pMHC Structural Comparisons as a Pivotal Element to Detect and Validate T-Cell Targets for Vaccine Development and Immunotherapy—A New Methodological Proposal
Abstract
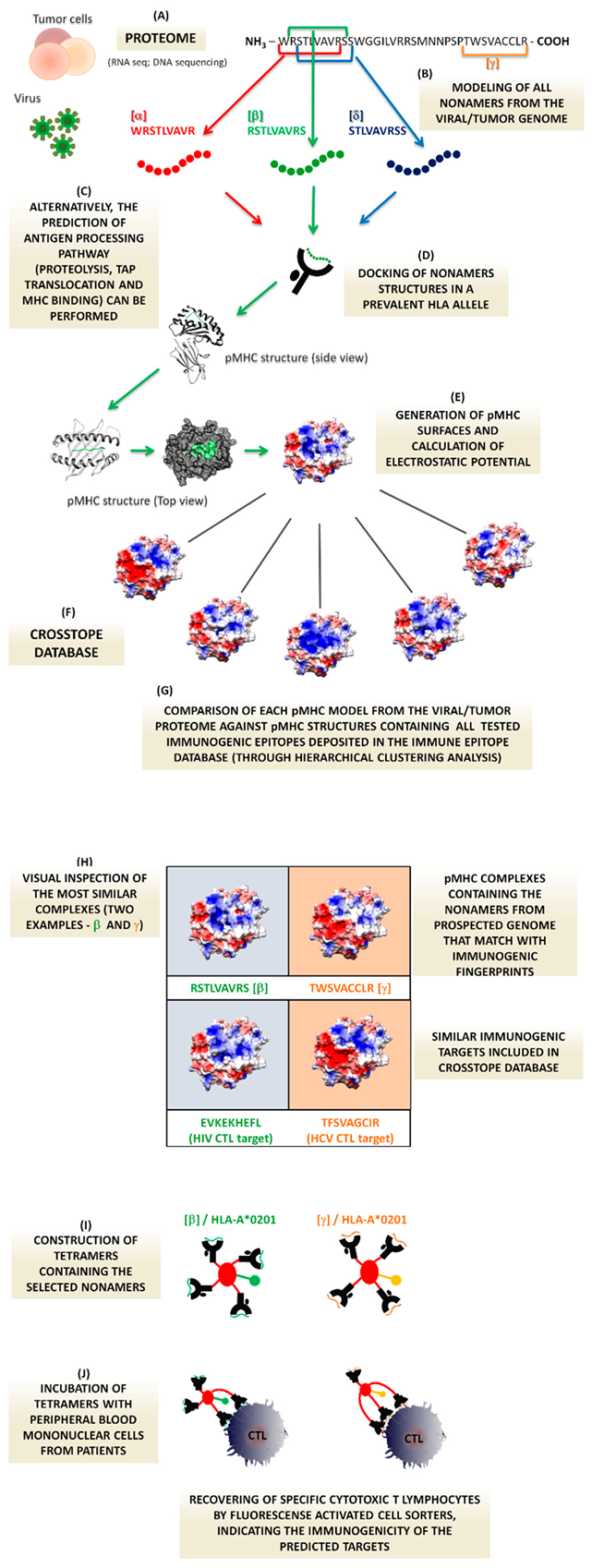
1. Background
1.1. Structural Immunoinformatica in Viral Infections—The Rational Basis
1.2. In Immunogenic Tumor Peptides
1.3. A Proof of Concept to Test the Putative T-Cell Targets: Tetramers Technology
1.4. Target Prediction and Cross-Reactivity
1.5. The Proposed Method Applied to Viral Vaccine Development
1.6. On Immunotherapy
2. Discussion and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Biek, R.; Pybus, O.G.; Lloyd-Smith, J.O.; Didelot, X. Measurably evolving pathogens in the genomic era. Trends Ecol. Evol. 2015, 30, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Cuevas, J.M.; Geller, R.; Garijo, R.; Lopez-Aldeguer, J.; Sanjuan, R. Extremely High Mutation Rate of HIV-1 in Vivo. PLoS Biol. 2015, 13, e1002251. [Google Scholar] [CrossRef] [PubMed]
- Lauring, A.S.; Frydman, J.; Andino, R. The role of mutational robustness in RNA virus evolution. Nat. Rev. Microbiol. 2013, 11, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, C.J.; Birger, R.B.; Funk, S.; Kouyos, R.D.; Lloyd-Smith, J.O.; Jansen, V.A. Five challenges in evolution and infectious diseases. Epidemics 2015, 10, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Pauly, M.D.; Procario, M.C.; Lauring, A.S. A novel twelve class fluctuation test reveals higher than expected mutation rates for influenza A viruses. Elife 2017, 6, e26437. [Google Scholar] [CrossRef] [PubMed]
- Volz, E.M.; Koelle, K.; Bedford, T. Viral phylodynamics. PLoS Comput. Biol. 2013, 9, e1002947. [Google Scholar] [CrossRef]
- Rosendahl Huber, S.; van Beek, J.; de Jonge, J.; Luytjes, W.; van Baarle, D. T cell responses to viral infections—Opportunities for Peptide vaccination. Front. Immunol. 2014, 5, 171. [Google Scholar] [CrossRef]
- Backert, L.; Kohlbacher, O. Immunoinformatics and epitope prediction in the age of genomic medicine. Genome Med. 2015, 7, 119. [Google Scholar] [CrossRef]
- Calis, J.J.; Maybeno, M.; Greenbaum, J.A.; Weiskopf, D.; De Silva, A.D.; Sette, A.; Kesmir, C.; Peters, B. Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Comput. Biol. 2013, 9, e1003266. [Google Scholar] [CrossRef]
- Kim, Y.; Sidney, J.; Buus, S.; Sette, A.; Nielsen, M.; Peters, B. Dataset size and composition impact the reliability of performance benchmarks for peptide-MHC binding predictions. BMC Bioinform. 2014, 15, 241. [Google Scholar] [CrossRef]
- Kuksa, P.P.; Min, M.R.; Dugar, R.; Gerstein, M. High-order neural networks and kernel methods for peptide-MHC binding prediction. Bioinformatics 2015, 31, 3600–3607. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, A.K.; Weiss, A. Insights into the initiation of TCR signaling. Nat. Immunol. 2014, 15, 798–807. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Shi, Y.; Cheng, H.; An, Y.Q.; Gao, G.F. Structural immunology and crystallography help immunologists see the immune system in action: How T and NK cells touch their ligands. IUBMB Life 2009, 61, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Hawse, W.F.; De, S.; Greenwood, A.I.; Nicholson, L.K.; Zajicek, J.; Kovrigin, E.L.; Kranz, D.M.; Garcia, K.C.; Baker, B.M. TCR scanning of peptide/MHC through complementary matching of receptor and ligand molecular flexibility. J. Immunol. 2014, 192, 2885–2891. [Google Scholar] [CrossRef] [PubMed]
