectin peptides. Cell Microbiol. 2012, 14, 914–936. [Google Scholar] [CrossRef]
- Griffeth, G.C.; Morris, D.O.; Abraham, J.L.; Shofer, F.S.; Rankin, S.C. Screening for skin carriage of methicillin-resistant coagulase-positive staphylococci and Staphylococcus schleiferi in dogs with healthy and inflamed skin. Vet. Dermatol. 2008, 19, 142–149. [Google Scholar] [CrossRef]
- Bannoehr, J.; Guardabassi, L. Staphylococcus pseudintermedius in the dog: taxonomy, diagnostics, ecology, epidemiology and pathogenicity. Vet. Dermatol. 2012, 23, 253–266. [Google Scholar] [CrossRef]
- Pietrocola, G.; Geoghegan, J.A.; Rindi, S.; Di Poto, A.; Missineo, A.; Consalvi, V.; Foster, T.J.; Speziale, P. Molecular Characterization of the Multiple Interactions of SpsD, a Surface Protein from Staphylococcus pseudintermedius, with Host Extracellular Matrix Proteins. PLoS ONE 2013, 8, e66901. [Google Scholar] [CrossRef]
- Pickering, A.C.; Vitry, P.; Prystopiuk, V.; Garcia, B.; Höök, M.; Schoenebeck, J.; Geoghegan, J.A.; Dufrêne, Y.F.; Fitzgerald, J.R. Host-specialized fibrinogen-binding by a bacterial surface protein promotes biofilm formation and innate immune evasion. PLoS Pathog. 2019, 15, e1007816. [Google Scholar] [CrossRef] [PubMed]
- Pietrocola, G.; Gianotti, V.; Richards, A.; Nobile, G.; Geoghegan, J.A.; Rindi, S.; Monk, I.R.; Bordt, A.S.; Foster, T.J.; Fitzgerald, J.R.; et al. Fibronectin Binding Proteins SpsD and SpsL Both Support Invasion of Canine Epithelial Cells by Staphylococcus pseudintermedius. Infect. Immun. 2015, 83, 4093–4102. [Google Scholar] [CrossRef] [PubMed]
- Stevens, D.L. Invasive group A streptococcus infections. Clin. Infect. Dis. 1992, 14, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, M.W. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 2000, 13, 470–511. [Google Scholar] [CrossRef] [PubMed]
- Carapetis, J.R.; Steer, A.C.; Mulholland, E.K.; Weber, M. The global burden of group A streptococcal diseases. Lancet Infect. Dis. 2005, 5, 685–694. [Google Scholar] [CrossRef]
- Lamagni, T.L.; Darenberg, J.; Luca-Harari, B.; Siljander, T.; Efstratiou, A.; Henriques-Normark, B.; Vuopio-Varkila, J.; Bouvet, A.; Creti, R.; Ekelund, K.; et al. Epidemiology of severe Streptococcus pyogenes disease in Europe. J. Clin. Microbiol. 2008, 46, 2359–2367. [Google Scholar] [CrossRef]
- O’Brien, K.L.; Beall, B.; Barrett, N.L.; Cieslak, P.R.; Reingold, A.; Farley, M.M.; Danila, R.; Zell, E.R.; Facklam, R.; Schwartz, B.; et al. Epidemiology of invasive group a streptococcus disease in the United States, 1995-1999. Clin. Infect. Dis. 2002, 35, 268–276. [Google Scholar] [CrossRef]
- Cunningham, M.W. Molecular Mimicry, Autoimmunity, and Infection: The Cross-Reactive Antigens of Group A Streptococci and their Sequelae. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Terao, Y.; Kawabata, S. Pleiotropic virulence factor - Streptococcus pyogenes fibronectin-binding proteins. Cell Microbiol. 2013, 15, 503–511. [Google Scholar] [CrossRef]
- Talay, S.R.; Valentin-Weigand, P.; Jerlström, P.G.; Timmis, K.N.; Chhatwal, G.S. Fibronectin-binding protein of Streptococcus pyogenes: Sequence of the binding domain involved in adherence of streptococci to epithelial cells. Infect. Immun. 1992, 60, 3837–3844. [Google Scholar]
- Hanski, E.; Caparon, M. Protein F, a fibronectin-binding protein, is an adhesin of the group A streptococcus Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 1992, 89, 6172–6176. [Google Scholar] [CrossRef] [PubMed]
- Hanski, E.; Horwitz, P.A.; Caparon, M.G. Expression of protein F, the fibronectin-binding protein of Streptococcus pyogenes JRS4, in heterologous streptococcal and enterococcal strains promotes their adherence to respiratory epithelial cells. Infect. Immun. 1992, 60, 5119–5125. [Google Scholar] [PubMed]
- Norris, N.C.; Bingham, R.J.; Harris, G.; Speakman, A.; Jones, R.P.; Leech, A.; Turkenburg, J.P.; Potts, J.R. Structural and functional analysis of the tandem β-zipper interaction of a Streptococcal protein with human fibronectin. J. Biol. Chem. 2011, 286, 38311–38320. [Google Scholar] [CrossRef]
- Sela, S.; Aviv, A.; Tovi, A.; Burstein, I.; Caparon, M.G.; Hanski, E. Protein F: An adhesin of Streptococcus pyogenes binds fibronectin via two distinct domains. Mol. Microbiol. 1993, 10, 1049–1055. [Google Scholar] [CrossRef] [PubMed]
- Ozeri, V.; Tovi, A.; Burstein, I.; Natanson-Yaron, S.; Caparon, M.G.; Yamada, K.M.; Akiyama, S.K.; Vlodavsky, I.; Hanski, E. A two-domain mechanism for group A streptococcal adherence through protein F to the extracellular matrix. EMBO J. 1996, 15, 989–998. [Google Scholar] [CrossRef]
