Differential Roles of Lipin1 and Lipin2 in the Hepatitis C Virus Replication Cycle
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
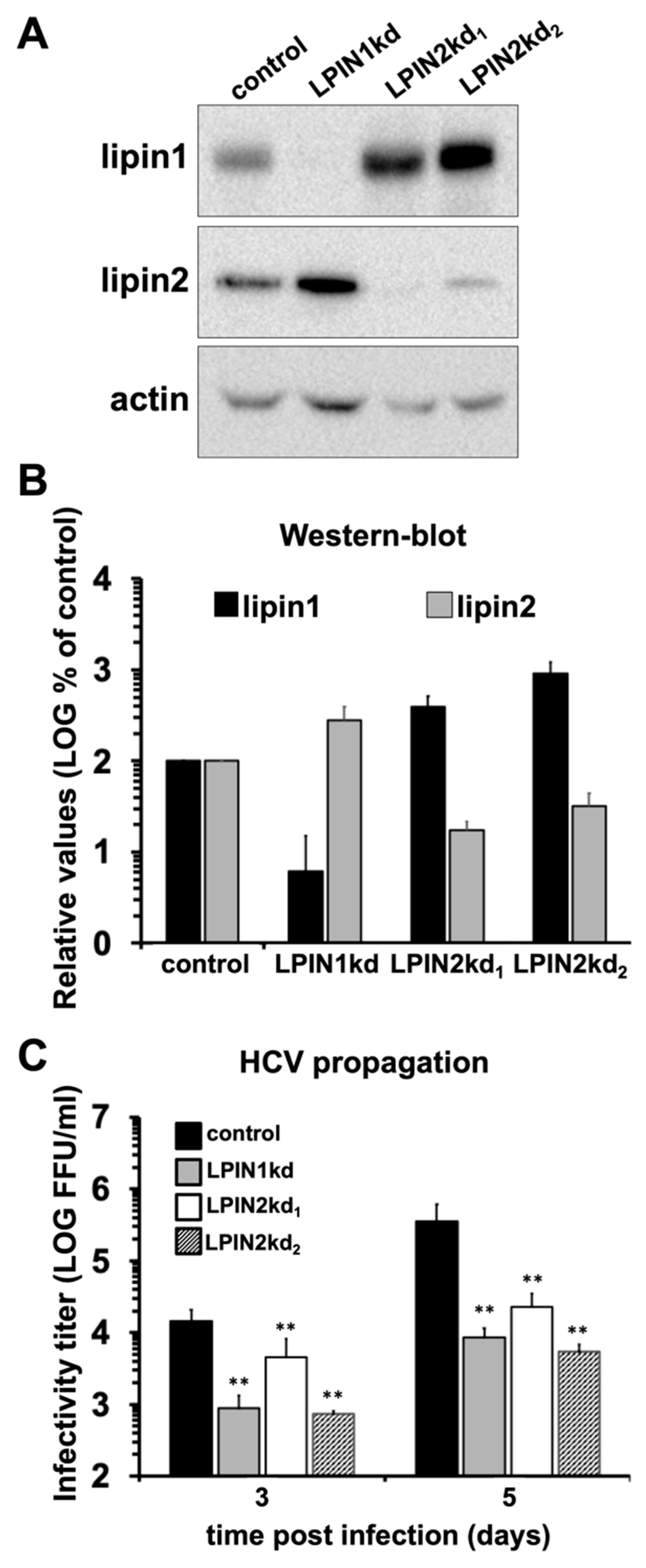
2.2. Silencing Experiments
2.3. Western-Blot
2.4. HCV Infection Experiments
2.5. Other Infection Experiments
2.6. Analysis of Viral Entry Using Retroviral Pseudotypes
2.7. In Vitro Transcription and HCV RNA Transfection
2.8. Gaussia Secretion Experiments
2.9. Confocal Immunofluorescence Microscopy
2.10. Statistical Analysis
3. Results
3.1. Lipin2 Silencing Interferes with HCV Propagation
3.2. Impact of Lipin2 Silencing on Early Aspects of HCV Infection
3.3. Lipin2 Interferes with Virion Secretion During HCV Infection
3.4. Lipin2 Silencing Causes Golgi Apparatus Fragmentation
3.5. Lipin2 Silencing Causes Mitochondria Elongation
3.6. Lipin2, but not Lipin1, Silencing Interferes with Flavivirus Spread
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Simmonds, P.; Becher, P.; Bukh, J.; Gould, E.A.; Meyers, G.; Monath, T.; Muerhoff, S.; Pletnev, A.; Rico-Hesse, R.; Smith, D.B.; et al. ICTV Report Consortium ICTV Virus Taxonomy Profile: Flaviviridae. J. Gen. Virol. 2017, 98, 2–3. [Google Scholar]
- Bartenschlager, R.; Lohmann, V.; Penin, F. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nat. Rev. Microbiol. 2013, 11, 482–496. [Google Scholar] [CrossRef]
- Moradpour, D.; Penin, F. Hepatitis C virus proteins: From structure to function. Curr. Top. Microbiol. Immunol. 2013, 369, 113–142. [Google Scholar]
- Lohmann, V.; Körner, F.; Koch, J.; Herian, U.; Theilmann, L.; Bartenschlager, R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 1999, 285, 110–113. [Google Scholar] [CrossRef]
- Romero-Brey, I.; Merz, A.; Chiramel, A.; Lee, J.-Y.; Chlanda, P.; Haselman, U.; Santarella-Mellwig, R.; Habermann, A.; Hoppe, S.; Kallis, S.; et al. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 2012, 8, e1003056. [Google Scholar] [CrossRef]
- Lindenbach, B.D.; Rice, C.M. The ins and outs of hepatitis C virus entry and assembly. Nat. Rev. Microbiol. 2013, 11, 688–700. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Cortese, M.; Haselmann, U.; Tabata, K.; Romero-Brey, I.; Funaya, C.; Schieber, N.L.; Qiang, Y.; Bartenschlager, M.; Kallis, S.; et al. Spatiotemporal Coupling of the Hepatitis C Virus Replication Cycle by Creating a Lipid Droplet- Proximal Membranous Replication Compartment. Cell. Rep. 2019, 27, 3602–3617. [Google Scholar] [CrossRef]