- Kass, I.; Buckle, A.M.; Borg, N.A. Understanding the structural dynamics of TCR-pMHC complex interactions. Trends Immunol. 2014, 35, 604–612. [Google Scholar] [CrossRef]
- Singh, N.K.; Riley, T.P.; Baker, S.C.B.; Borrman, T.; Weng, Z.; Baker, B.M. Emerging Concepts in TCR Specificity: Rationalizing and (Maybe) Predicting Outcomes. J. Immunol. 2017, 199, 2203–2213. [Google Scholar] [CrossRef]
- Xia, Z.; Chen, H.; Kang, S.G.; Huynh, T.; Fang, J.W.; Lamothe, P.A.; Walker, B.D.; Zhou, R. The complex and specific pMHC interactions with diverse HIV-1 TCR clonotypes reveal a structural basis for alterations in CTL function. Sci. Rep. 2014, 4, 4087. [Google Scholar] [CrossRef]
- Antunes, D.A.; Rigo, M.M.; Silva, J.P.; Cibulski, S.P.; Sinigaglia, M.; Chies, J.A.; Vieira, G.F. Structural in silico analysis of cross-genotype-reactivity among naturally occurring HCV NS3-1073-variants in the context of HLA-A*02:01 allele. Mol. Immunol. 2011, 48, 1461–1467. [Google Scholar] [CrossRef]
- Mendes, M.F.; Antunes, D.A.; Rigo, M.M.; Sinigaglia, M.; Vieira, G.F. Improved structural method for T-cell cross-reactivity prediction. Mol. Immunol. 2015, 67, 303–310. [Google Scholar] [CrossRef]
- Sandalova, T.; Michaelsson, J.; Harris, R.A.; Odeberg, J.; Schneider, G.; Karre, K.; Achour, A. A structural basis for CD8+ T cell-dependent recognition of non-homologous peptide ligands: Implications for molecular mimicry in autoreactivity. J. Biol. Chem. 2005, 280, 27069–27075. [Google Scholar] [CrossRef]
- Rigo, M.M.; Antunes, D.A.; Vaz de Freitas, M.; Fabiano de Almeida Mendes, M.; Meira, L.; Sinigaglia, M.; Vieira, G.F. DockTope: A Web-based tool for automated pMHC-I modelling. Sci. Rep. 2015, 5, 18413. [Google Scholar] [CrossRef] [PubMed]
- Vita, R.; Overton, J.A.; Greenbaum, J.A.; Ponomarenko, J.; Clark, J.D.; Cantrell, J.R.; Wheeler, D.K.; Gabbard, J.L.; Hix, D.; Sette, A.; et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015, 43, D405–D412. [Google Scholar] [CrossRef] [PubMed]
- Sinigaglia, M.; Antunes, D.A.; Rigo, M.M.; Chies, J.A.; Vieira, G.F. CrossTope: A curate repository of 3D structures of immunogenic peptide: MHC complexes. Database 2013, 2013, bat002. [Google Scholar] [CrossRef] [PubMed]
- Hansen, S.G.; Sacha, J.B.; Hughes, C.M.; Ford, J.C.; Burwitz, B.J.; Scholz, I.; Gilbride, R.M.; Lewis, M.S.; Gilliam, A.N.; Ventura, A.B.; et al. Cytomegalovirus vectors violate CD8+ T cell epitope recognition paradigms. Science 2013, 340, 1237874. [Google Scholar] [CrossRef] [PubMed]
- Morens, D.M.; Folkers, G.K.; Fauci, A.S. The challenge of emerging and re-emerging infectious diseases. Nature 2004, 430, 242–249. [Google Scholar] [CrossRef]
- Bordner, A.J. Towards universal structure-based prediction of class II MHC epitopes for diverse allotypes. PLoS ONE 2010, 5, e14383. [Google Scholar] [CrossRef][Green Version]
- Khan, J.M.; Ranganathan, S. pDOCK: A new technique for rapid and accurate docking of peptide ligands to Major Histocompatibility Complexes. Immunome Res. 2010, 6, S2. [Google Scholar] [CrossRef]
- Tong, J.C.; Tan, T.W.; Ranganathan, S. Modeling the structure of bound peptide ligands to major histocompatibility complex. Protein Sci. 2004, 13, 2523–2532. [Google Scholar] [CrossRef]