- Talay, S.R.; Zock, A.; Rohde, M.; Molinari, G.; Oggioni, M.; Pozzi, G.; Guzman, C.A.; Chhatwal, G.S. Co-operative binding of human fibronectin to Sfbl protein triggers streptococcal invasion into respiratory epithelial cells. Cell Microbiol. 2000, 2, 521–535. [Google Scholar] [CrossRef]
- Tomasini-Johansson, B.R.; Kaufman, N.R.; Ensenberger, M.G.; Ozeri, V.; Hanski, E.; Mosher, D.F. A 49-residue peptide from adhesin F1 of Streptococcus pyogenes inhibits fibronectin matrix assembly. J. Biol. Chem. 2001, 276, 23430–23439. [Google Scholar] [CrossRef]
- Kreikemeyer, B.; Oehmcke, S.; Nakata, M.; Hoffrogge, R.; Podbielski, A. Streptococcus pyogenes fibronectin-binding protein F2: Expression profile, binding characteristics, and impact on eukaryotic cell interactions. J. Biol. Chem. 2004, 279, 15850–15859. [Google Scholar] [CrossRef] [PubMed]
- Terao, Y.; Kawabata, S.; Kunitomo, E.; Murakami, J.; Nakagawa, I.; Hamada, S. Fba, a novel fibronectin-binding protein from Streptococcus pyogenes, promotes bacterial entry into epithelial cells, and the fba gene is positively transcribed under the Mga regulator. Mol. Microbiol. 2001, 42, 75–86. [Google Scholar] [CrossRef]
- Terao, Y.; Kawabata, S.; Nakata, M.; Nakagawa, I.; Hamada, S. Molecular characterization of a novel fibronectin-binding protein of Streptococcus pyogenes strains isolated from toxic shock-like syndrome patients. J. Biol. Chem. 2002, 277, 47428–47435. [Google Scholar] [CrossRef]
- Amelung, S.; Nerlich, A.; Rohde, M.; Spellerberg, B.; Cole, J.N.; Nizet, V.; Chhatwal, G.S.; Talay, S.R. The FbaB-type fibronectin-binding protein of Streptococcus pyogenes promotes specific invasion into endothelial cells. Cell Microbiol. 2011, 13, 1200–1211. [Google Scholar] [CrossRef] [PubMed]
- Oehmcke, S.; Podbielski, A.; Kreikemeyer, B. Function of the fibronectin-binding serum opacity factor of Streptococcus pyogenes in adherence to epithelial cells. Infect. Immun. 2004, 72, 4302–4308. [Google Scholar] [CrossRef]
- Jeng, A.; Sakota, V.; Li, Z.; Datta, V.; Beall, B.; Nizet, V. Molecular genetic analysis of a group A Streptococcus operon encoding serum opacity factor and a novel fibronectin-binding protein, SfbX. J. Bacteriol. 2003, 185, 1208–1217. [Google Scholar] [CrossRef] [PubMed]
- Cue, D.; Dombek, P.E.; Lam, H.; Cleary, P.P. Streptococcus pyogenes serotype M1 encodes multiple pathways for entry into human epithelial cells. Infect. Immun. 1998, 66, 4593–4601. [Google Scholar]
- Cue, D.; Southern, S.O.; Southern, P.J.; Prabhakar, J.; Lorelli, W.; Smallheer, J.M.; Mousa, S.A.; Cleary, P.P. A nonpeptide integrin antagonist can inhibit epithelial cell ingestion of Streptococcus pyogenes by blocking formation of integrin alpha 5beta 1-fibronectin-M1 protein complexes. Proc. Natl. Acad. Sci. USA 2000, 97, 2858–2863. [Google Scholar] [CrossRef] [PubMed]
- Frick, I.M.; Crossin, K.L.; Edelman, G.M.; Björck, L. Protein H--a bacterial surface protein with affinity for both immunoglobulin and fibronectin type III domains. EMBO J. 1995, 14, 1674–1679. [Google Scholar] [CrossRef]
- Bates, C.S.; Montañez, G.E.; Woods, C.R.; Vincent, R.M.; Eichenbaum, Z. Identification and characterization of a Streptococcus pyogenes operon involved in binding of hemoproteins and acquisition of iron. Infect. Immun. 2003, 71, 1042–1055. [Google Scholar] [CrossRef]
- Fisher, M.; Huang, Y.S.; Li, X.; McIver, K.S.; Toukoki, C.; Eichenbaum, Z. Shr is a broad-spectrum surface receptor that contributes to adherence and virulence in group A streptococcus. Infect. Immun. 2008, 76, 5006–5015. [Google Scholar] [CrossRef]
- McNitt, D.H.; De Water, L.V.; Marasco, D.; Berisio, R.; Lukomski, S. Streptococcal Collagen-like Protein 1 Binds Wound Fibronectin: Implications in Pathogen Targeting. Curr. Med. Chem. 2019, 26, 1933–1945. [Google Scholar] [CrossRef]
- McNitt, D.H.; Choi, S.J.; Allen, J.L.; Hames, R.A.; Weed, S.A.; Van De Water, L.; Berisio, R.; Lukomski, S. Adaptation of the group A Streptococcus adhesin Scl1 to bind fibronectin type III repeats within wound-associated extracellular matrix: Implications for cancer therapy. Mol. Microbiol. 2019, 112, 800–819. [Google Scholar] [CrossRef]
- Oliver-Kozup, H.; Martin, K.H.; Schwegler-Berry, D.; Green, B.J.; Betts, C.; Shinde, A.V.; Van De Water, L.; Lukomski, S. The group A streptococcal collagen-like protein-1, Scl1, mediates biofilm formation by targeting the extra domain A-containing variant of cellular fibronectin expressed in wounded tissue. Mol. Microbiol. 2013, 87, 672–689. [Google Scholar] [CrossRef] [PubMed]