- Hofmann, S.; Krajewski, M.; Scherer, C.; Scholz, V.; Mordhorst, V.; Truschow, P.; Schöbel, A.; Reimer, R.; Schwudke, D.; Herker, E. Complex lipid metabolic remodeling is required for efficient hepatitis C virus replication. BBA- Mol. Cell Biol. Lipids 2018, 1863, 1041–1056. [Google Scholar] [CrossRef]
- Gerold, G.; Moeller, R.; Pietschmann, T. Hepatitis C Virus Entry: Protein Interactions and Fusion Determinants Governing Productive Hepatocyte Invasion. Cold. Spring. Harb. Perspect. Med. 2019, 9, a036830. [Google Scholar] [CrossRef]
- Paul, D.; Madan, V.; Bartenschlager, R. Hepatitis C virus RNA replication and assembly: Living on the fat of the land. Cell. Host. Microbe 2014, 16, 569–579. [Google Scholar] [CrossRef]
- Catanese, M.T.; Uryu, K.; Kopp, M.; Edwards, T.J.; Andrus, L.; Rice, W.J.; Silvestry, M.; Kuhn, R.J.; Rice, C.M. Ultrastructural analysis of hepatitis C virus particles. Proc. Natl. Acad. Sci. USA 2013, 110, 9505–9510. [Google Scholar] [CrossRef]
- Merz, A.; Long, G.; Hiet, M.-S.; Brügger, B.; Chlanda, P.; André, P.; Wieland, F.; Krijnse-Locker, J.; Bartenschlager, R. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J. Biol. Chem. 2011, 286, 3018–3032. [Google Scholar] [CrossRef]
- Gastaminza, P.; Dryden, K.A.; Boyd, B.; Wood, M.R.; Law, M.; Yeager, M.; Chisari, F.V. Ultrastructural and biophysical characterization of hepatitis C virus particles produced in cell culture. J. Virol. 2010, 84, 10999–11009. [Google Scholar] [CrossRef]
- Wrensch, F.; Crouchet, E.; Ligat, G.; Zeisel, M.B.; Keck, Z.-Y.; Foung, S.K.H.; Schuster, C.; Baumert, T.F. Hepatitis C Virus (HCV)-Apolipoprotein Interactions and Immune Evasion and Their Impact on HCV Vaccine Design. Front. Immunol. 2018, 9, 1436. [Google Scholar] [CrossRef]
- Egger, D.; Wölk, B.; Gosert, R.; Bianchi, L.; Blum, H.E.; Moradpour, D.; Bienz, K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 2002, 76, 5974–5984. [Google Scholar] [CrossRef]