- Antunes, D.A.; Devaurs, D.; Moll, M.; Lizée, G.; Kavraki, L.E. General Prediction of Peptide-MHC Binding Modes Using Incremental Docking: A Proof of Concept. Sci. Rep. 2018, 8, 4327. [Google Scholar] [CrossRef]
- ACS Cancer Statistics Center. Available online: https://cancerstatisticscenter.cancer.org/#!/ (accessed on 18 November 2019).
- INCA Estimative 2018, Incidence of Cancer in Brazil. Available online: http://www1.inca.gov.br/estimativa/2018/ (accessed on 18 November 2019).
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Borresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef]
- Chen, Y.; McGee, J.; Chen, X.; Doman, T.N.; Gong, X.; Zhang, Y.; Hamm, N.; Ma, X.; Higgs, R.E.; Bhagwat, S.V.; et al. Identification of druggable cancer driver genes amplified across TCGA datasets. PLoS ONE 2014, 9, e98293. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A., Jr.; Kinzler, K.W. Cancer genome landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef] [PubMed]
- Sjoblom, T.; Jones, S.; Wood, L.D.; Parsons, D.W.; Lin, J.; Barber, T.D.; Mandelker, D.; Leary, R.J.; Ptak, J.; Silliman, N.; et al. The consensus coding sequences of human breast and colorectal cancers. Science 2006, 314, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Boon, T.; van der Bruggen, P. Human tumor antigens recognized by T lymphocytes. J. Exp. Med. 1996, 183, 725–729. [Google Scholar] [CrossRef] [PubMed]
- Coulie, P.G.; Lehmann, F.; Lethe, B.; Herman, J.; Lurquin, C.; Andrawiss, M.; Boon, T. A mutated intron sequence codes for an antigenic peptide recognized by cytolytic T lymphocytes on a human melanoma. Proc. Natl. Acad. Sci. USA 1995, 92, 7976–7980. [Google Scholar] [CrossRef]
- Mandruzzato, S.; Brasseur, F.; Andry, G.; Boon, T.; van der Bruggen, P. A CASP-8 mutation recognized by cytolytic T lymphocytes on a human head and neck carcinoma. J. Exp. Med. 1997, 186, 785–793. [Google Scholar] [CrossRef]
- Beatty, G.L.; Gladney, W.L. Immune escape mechanisms as a guide for cancer immunotherapy. Clin. Cancer Res. 2015, 21, 687–692. [Google Scholar] [CrossRef]
- Messerschmidt, J.L.; Prendergast, G.C.; Messerschmidt, G.L. How Cancers Escape Immune Destruction and Mechanisms of Action for the New Significantly Active Immune Therapies: Helping Nonimmunologists Decipher Recent Advances. Oncologist 2016, 21, 233–243. [Google Scholar] [CrossRef]
- Kreiter, S.; Vormehr, M.; van de Roemer, N.; Diken, M.; Lower, M.; Diekmann, J.; Boegel, S.; Schrors, B.; Vascotto, F.; Castle, J.C.; et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature 2015, 520, 692–696. [Google Scholar] [CrossRef]
- Li, D.; Bentley, C.; Yates, J.; Salimi, M.; Greig, J.; Wiblin, S.; Hassanali, T.; Banham, A.H. Engineering chimeric human and mouse major histocompatibility complex (MHC) class I tetramers for the production of T-cell receptor (TCR) mimic antibodies. PLoS ONE 2017, 12, e0176642. [Google Scholar] [CrossRef]
- Altman, J.D.; Moss, P.A.; Goulder, P.J.; Barouch, D.H.; McHeyzer-Williams, M.G.; Bell, J.I.; McMichael, A.J.; Davis, M.M. Phenotypic analysis of antigen-specific T lymphocytes. Science 1996, 274, 94–96. [Google Scholar] [CrossRef] [PubMed]