- McNitt, D.H.; Choi, S.J.; Keene, D.R.; Van De Water, L.; Squeglia, F.; Berisio, R.; Lukomski, S. Surface-exposed loops and an acidic patch in the Scl1 protein of group A Streptococcus enable Scl1 binding to wound-associated fibronectin. J. Biol. Chem. 2018, 293, 7796–7810. [Google Scholar] [CrossRef] [PubMed]
- Pancholi, V.; Fischetti, V.A. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 1992, 176, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Courtney, H.S.; Li, Y.; Dale, J.B.; Hasty, D.L. Cloning, sequencing, and expression of a fibronectin/fibrinogen-binding protein from group A streptococci. Infect. Immun. 1994, 62, 3937–3946. [Google Scholar]
- Courtney, H.S.; Dale, J.B.; Hasty, D.I. Differential effects of the streptococcal fibronectin-binding protein, FBP54, on adhesion of group A streptococci to human buccal cells and HEp-2 tissue culture cells. Infect. Immun. 1996, 64, 2415–2419. [Google Scholar]
- Austrian, R. Some aspects of the pneumococcal carrier state. J. Antimicrob. Chemother. 1986, 18, 35–45. [Google Scholar] [CrossRef]
- Musher, D.M. How contagious are common respiratory tract infections? N. Engl. J. Med. 2003, 348, 1256–1266. [Google Scholar] [CrossRef]
- Holmes, A.R.; McNab, R.; Millsap, K.W.; Rohde, M.; Hammerschmidt, S.; Mawdsley, J.L.; Jenkinson, H.F. The pavA gene of Streptococcus pneumoniae encodes a fibronectin-binding protein that is essential for virulence. Mol. Microbiol. 2001, 41, 1395–1408. [Google Scholar] [CrossRef]
- Voss, S.; Gámez, G.; Hammerschmidt, S. Impact of pneumococcal microbial surface components recognizing adhesive matrix molecules on colonization. Mol. Oral Microbiol. 2012, 27, 246–256. [Google Scholar] [CrossRef]
- Bumbaca, D.; Littlejohn, J.E.; Nayakanti, H.; Rigden, D.J.; Galperin, M.Y.; Jedrzejas, M.J. Sequence analysis and characterization of a novel fibronectin-binding repeat domain from the surface of Streptococcus pneumoniae. OMICS 2004, 8, 341–356. [Google Scholar] [CrossRef]
- Jensch, I.; Gámez, G.; Rothe, M.; Ebert, S.; Fulde, M.; Somplatzki, D.; Bergmann, S.; Petruschka, L.; Rohde, M.; Nau, R.; et al. PavB is a surface-exposed adhesin of Streptococcus pneumoniae contributing to nasopharyngeal colonization and airways infections. Mol. Microbiol. 2010, 77, 22–43. [Google Scholar] [CrossRef]
- Jedrzejas, M.J. Unveiling molecular mechanisms of bacterial surface proteins: Streptococcus pneumoniae as a model organism for structural studies. Cell Mol. Life Sci. 2007, 64, 2799–2822. [Google Scholar] [CrossRef]
- Binsker, U.; Kohler, T.P.; Krauel, K.; Kohler, S.; Schwertz, H.; Hammerschmidt, S. Pneumococcal Adhesins PavB and PspC Are Important for the Interplay with Human Thrombospondin-1. J. Biol. Chem. 2015, 290, 14542–14555. [Google Scholar] [CrossRef] [PubMed]
- Kanwal, S.; Jensch, I.; Palm, G.J.; Brönstrup, M.; Rohde, M.; Kohler, T.P.; Somplatzki, D.; Tegge, W.; Jenkinson, H.F.; Hammerschmidt, S. Mapping the recognition domains of pneumococcal fibronectin-binding proteins PavA and PavB demonstrates a common pattern of molecular interactions with fibronectin type III repeats. Mol. Microbiol. 2017, 105, 839–859. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M.; Terao, Y.; Mori, Y.; Hamada, S.; Kawabata, S. PfbA, a novel plasmin- and fibronectin-binding protein of Streptococcus pneumoniae, contributes to fibronectin-dependent adhesion and antiphagocytosis. J. Biol. Chem. 2008, 283, 36272–36279. [Google Scholar] [CrossRef]
- Beulin, D.S.J.; Radhakrishnan, D.; Suresh, S.; Sadasivan, C.; Yamaguchi, M.; Kawabata, S.; Ponnuraj, K. Streptococcus pneumoniae surface protein PfbA is a versatile multidomain and multiligand-binding adhesin employing different binding mechanisms. FEBS J. 2017, 284, 3404–3421. [Google Scholar] [CrossRef] [PubMed]
- Papasergi, S.; Garibaldi, M.; Tuscano, G.; Signorino, G.; Ricci, S.; Peppoloni, S.; Pernice, I.; Lo Passo, C.; Teti, G.; Felici, F.; et al. Plasminogen- and fibronectin-binding protein B is involved in the adherence of Streptococcus pneumoniae to human epithelial cells. J. Biol. Chem. 2010, 285, 7517–7524. [Google Scholar] [CrossRef]
- Izoré, T.; Contreras-Martel, C.; El Mortaji, L.; Manzano, C.; Terrasse, R.; Vernet, T.; Di Guilmi, A.M.; Dessen, A. Structural basis of host cell recognition by the pilus adhesin from Streptococcus pneumoniae. Structure 2010, 18, 106–115. [Google Scholar] [CrossRef]
- Nelson, A.L.; Ries, J.; Bagnoli, F.; Dahlberg, S.; Fälker, S.; Rounioja, S.; Tschöp, J.; Morfeldt, E.; Ferlenghi, I.; Hilleringmann, M.; et al. RrgA is a pilus-associated adhesin in Streptococcus pneumoniae. Mol. Microbiol. 2007, 66, 329–340. [Google Scholar] [CrossRef]
- Hilleringmann, M.; Giusti, F.; Baudner, B.C.; Masignani, V.; Covacci, A.; Rappuoli, R.; Barocchi, M.A.; Ferlenghi, I. Pneumococcal pili are composed of protofilaments exposing adhesive clusters of Rrg A. PLoS Pathog. 2008, 4, e1000026. [Google Scholar] [CrossRef]
- Becke, T.D.; Ness, S.; Gürster, R.; Schilling, A.F.; di Guilmi, A.M.; Sudhop, S.; Hilleringmann, M.; Clausen-Schaumann, H. Single Molecule Force Spectroscopy Reveals Two-Domain Binding Mode of Pilus-1 Tip Protein RrgA of Streptococcus pneumoniae to Fibronectin. ACS Nano 2018, 12, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Rosan, B.; Lamont, R.J. Dental plaque formation. Microbes. Infect. 2000, 2, 1599–1607. [Google Scholar] [CrossRef]
- Christie, J.; McNab, R.; Jenkinson, H.F. Expression of fibronectin-binding protein FbpA modulates adhesion in Streptococcus gordonii. Microbiology 2002, 148, 1615–1625. [Google Scholar] [CrossRef] [PubMed]
- Giomarelli, B.; Visai, L.; Hijazi, K.; Rindi, S.; Ponzio, M.; Iannelli, F.; Speziale, P.; Pozzi, G. Binding of Streptococcus gordonii to extracellular matrix proteins. FEMS Microbiol. Lett. 2006, 265, 172–177. [Google Scholar] [CrossRef] [PubMed]
- McNab, R.; Jenkinson, H.F.; Loach, D.M.; Tannock, G.W. Cell-surface-associated polypeptides CshA and CshB of high molecular mass are colonization determinants in the oral bacterium Streptococcus gordonii. Mol. Microbiol. 1994, 14, 743–754. [Google Scholar] [CrossRef] [PubMed]
- McNab, R.; Holmes, A.R.; Clarke, J.M.; Tannock, G.W.; Jenkinson, H.F. Cell surface polypeptide CshA mediates binding of Streptococcus gordonii to other oral bacteria and to immobilized fibronectin. Infect. Immun. 1996, 64, 4204–4210. [Google Scholar] [PubMed]
- Back, C.R.; Sztukowska, M.N.; Till, M.; Lamont, R.J.; Jenkinson, H.F.; Nobbs, A.H.; Rac, P.R. The Streptococcus gordonii Adhesin CshA Protein Binds Host Fibronectin via a Catch-Clamp Mechanism. J. Biol. Chem. 2017, 292, 1538–1549. [Google Scholar] [CrossRef]
- Miller-Torbert, T.A.; Sharma, S.; Holt, R.G. Inactivation of a gene for a fibronectin-binding protein of the oral bacterium Streptococcus mutans partially impairs its adherence to fibronectin. Microb. Pathog. 2008, 45, 53–59. [Google Scholar] [CrossRef]
- Staats, J.J.; Feder, I.; Okwumabua, O.; Chengappa, M.M. Streptococcus suis: Past and present. Vet. Res. Commun. 1997, 21, 381–407. [Google Scholar] [CrossRef]
- de Greeff, A.; Buys, H.; Verhaar, R.; Dijkstra, J.; van Alphen, L.; Smith, H.E. Contribution of fibronectin-binding protein to pathogenesis of Streptococcus suis serotype 2. Infect. Immun. 2002, 70, 1319–1325. [Google Scholar] [CrossRef]
- Sun, L.Y.; Fan, H.J.; Lu, C.P. Determination of fibronectin-binding region of FBPS of Streptococcus suis type 2. Wei Sheng Wu Xue Bao 2005, 45, 753–756. [Google Scholar] [PubMed]
- Esgleas, M.; Li, Y.; Hancock, M.A.; Harel, J.; Dubreuil, J.D.; Gottschalk, M. Isolation and characterization of alpha-enolase, a novel fibronectin-binding protein from Streptococcus suis. Microbiology 2008, 154, 2668–2679. [Google Scholar] [CrossRef] [PubMed]
- Baums, C.G.; Kaim, U.; Fulde, M.; Ramachandran, G.; Goethe, R.; Valentin-Weigand, P. Identification of a novel virulence determinant with serum opacification activity in Streptococcus suis. Infect. Immun. 2006, 74, 6154–6162. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.K.; Gu, Z.Y.; Matsuka, Y.V.; Purushothaman, S.S.; Winter, L.A.; Cleary, P.P.; Olmsted, S.B.; Ohlendorf, D.H.; Earhart, C.A. Structure of the streptococcal cell wall C5a peptidase. Proc. Natl. Acad. Sci. USA 2005, 102, 18391–18396. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Stafslien, D.; Purushothaman, S.S.; Cleary, P. The group B streptococcal C5a peptidase is both a specific protease and an invasin. Infect. Immun. 2002, 70, 2408–2413. [Google Scholar] [CrossRef] [PubMed]
- Hull, J.R.; Tamura, G.S.; Castner, D.G. Interactions of the streptococcal C5a peptidase with human fibronectin. Acta Biomater. 2008, 4, 504–513. [Google Scholar] [CrossRef][Green Version]
- Tamura, G.S.; Hull, J.R.; Oberg, M.D.; Castner, D.G. High-affinity interaction between fibronectin and the group B streptococcal C5a peptidase is unaffected by a naturally occurring four-amino-acid deletion that eliminates peptidase activity. Infect. Immun. 2006, 74, 5739–5746. [Google Scholar] [CrossRef]
- Murray, B.E. Vancomycin-resistant enterococcal infections. N. Engl. J. Med. 2000, 342, 710–721. [Google Scholar] [CrossRef]
- Teng, F.; Kawalec, M.; Weinstock, G.M.; Hryniewicz, W.; Murray, B.E. An Enterococcus faecium secreted antigen, SagA, exhibits broad-spectrum binding to extracellular matrix proteins and appears essential for E. faecium growth. Infect. Immun. 2003, 71, 5033–5041. [Google Scholar] [CrossRef]