- Bodinier, M.; Peyrat, M.A.; Tournay, C.; Davodeau, F.; Romagne, F.; Bonneville, M.; Lang, F. Efficient detection and immunomagnetic sorting of specific T cells using multimers of MHC class I and peptide with reduced CD8 binding. Nat. Med. 2000, 6, 707–710. [Google Scholar] [CrossRef] [PubMed]
- Fehlings, M.; Chakarov, S.; Simoni, Y.; Sivasankar, B.; Ginhoux, F.; Newell, E.W. Multiplex peptide-MHC tetramer staining using mass cytometry for deep analysis of the influenza-specific T-cell response in mice. J. Immunol. Methods 2018, 453, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Bharathan, M.; Trebska-McGowan, K.; Anna, P.; Deniger, D.C.; Hanada, K.; Gartner, J.J.; Yang, J.C.; Rosenberg, S.A.; Robbins, P.F. Tetramer based approach for efficient identification and isolation of neo-antigen specific CD8 T cells from peripheral blood (PBL) of patients with metastatic cancers. J. Immunother. Cancer 2015, 3, 47. [Google Scholar] [CrossRef][Green Version]
- Nitschke, K.; Flecken, T.; Schmidt, J.; Gostick, E.; Marget, M.; Neumann-Haefelin, C.; Blum, H.E.; Price, D.A.; Thimme, R. Tetramer enrichment reveals the presence of phenotypically diverse hepatitis C virus-specific CD8+ T cells in chronic infection. J. Virol. 2015, 89, 25–34. [Google Scholar] [CrossRef][Green Version]
- Romero, P.; Dunbar, P.R.; Valmori, D.; Pittet, M.; Ogg, G.S.; Rimoldi, D.; Chen, J.L.; Lienard, D.; Cerottini, J.C.; Cerundolo, V. Ex vivo staining of metastatic lymph nodes by class I major histocompatibility complex tetramers reveals high numbers of antigen-experienced tumor-specific cytolytic T lymphocytes. J. Exp. Med. 1998, 188, 1641–1650. [Google Scholar] [CrossRef]
- Borrman, T.; Cimons, J.; Cosiano, M.; Purcaro, M.; Pierce, B.G.; Baker, B.M.; Weng, Z. ATLAS: A database linking binding affinities with structures for wild-type and mutant TCR-pMHC complexes. Proteins 2017, 85, 908–916. [Google Scholar] [CrossRef]
- Chowell, D.; Krishna, S.; Becker, P.D.; Cocita, C.; Shu, J.; Tan, X.; Greenberg, P.D.; Klavinskis, L.S.; Blattman, J.N.; Anderson, K.S. TCR contact residue hydrophobicity is a hallmark of immunogenic CD8+ T cell epitopes. Proc. Natl. Acad. Sci. USA 2015, 112, E1754–E1762. [Google Scholar] [CrossRef]
- Holt, R.A. Interpreting the T-cell receptor repertoire. Nat. Biotechnol. 2017, 35, 829–830. [Google Scholar] [CrossRef]
- Vieira, G.F.; Chies, J.A. Immunodominant viral peptides as determinants of cross-reactivity in the immune system—Can we develop wide spectrum viral vaccines? Med. Hypotheses 2005, 65, 873–879. [Google Scholar] [CrossRef]
- Wooldridge, L.; Ekeruche-Makinde, J.; van den Berg, H.A.; Skowera, A.; Miles, J.J.; Tan, M.P.; Dolton, G.; Clement, M.; Llewellyn-Lacey, S.; Price, D.A.; et al. A single autoimmune T cell receptor recognizes more than a million different peptides. J. Biol. Chem. 2012, 287, 1168–1177. [Google Scholar] [CrossRef] [PubMed]
- Fytili, P.; Dalekos, G.N.; Schlaphoff, V.; Suneetha, P.V.; Sarrazin, C.; Zauner, W.; Zachou, K.; Berg, T.; Manns, M.P.; Klade, C.S.; et al. Cross-genotype-reactivity of the immunodominant HCV CD8 T-cell epitope NS3–1073. Vaccine 2008, 26, 3818–3826. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Sefah, K.; Tang, L.; Zhao, Z.; Zhu, G.; Ye, M.; Sun, W.; Goodison, S.; Tan, W. A novel aptamer developed for breast cancer cell internalization. ChemMedChem 2012, 7, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Antunes, D.A.; Rigo, M.M.; Freitas, M.V.; Mendes, M.F.A.; Sinigaglia, M.; Lizee, G.; Kavraki, L.E.; Selin, L.K.; Cornberg, M.; Vieira, G.F. Interpreting T-Cell Cross-reactivity through Structure: Implications for TCR-Based Cancer Immunotherapy. Front. Immunol. 2017, 8, 1210. [Google Scholar] [CrossRef] [PubMed]