- Somarajan, S.R.; La Rosa, S.L.; Singh, K.V.; Roh, J.H.; Höök, M.; Murray, B.E. The fibronectin-binding protein Fnm contributes to adherence to extracellular matrix components and virulence of Enterococcus faecium. Infect. Immun. 2015, 83, 4653–4661. [Google Scholar] [CrossRef]
- Galloway-Peña, J.R.; Liang, X.; Singh, K.V.; Yadav, P.; Chang, C.; La Rosa, S.L.; Shelburne, S.; Ton-That, H.; Höök, M.; Murray, B.E. The identification and functional characterization of WxL proteins from Enterococcus faecium reveal surface proteins involved in extracellular matrix interactions. J. Bacteriol. 2015, 197, 882–892. [Google Scholar] [CrossRef] [PubMed]
- Guzmán Prieto, A.M.; Urbanus, R.T.; Zhang, X.; Bierschenk, D.; Koekman, C.A.; van Luit-Asbroek, M.; Ouwerkerk, J.P.; Pape, M.; Paganelli, F.L.; Wobser, D.; et al. The N-terminal domain of the thermo-regulated surface protein PrpA of Enterococcus faecium binds to fibrinogen, fibronectin and platelets. Sci Rep. 2015, 5, 18255. [Google Scholar] [CrossRef]
- Murray, B.E. The life and times of the Enterococcus. Clin. Microbiol. Rev. 1990, 3, 46–65. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, J.A.; Teng, F.; Nallapareddy, S.R.; Murray, B.E. Pleiotrophic effects of 2 Enterococcus faecalis sagA-like genes, salA and salB, which encode proteins that are antigenic during human infection, on biofilm formation and binding to collagen type i and fibronectin. J. Infect. Dis. 2006, 193, 231–240. [Google Scholar] [CrossRef]
- Torelli, R.; Serror, P.; Bugli, F.; Paroni Sterbini, F.; Florio, A.R.; Stringaro, A.; Colone, M.; De Carolis, E.; Martini, C.; Giard, J.C.; et al. The PavA-like fibronectin-binding protein of Enterococcus faecalis, EfbA, is important for virulence in a mouse model of ascending urinary tract infection. J. Infect. Dis. 2012, 206, 952–960. [Google Scholar] [CrossRef][Green Version]
- Singh, K.V.; La Rosa, S.L.; Somarajan, S.R.; Roh, J.H.; Murray, B.E. The fibronectin-binding protein EfbA contributes to pathogenesis and protects against infective endocarditis caused by Enterococcus faecalis. Infect. Immun. 2015, 83, 4487–4494. [Google Scholar] [CrossRef][Green Version]
- Peake, P.; Gooley, A.; Britton, W.J. Mechanism of interaction of the 85B secreted protein of Mycobacterium bovis with fibronectin. Infect. Immun. 1993, 61, 4828–4834. [Google Scholar] [PubMed]
- Naito, M.; Fukuda, T.; Sekiguchi, K.; Yamada, T. The domains of human fibronectin mediating the binding of alpha antigen, the most immunopotent antigen of mycobacteria that induces protective immunity against mycobacterial infection. Biochem. J. 2000, 347 Pt 3, 725–731. [Google Scholar] [CrossRef]
- Ohara, N.; Kitaura, H.; Hotokezaka, H.; Nishiyama, T.; Wada, N.; Matsumoto, S.; Matsuo, T.; Naito, M.; Yamada, T. Characterization of the gene encoding the MPB51, one of the major secreted protein antigens of Mycobacterium bovis BCG, and identification of the secreted protein closely related to the fibronectin binding 85 complex. Scand J. Immunol. 1995, 41, 433–442. [Google Scholar] [CrossRef]
- Schorey, J.S.; Li, Q.; McCourt, D.W.; Bong-Mastek, M.; Clark-Curtiss, J.E.; Ratliff, T.L.; Brown, E.J. A Mycobacterium leprae gene encoding a fibronectin binding protein is used for efficient invasion of epithelial cells and Schwann cells. Infect. Immun. 1995, 63, 2652–2657. [Google Scholar]
- Schorey, J.S.; Holsti, M.A.; Ratliff, T.L.; Allen, P.M.; Brown, E.J. Characterization of the fibronectin-attachment protein of Mycobacterium avium reveals a fibronectin-binding motif conserved among mycobacteria. Mol. Microbiol. 1996, 21, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Schorey, J.S.; Groger, R.; Allen, P.M.; Brown, E.J.; Ratliff, T.L. Characterization of the fibronectin binding motif for a unique mycobacterial fibronectin attachment protein, FAP. J. Biol. Chem. 1999, 274, 4521–4526. [Google Scholar] [CrossRef] [PubMed]
- Kinhikar, A.G.; Vargas, D.; Li, H.; Mahaffey, S.B.; Hinds, L.; Belisle, J.T.; Laal, S. Mycobacterium tuberculosis malate synthase is a laminin-binding adhesin. Mol. Microbiol. 2006, 60, 999–1013. [Google Scholar] [CrossRef] [PubMed]
- Gilot, P.; André, P.; Content, J. Listeria monocytogenes possesses adhesins for fibronectin. Infect. Immun. 1999, 67, 6698–6701. [Google Scholar]
- Gilot, P.; Jossin, Y.; Content, J. Cloning, sequencing and characterisation of a Listeria monocytogenes gene encoding a fibronectin-binding protein. J. Med. Microbiol. 2000, 49, 887–896. [Google Scholar] [CrossRef][Green Version]
- Dramsi, S.; Bourdichon, F.; Cabanes, D.; Lecuit, M.; Fsihi, H.; Cossart, P. FbpA, a novel multifunctional Listeria monocytogenes virulence factor. Mol. Microbiol. 2004, 53, 639–649. [Google Scholar] [CrossRef]