- Decker, W.K.; da Silva, R.F.; Sanabria, M.H.; Angelo, L.S.; Guimaraes, F.; Burt, B.M.; Kheradmand, F.; Paust, S. Cancer Immunotherapy: Historical Perspective of a Clinical Revolution and Emerging Preclinical Animal Models. Front. Immunol. 2017, 8, 829. [Google Scholar] [CrossRef] [PubMed]
- Burdick, C.G. William Bradley Coley 1862–1936. Ann. Surg. 1937, 105, 152–155. [Google Scholar]
- Hoption Cann, S.A.; van Netten, J.P.; van Netten, C. Dr William Coley and tumour regression: A place in history or in the future. Postgr. Med. J. 2003, 79, 672–680. [Google Scholar]
- Starnes, C.O. Coley’s toxins in perspective. Nature 1992, 357, 11–12. [Google Scholar] [CrossRef]
- D’Errico, G.; Machado, H.L.; Sainz, B., Jr. A current perspective on cancer immune therapy: Step-by-step approach to constructing the magic bullet. Clin. Transl. Med. 2017, 6, 3. [Google Scholar]
- Restifo, N.P.; Smyth, M.J.; Snyder, A. Acquired resistance to immunotherapy and future challenges. Nat. Rev. Cancer 2016, 16, 121–126. [Google Scholar] [CrossRef]
- Kranz, L.M.; Diken, M.; Haas, H.; Kreiter, S.; Loquai, C.; Reuter, K.C.; Meng, M.; Fritz, D.; Vascotto, F.; Hefesha, H.; et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 2016, 534, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Desrichard, A.; Snyder, A.; Chan, T.A. Cancer Neoantigens and Applications for Immunotherapy. Clin. Cancer Res. 2016, 22, 807–812. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Ott, P.A.; Wu, C.J. Towards personalized, tumour-specific, therapeutic vaccines for cancer. Nat. Rev. Immunol. 2018, 18, 168–182. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.S.; Stojanov, P.; Polak, P.; Kryukov, G.V.; Cibulskis, K.; Sivachenko, A.; Carter, S.L.; Stewart, C.; Mermel, C.H.; Roberts, S.A.; et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013, 499, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lee, S.W. The Roles of Carcinoembryonic Antigen in Liver Metastasis and Therapeutic Approaches. Gastroenterol. Res. Pract. 2017, 2017, 7521987. [Google Scholar] [CrossRef] [PubMed]
- Park, T.S.; Groh, E.M.; Patel, K.; Kerkar, S.P.; Lee, C.C.; Rosenberg, S.A. Expression of MAGE-A and NY-ESO-1 in Primary and Metastatic Cancers. J. Immunother. 2016, 39, 1–7. [Google Scholar] [CrossRef]
- Parle-McDermott, A.; McWilliam, P.; Tighe, O.; Dunican, D.; Croke, D.T. Serial analysis of gene expression identifies putative metastasis-associated transcripts in colon tumour cell lines. Br. J. Cancer 2000, 83, 725–728. [Google Scholar] [CrossRef]
- Prager, G.W.; Braemswig, K.H.; Martel, A.; Unseld, M.; Heinze, G.; Brodowicz, T.; Scheithauer, W.; Kornek, G.; Zielinski, C.C. Baseline carcinoembryonic antigen (CEA) serum levels predict bevacizumab-based treatment response in metastatic colorectal cancer. Cancer Sci. 2014, 105, 996–1001. [Google Scholar] [CrossRef]