- Hennequin, C.; Janoir, C.; Barc, M.C.; Collignon, A.; Karjalainen, T. Identification and characterization of a fibronectin-binding protein from Clostridium difficile. Microbiology 2003, 149, 2779–2787. [Google Scholar] [CrossRef]
- Katayama, S.; Nozu, N.; Okuda, M.; Hirota, S.; Yamasaki, T.; Hitsumoto, Y. Characterization of two putative fibronectin-binding proteins of Clostridium perfringens. Anaerobe 2009, 15, 155–159. [Google Scholar] [CrossRef]
- Katayama, S.; Tagomori, M.; Morita, N.; Yamasaki, T.; Nariya, H.; Okada, M.; Watanabe, M.; Hitsumoto, Y. Determination of the Clostridium perfringens-binding site on fibronectin. Anaerobe 2015, 34, 174–181. [Google Scholar] [CrossRef]
- Fröman, G.; Switalski, L.M.; Faris, A.; Wadström, T.; Höök, M. Binding of Escherichia coli to fibronectin. A mechanism of tissue adherence. J. Biol. Chem. 1984, 259, 14899–14905. [Google Scholar]
- Sokurenko, E.V.; Courtney, H.S.; Ohman, D.E.; Klemm, P.; Hasty, D.L. FimH family of type 1 fimbrial adhesins: Functional heterogeneity due to minor sequence variations among fimH genes. J. Bacteriol. 1994, 176, 748–755. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Valle, J.; Mabbett, A.N.; Ulett, G.C.; Toledo-Arana, A.; Wecker, K.; Totsika, M.; Schembri, M.A.; Ghigo, J.M.; Beloin, C. UpaG, a new member of the trimeric autotransporter family of adhesins in uropathogenic Escherichia coli. J. Bacteriol. 2008, 190, 4147–4161. [Google Scholar] [CrossRef] [PubMed]
- Gophna, U.; hlaeger, T.A.; Hacker, J.; Ron, E.Z. Role of fibronectin in curli-mediated internalization. FEMS Microbiol. Lett. 2002, 212, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Xicohtencatl-Cortes, J.; Monteiro-Neto, V.; Saldaña, Z.; Ledesma, M.A.; Puente, J.L.; Girón, J.A. The type 4 pili of enterohemorrhagic Escherichia coli O157:H7 are multipurpose structures with pathogenic attributes. J. Bacteriol. 2009, 191, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Konkel, M.E.; Christensen, J.E.; Keech, A.M.; Monteville, M.R.; Klena, J.D.; Garvis, S.G. Identification of a fibronectin-binding domain within the Campylobacter jejuni CadF protein. Mol. Microbiol. 2005, 57, 1022–1035. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, R.C.; Neal-McKinney, J.M.; Dhillon, A.S.; Miller, W.G.; Konkel, M.E. Examination of Campylobacter jejuni putative adhesins leads to the identification of a new protein, designated FlpA, required for chicken colonization. Infect. Immun. 2009, 77, 2399–2407. [Google Scholar] [CrossRef]
- Konkel, M.E.; Larson, C.L.; Flanagan, R.C. Campylobacter jejuni FlpA binds fibronectin and is required for maximal host cell adherence. J. Bacteriol. 2010, 192, 68–76. [Google Scholar] [CrossRef]
- Eucker, T.P.; Konkel, M.E. The cooperative action of bacterial fibronectin-binding proteins and secreted proteins promote maximal Campylobacter jejuni invasion of host cells by stimulating membrane ruffling. Cell Microbiol. 2012, 14, 226–238. [Google Scholar] [CrossRef]
- Kingsley, R.A.; Santos, R.L.; Keestra, A.M.; Adams, L.G.; Bäumler, A.J. Salmonella enterica serotype Typhimurium ShdA is an outer membrane fibronectin-binding protein that is expressed in the intestine. Mol. Microbiol. 2002, 43, 895–905. [Google Scholar] [CrossRef]
- Kingsley, R.A.; Abi Ghanem, D.; Puebla-Osorio, N.; Keestra, A.M.; Berghman, L.; Bäumler, A.J. Fibronectin binding to the Salmonella enterica serotype Typhimurium ShdA autotransporter protein is inhibited by a monoclonal antibody recognizing the A3 repeat. J. Bacteriol. 2004, 186, 4931–4939. [Google Scholar] [CrossRef]
- Kingsley, R.A.; Keestra, A.M.; de Zoete, M.R.; Bäumler, A.J. The ShdA adhesin binds to the cationic cradle of the fibronectin 13FnIII repeat module: Evidence for molecular mimicry of heparin binding. Mol. Microbiol. 2004, 52, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Dorsey, C.W.; Laarakker, M.C.; Humphries, A.D.; Weening, E.H.; Bäumler, A.J. Salmonella enterica serotype Typhimurium MisL is an intestinal colonization factor that binds fibronectin. Mol. Microbiol. 2005, 57, 196–211. [Google Scholar] [CrossRef] [PubMed]
- Stanek, G.; Wormser, G.P.; Gray, J.; Strle, F. Lyme borreliosis. Lancet 2012, 379, 461–473. [Google Scholar] [CrossRef]
- Radolf, J.D.; Caimano, M.J.; Stevenson, B.; Hu, L.T. Of ticks, mice and men: Understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat. Rev. Microbiol. 2012, 10, 87–99. [Google Scholar] [CrossRef]
- Hyde, J.A.; Weening, E.H.; Chang, M.; Trzeciakowski, J.P.; Höök, M.; Cirillo, J.D.; Skare, J.T. Bioluminescent imaging of Borrelia burgdorferi in vivo demonstrates that the fibronectin-binding protein BBK32 is required for optimal infectivity. Mol. Microbiol. 2011, 82, 99–113. [Google Scholar] [CrossRef]