- Suzuki, S.; Sasajima, K.; Sato, Y.; Watanabe, H.; Matsutani, T.; Iida, S.; Hosone, M.; Tsukui, T.; Maeda, S.; Shimizu, K.; et al. MAGE-A protein and MAGE-A10 gene expressions in liver metastasis in patients with stomach cancer. Br. J. Cancer 2008, 99, 350–356. [Google Scholar] [CrossRef][Green Version]
- Mendonca, B.D.S.; Agostini, M.; Aquino, I.G.; Dias, W.B.; Bastos, D.C.; Rumjanek, F.D. Suppression of MAGE-A10 alters the metastatic phenotype of tongue squamous cell carcinoma cells. Biochem. Biophys. Rep. 2017, 10, 267–275. [Google Scholar] [CrossRef]
- Fratta, E.; Coral, S.; Covre, A.; Parisi, G.; Colizzi, F.; Danielli, R.; Nicolay, H.J.; Sigalotti, L.; Maio, M. The biology of cancer testis antigens: Putative function, regulation and therapeutic potential. Mol. Oncol. 2011, 5, 164–182. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Ousati Ashtiani, Z.; Sabah Golian, B.; Hasheminasab, S.M.; Modarressi, M.H. Expression of two testis-specific genes, SPATA19 and LEMD1, in prostate cancer. Arch. Med. Res. 2010, 41, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Scanlan, M.J.; Gure, A.O.; Jungbluth, A.A.; Old, L.J.; Chen, Y.T. Cancer/testis antigens: An expanding family of targets for cancer immunotherapy. Immunol. Rev. 2002, 188, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Odunsi, K.; Qian, F.; Matsuzaki, J.; Mhawech-Fauceglia, P.; Andrews, C.; Hoffman, E.W.; Pan, L.; Ritter, G.; Villella, J.; Thomas, B.; et al. Vaccination with an NY-ESO-1 peptide of HLA class I/II specificities induces integrated humoral and T cell responses in ovarian cancer. Proc. Natl. Acad. Sci. USA 2007, 104, 12837–12842. [Google Scholar] [CrossRef] [PubMed]
- Chianese-Bullock, K.A.; Pressley, J.; Garbee, C.; Hibbitts, S.; Murphy, C.; Yamshchikov, G.; Petroni, G.R.; Bissonette, E.A.; Neese, P.Y.; Grosh, W.W.; et al. MAGE-A1-, MAGE-A10-, and gp100-derived peptides are immunogenic when combined with granulocyte-macrophage colony-stimulating factor and montanide ISA-51 adjuvant and administered as part of a multipeptide vaccine for melanoma. J. Immunol. 2005, 174, 3080–3086. [Google Scholar] [CrossRef]
- Goodison, S.; Chang, M.; Dai, Y.; Urquidi, V.; Rosser, C.J. A multi-analyte assay for the non-invasive detection of bladder cancer. PLoS ONE 2012, 7, e47469. [Google Scholar] [CrossRef]
- De, S.; Ganesan, S. Looking beyond drivers and passengers in cancer genome sequencing data. Ann. Oncol. 2017, 28, 938–945. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Stratton, M.R.; Campbell, P.J.; Futreal, P.A. The cancer genome. Nature 2009, 458, 719–724. [Google Scholar] [CrossRef]
- McFarland, C.D.; Korolev, K.S.; Kryukov, G.V.; Sunyaev, S.R.; Mirny, L.A. Impact of deleterious passenger mutations on cancer progression. Proc. Natl. Acad. Sci. USA 2013, 110, 2910–2915. [Google Scholar] [CrossRef]
- McFarland, C.D.; Mirny, L.A.; Korolev, K.S. Tug-of-war between driver and passenger mutations in cancer and other adaptive processes. Proc. Natl. Acad. Sci. USA 2014, 111, 15138–15143. [Google Scholar] [CrossRef] [PubMed]
- McFarland, C.D.; Yaglom, J.A.; Wojtkowiak, J.W.; Scott, J.G.; Morse, D.L.; Sherman, M.Y.; Mirny, L.A. The Damaging Effect of Passenger Mutations on Cancer Progression. Cancer Res. 2017, 77, 4763–4772. [Google Scholar] [CrossRef] [PubMed]
- Gubin, M.M.; Zhang, X.; Schuster, H.; Caron, E.; Ward, J.P.; Noguchi, T.; Ivanova, Y.; Hundal, J.; Arthur, C.D.; Krebber, W.J.; et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 2014, 515, 577–581. [Google Scholar] [CrossRef] [PubMed]
- Rooney, M.S.; Shukla, S.A.; Wu, C.J.; Getz, G.; Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015, 160, 48–61. [Google Scholar] [CrossRef]
- Snyder, A.; Makarov, V.; Merghoub, T.; Yuan, J.; Zaretsky, J.M.; Desrichard, A.; Walsh, L.A.; Postow, M.A.; Wong, P.; Ho, T.S.; et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 2014, 371, 2189–2199. [Google Scholar] [CrossRef]
- Yadav, M.; Jhunjhunwala, S.; Phung, Q.T.; Lupardus, P.; Tanguay, J.; Bumbaca, S.; Franci, C.; Cheung, T.K.; Fritsche, J.; Weinschenk, T.; et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 2014, 515, 572–576. [Google Scholar] [CrossRef]
- Weaver, B.A.; Silk, A.D.; Montagna, C.; Verdier-Pinard, P.; Cleveland, D.W. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell 2007, 11, 25–36. [Google Scholar] [CrossRef]
- Williams, B.R.; Prabhu, V.R.; Hunter, K.E.; Glazier, C.M.; Whittaker, C.A.; Housman, D.E.; Amon, A. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 2008, 322, 703–709. [Google Scholar] [CrossRef]
- Andor, N.; Graham, T.A.; Jansen, M.; Xia, L.C.; Aktipis, C.A.; Petritsch, C.; Ji, H.P.; Maley, C.C. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat. Med. 2016, 22, 105–113. [Google Scholar] [CrossRef]
- Birkbak, N.J.; Eklund, A.C.; Li, Q.; McClelland, S.E.; Endesfelder, D.; Tan, P.; Tan, I.B.; Richardson, A.L.; Szallasi, Z.; Swanton, C. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 2011, 71, 3447–3452. [Google Scholar] [CrossRef]
- Sheltzer, J.M.; Amon, A. The aneuploidy paradox: Costs and benefits of an incorrect karyotype. Trends Genet. 2011, 27, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, S.A.; Dudley, M.E. Adoptive cell therapy for the treatment of patients with metastatic melanoma. Curr. Opin. Immunol. 2009, 21, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, T.N.; Schreiber, R.D. Neoantigens in cancer immunotherapy. Science 2015, 348, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.C.; Yao, X.; Crystal, J.S.; Li, Y.F.; El-Gamil, M.; Gross, C.; Davis, L.; Dudley, M.E.; Yang, J.C.; Samuels, Y.; et al. Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clin. Cancer Res. 2014, 20, 3401–3410. [Google Scholar] [CrossRef] [PubMed]
- Robbins, P.F.; Lu, Y.C.; El-Gamil, M.; Li, Y.F.; Gross, C.; Gartner, J.; Lin, J.C.; Teer, J.K.; Cliften, P.; Tycksen, E.; et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat. Med. 2013, 19, 747–752. [Google Scholar] [CrossRef]
- Van Rooij, N.; van Buuren, M.M.; Philips, D.; Velds, A.; Toebes, M.; Heemskerk, B.; van Dijk, L.J.; Behjati, S.; Hilkmann, H.; El Atmioui, D.; et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J. Clin. Oncol. 2013, 31, e439–e442. [Google Scholar] [CrossRef]
- Linnemann, C.; van Buuren, M.M.; Bies, L.; Verdegaal, E.M.; Schotte, R.; Calis, J.J.; Behjati, S.; Velds, A.; Hilkmann, H.; Atmioui, D.E.; et al. High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4+ T cells in human melanoma. Nat. Med. 2015, 21, 81–85. [Google Scholar] [CrossRef]
- McGranahan, N.; Furness, A.J.; Rosenthal, R.; Ramskov, S.; Lyngaa, R.; Saini, S.K.; Jamal-Hanjani, M.; Wilson, G.A.; Birkbak, N.J.; Hiley, C.T.; et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016, 351, 1463–1469. [Google Scholar] [CrossRef]