- Caine, J.A.; Coburn, J. Multifunctional and Redundant Roles of Borrelia burgdorferi Outer Surface Proteins in Tissue Adhesion, Colonization, and Complement Evasion. Front. Immunol. 2016, 7, 442. [Google Scholar] [CrossRef]
- Kim, J.H.; Singvall, J.; Schwarz-Linek, U.; Johnson, B.J.; Potts, J.R.; Höök, M. BBK32, a fibronectin binding MSCRAMM from Borrelia burgdorferi, contains a disordered region that undergoes a conformational change on ligand binding. J. Biol. Chem. 2004, 279, 41706–41714. [Google Scholar] [CrossRef]
- Prabhakaran, S.; Liang, X.; Skare, J.T.; Potts, J.R.; Höök, M. A novel fibronectin binding motif in MSCRAMMs targets F3 modules. PLoS ONE 2009, 4, e5412. [Google Scholar] [CrossRef]
- Wu, J.; Weening, E.H.; Faske, J.B.; Höök, M.; Skare, J.T. Invasion of eukaryotic cells by Borrelia burgdorferi requires β(1) integrins and Src kinase activity. Infect. Immun. 2011, 79, 1338–1348. [Google Scholar] [CrossRef]
- Gaultney, R.A.; Gonzalez, T.; Floden, A.M.; Brissette, C.A. BB0347, from the lyme disease spirochete Borrelia burgdorferi, is surface exposed and interacts with the CS1 heparin-binding domain of human fibronectin. PLoS ONE 2013, 8, e75643. [Google Scholar] [CrossRef]
- Floden, A.M.; Gonzalez, T.; Gaultney, R.A.; Brissette, C.A. Evaluation of RevA, a fibronectin-binding protein of Borrelia burgdorferi, as a potential vaccine candidate for lyme disease. Clin. Vaccine Immunol. 2013, 20, 892–899. [Google Scholar] [CrossRef]
- Kuypers, J.M.; Proctor, R.A. Reduced adherence to traumatized rat heart valves by a low-fibronectin-binding mutant of Staphylococcus aureus. Infect. Immun. 1989, 57, 2306–2312. [Google Scholar]
- Que, Y.A.; François, P.; Haefliger, J.A.; Entenza, J.M.; Vaudaux, P.; Moreillon, P. Reassessing the role of Staphylococcus aureus clumping factor and fibronectin-binding protein by expression in Lactococcus lactis. Infect. Immun. 2001, 69, 6296–6302. [Google Scholar] [CrossRef]
- Heying, R.; van de Gevel, J.; Que, Y.A.; Moreillon, P.; Beekhuizen, H. Fibronectin-binding proteins and clumping factor A in Staphylococcus aureus experimental endocarditis: FnBPA is sufficient to activate human endothelial cells. Thromb. Haemost. 2007, 97, 617–626. [Google Scholar] [CrossRef]
- Piroth, L.; Que, Y.A.; Widmer, E.; Panchaud, A.; Piu, S.; Entenza, J.M.; Moreillon, P. The fibrinogen- and fibronectin-binding domains of Staphylococcus aureus fibronectin-binding protein A synergistically promote endothelial invasion and experimental endocarditis. Infect. Immun. 2008, 76, 3824–3831. [Google Scholar] [CrossRef]
- Heying, R.; van de Gevel, J.; Que, Y.A.; Piroth, L.; Moreillon, P.; Beekhuizen, H. Contribution of (sub)domains of Staphylococcus aureus fibronectin-binding protein to the proinflammatory and procoagulant response of human vascular endothelial cells. Thromb. Haemost. 2009, 101, 495–504. [Google Scholar] [CrossRef]
- Courtney, H.S.; Hasty, D.L.; Li, Y.; Chiang, H.C.; Thacker, J.L.; Dale, J.B. Serum opacity factor is a major fibronectin-binding protein and a virulence determinant of M type 2 Streptococcus pyogenes. Mol. Microbiol. 1999, 32, 89–98. [Google Scholar] [CrossRef]
- Lowrance, J.H.; Baddour, L.M.; Simpson, W.A. The role of fibronectin binding in the rat model of experimental endocarditis caused by Streptococcus sanguis. J. Clin. Investig. 1990, 86, 7–13. [Google Scholar] [CrossRef]
- Seshu, J.; Esteve-Gassent, M.D.; Labandeira-Rey, M.; Kim, J.H.; Trzeciakowski, J.P.; Höök, M.; Skare, J.T. Inactivation of the fibronectin-binding adhesin gene bbk32 significantly attenuates the infectivity potential of Borrelia burgdorferi. Mol. Microbiol. 2006, 59, 1591–1601. [Google Scholar] [CrossRef]
- Arciola, C.R.; Radin, L.; Alvergna, P.; Cenni, E.; Pizzoferrato, A. Heparin surface treatment of poly(methylmethacrylate) alters adhesion of a Staphylococcus aureus strain: Utility of bacterial fatty acid analysis. Biomaterials 1993, 14, 1161–1164. [Google Scholar] [CrossRef]
- Arciola, C.R.; Bustanji, Y.; Conti, M.; Campoccia, D.; Baldassarri, L.; Samorì, B.; Montanaro, L. Staphylococcus epidermidis-fibronectin binding and its inhibition by heparin. Biomaterials 2003, 24, 3013–3019. [Google Scholar] [CrossRef]
- Arciola, C.R.; Campoccia, D.; Gamberini, S.; Baldassarri, L.; Montanaro, L. Prevalence of cna, fnbA and fnbB adhesin genes among Staphylococcus aureus isolates from orthopedic infections associated to different types of implant. FEMS Microbiol. Lett. 2005, 246, 81–86. [Google Scholar] [CrossRef]