- Alizadeh, A.A.; Aranda, V.; Bardelli, A.; Blanpain, C.; Bock, C.; Borowski, C.; Caldas, C.; Califano, A.; Doherty, M.; Elsner, M.; et al. Toward understanding and exploiting tumor heterogeneity. Nat. Med. 2015, 21, 846–853. [Google Scholar] [CrossRef]
- Hemelaar, J. The origin and diversity of the HIV-1 pandemic. Trends Mol. Med. 2012, 18, 182–192. [Google Scholar] [CrossRef]
- Castle, J.C.; Kreiter, S.; Diekmann, J.; Lower, M.; van de Roemer, N.; de Graaf, J.; Selmi, A.; Diken, M.; Boegel, S.; Paret, C.; et al. Exploiting the mutanome for tumor vaccination. Cancer Res. 2012, 72, 1081–1091. [Google Scholar] [CrossRef] [PubMed]
- Dudley, M.E.; Wunderlich, J.R.; Yang, J.C.; Sherry, R.M.; Topalian, S.L.; Restifo, N.P.; Royal, R.E.; Kammula, U.; White, D.E.; Mavroukakis, S.A.; et al. Adoptive cell transfer therapy following non-myeloablative but lymphodepleting chemotherapy for the treatment of patients with refractory metastatic melanoma. J. Clin. Oncol. 2005, 23, 2346–2357. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, S.A.; Restifo, N.P. Adoptive cell transfer as personalized immunotherapy for human cancer. Science 2015, 348, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Zacharakis, N.; Chinnasamy, H.; Black, M.; Xu, H.; Lu, Y.C.; Zheng, Z.; Pasetto, A.; Langhan, M.; Shelton, T.; Prickett, T.; et al. Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat. Med. 2018, 24, 724–730. [Google Scholar] [CrossRef] [PubMed]
- Cameron, B.J.; Gerry, A.B.; Dukes, J.; Harper, J.V.; Kannan, V.; Bianchi, F.C.; Grand, F.; Brewer, J.E.; Gupta, M.; Plesa, G.; et al. Identification of a Titin-derived HLA-A1-presented peptide as a cross-reactive target for engineered MAGE A3-directed T cells. Sci. Transl. Med. 2013, 5, 197ra103. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vianna, P.; Mendes, M.F.A.; Bragatte, M.A.; Ferreira, P.S.; Salzano, F.M.; Bonamino, M.H.; Vieira, G.F. pMHC Structural Comparisons as a Pivotal Element to Detect and Validate T-Cell Targets for Vaccine Development and Immunotherapy—A New Methodological Proposal. Cells 2019, 8, 1488. https://doi.org/10.3390/cells8121488
Vianna P, Mendes MFA, Bragatte MA, Ferreira PS, Salzano FM, Bonamino MH, Vieira GF. pMHC Structural Comparisons as a Pivotal Element to Detect and Validate T-Cell Targets for Vaccine Development and Immunotherapy—A New Methodological Proposal. Cells. 2019; 8(12):1488. https://doi.org/10.3390/cells8121488
Chicago/Turabian StyleVianna, Priscila, Marcus F.A. Mendes, Marcelo A. Bragatte, Priscila S. Ferreira, Francisco M. Salzano, Martin H. Bonamino, and Gustavo F. Vieira. 2019. "pMHC Structural Comparisons as a Pivotal Element to Detect and Validate T-Cell Targets for Vaccine Development and Immunotherapy—A New Methodological Proposal" Cells 8, no. 12: 1488. https://doi.org/10.3390/cells8121488
APA StyleVianna, P., Mendes, M. F. A., Bragatte, M. A., Ferreira, P. S., Salzano, F. M., Bonamino, M. H., & Vieira, G. F. (2019). pMHC Structural Comparisons as a Pivotal Element to Detect and Validate T-Cell Targets for Vaccine Development and Immunotherapy—A New Methodological Proposal. Cells, 8(12), 1488. https://doi.org/10.3390/cells8121488