- Arciola, C.R.; Campoccia, D.; Montanaro, L. Implant infections: Adhesion, biofilm formation and immune evasion. Nat. Rev. Microbiol. 2018, 16, 397–409. [Google Scholar] [CrossRef]
- Guzmán, C.A.; Talay, S.R.; Molinari, G.; Medina, E.; Chhatwal, G.S. Protective immune response against Streptococcus pyogenes in mice after intranasal vaccination with the fibronectin-binding protein SfbI. J. Infect. Dis. 1999, 179, 901–906. [Google Scholar] [CrossRef]
- Kawabata, S.; Kunitomo, E.; Terao, Y.; Nakagawa, I.; Kikuchi, K.; Totsuka, K.; Hamada, S. Systemic and mucosal immunizations with fibronectin-binding protein FBP54 induce protective immune responses against Streptococcus pyogenes challenge in mice. Infect. Immun. 2001, 69, 924–930. [Google Scholar] [CrossRef]
- Casolini, F.; Visai, L.; Joh, D.; Conaldi, P.G.; Toniolo, A.; Höök, M.; Speziale, P. Antibody response to fibronectin-binding adhesin FnbpA in patients with Staphylococcus aureus infections. Infect. Immun. 1998, 66, 5433–5442. [Google Scholar]
- Brown, E.L.; Kim, J.H.; Reisenbichler, E.S.; Höök, M. Multicomponent Lyme vaccine: Three is not a crowd. Vaccine 2005, 23, 3687–3696. [Google Scholar] [CrossRef]
- Abd El Ghany, M.; Jansen, A.; Clare, S.; Hall, L.; Pickard, D.; Kingsley, R.A.; Dougan, G. Candidate live, attenuated Salmonella enterica serotype Typhimurium vaccines with reduced fecal shedding are immunogenic and effective oral vaccines. Infect. Immun. 2007, 75, 1835–1842. [Google Scholar] [CrossRef][Green Version]
| Adhesin | Host | Mass (kDa) | Fn Site | Binding Mechanism | Refs |
|---|---|---|---|---|---|
| FnBPA-FnBPB | Staphylococcus aureus | 106–104 | N-terminal domain | β-zipper | [28,31] |
| Ebh | S. aureus | 1100 | ? | ? | [42] |
| Eap (MAP) | S. aureus | 15 | ? | ? | [44] |
| Emp | S. aureus | 36 | ? | ? | [46,47] |
| SpsD | Staphylococcus pseudintermedius | 114 | N-terminal domain | ? | [53] |
| SpsL | S. pseudintermedius | 102 | N-terminal domain | ? | [53] |
| F1/Sfb1 | Streptococcus pyogenes | 69 | N-terminal domain Gelatin-binding domain | β-zipper | [64,65,66] |
| F2 | S. pyogenes | 128 | N-terminal domain | ? | [69] |
| FbaA | S. pyogenes | 38 | ? | ? | [70] |
| FbaB | S. pyogenes | 81 | ? | ? | [71] |
| SOF | S. pyogenes | 112 | N-terminal domain | ? | [73] |
| SfbX | S. pyogenes | 82 | ? | ? | [74] |
| Protein H | S. pyogenes | 37 | Central-binding domain | ? | [77] |
| Shr | S. pyogenes | 143 | ? | ? | [79] |
| Scl1 | S. pyogenes | 46 | EDA-EDB | ? | [80,81] |
| Fbp54 | S. pyogenes | 54 | N-terminal domain | ? | [85] |
| PavA | Streptococcus pneumoniae | 63 | Central-binding domain | ? | [89,95] |
| PavB | S. pneumoniae | 107 | Central-binding domain | ? | [95] |
| PfbA | S. pneumoniae | 74 | ? | ? | [96,97] |
| PfbB | S. pneumoniae | 120 | ? | ? | [98] |
| RrgA | S. pneumoniae | 98 | ? | ? | [100,101] |
| FbpA | Streptococcus gordonii | 63 | ? | ? | [104] |
| CshA | S. gordonii | 265 | ? | catch-clamp | [108] |
| FbpS | Streptococcus suis | 64 | N-terminal domain | ? | [111,112] |
| ScpB | Streptococcus agalactiae | 126 | ? | ? | [116,117] |
| SagA | Enterococcus faecium | 52 | ? | ? | [120] |
| Antigen 85A Antigen 85B Antigen 85C | Mycobacteria spp. | 31 30 31 | N-terminal domain Gelatin-binding domain | ? | [128,129] |
| FAP | Mycobacteria spp. | 30 | N-terminal domain | ? | [131,132,133] |
| FbpB | Clostridium perfrigens | 66 | FnIII9-FnIII10 | ? | [139,140] |
| Adhesin | Host | Mass (kDa) | Fn Site | Binding Mechanism | Refs |
|---|---|---|---|---|---|
| Curli | Escherichia coli | ? | FnIII10 | ? | [144] |
| CadF | Campilobacter jejuni | 34 | ? | ? | [146] |
| FlpA | C. jejuni | 46 | ? | ? | [147] |
| ShdA | Salmonella enterica serovar Typhimurium | 207 | FnIII13 | ? | [150,151,152] |
| MisL | S. enterica serovar Typhimurium | 101 | ? | ? | [153] |
| BBK32 | Borrelia burgdorferi | 47 | N-terminal domain Gelatin-binding domain FIII1–3 | β-zipper | [158,159] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Speziale, P.; Arciola, C.R.; Pietrocola, G. Fibronectin and Its Role in Human Infective Diseases. Cells 2019, 8, 1516. https://doi.org/10.3390/cells8121516
Speziale P, Arciola CR, Pietrocola G. Fibronectin and Its Role in Human Infective Diseases. Cells. 2019; 8(12):1516. https://doi.org/10.3390/cells8121516
Chicago/Turabian StyleSpeziale, Pietro, Carla Renata Arciola, and Giampiero Pietrocola. 2019. "Fibronectin and Its Role in Human Infective Diseases" Cells 8, no. 12: 1516. https://doi.org/10.3390/cells8121516
APA StyleSpeziale, P., Arciola, C. R., & Pietrocola, G. (2019). Fibronectin and Its Role in Human Infective Diseases. Cells, 8(12), 1516. https://doi.org/10.3390/cells8121516