- Paul, D.; Hoppe, S.; Saher, G.; Krijnse-Locker, J.; Bartenschlager, R. Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment. J. Virol. 2013, 87, 10612–10627. [Google Scholar] [CrossRef]
- Miyanari, Y.; Atsuzawa, K.; Usuda, N.; Watashi, K.; Hishiki, T.; Zayas, M.; Bartenschlager, R.; Wakita, T.; Hijikata, M.; Shimotohno, K. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell. Biol. 2007, 9, 1089–1097. [Google Scholar] [CrossRef]
- Chang, K.-S.; Jiang, J.; Cai, Z.; Luo, G. Human apolipoprotein e is required for infectivity and production of hepatitis C virus in cell culture. J. Virol. 2007, 81, 13783–13793. [Google Scholar] [CrossRef]
- Herker, E.; Harris, C.; Hernandez, C.; Carpentier, A.; Kaehlcke, K.; Rosenberg, A.R.; Farese, R.V.; Ott, M. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nat. Med. 2010, 16, 1295–1298. [Google Scholar] [CrossRef]
- Fukuhara, T.; Wada, M.; Nakamura, S.; Ono, C.; Shiokawa, M.; Yamamoto, S.; Motomura, T.; Okamoto, T.; Okuzaki, D.; Yamamoto, M.; et al. Amphipathic α-Helices in Apolipoproteins Are Crucial to the Formation of Infectious Hepatitis C Virus Particles. PLoS Pathog. 2014, 10, e1004534. [Google Scholar] [CrossRef]
- Mankouri, J.; Walter, C.; Stewart, H.; Bentham, M.; Park, W.S.; Heo, W.D.; Fukuda, M.; Griffin, S.; Harris, M. Release of Infectious Hepatitis C Virus from Huh7 Cells Occurs via a trans-Golgi Network-to-Endosome Pathway Independent of Very-Low-Density Lipoprotein Secretion. J. Virol. 2016, 90, 7159–7170. [Google Scholar] [CrossRef]
- Coller, K.E.; Heaton, N.S.; Berger, K.L.; Cooper, J.D.; Saunders, J.L.; Randall, G. Molecular determinants and dynamics of hepatitis C virus secretion. PLoS Pathog. 2012, 8, e1002466. [Google Scholar] [CrossRef]
- Ariumi, Y.; Kuroki, M.; Maki, M.; Ikeda, M.; Dansako, H.; Wakita, T.; Kato, N. The ESCRT system is required for hepatitis C virus production. PLoS ONE 2011, 6, e14517. [Google Scholar] [CrossRef]
- Lai, C.-K.; Jeng, K.-S.; Machida, K.; Lai, M.M.C. Hepatitis C virus egress and release depend on endosomal trafficking of core protein. J. Virol. 2010, 84, 11590–11598. [Google Scholar] [CrossRef]
- Xiao, F.; Wang, S.; Barouch-Bentov, R.; Neveu, G.; Pu, S.; Beer, M.; Schor, S.; Kumar, S.; Nicolaescu, V.; Lindenbach, B.D.; et al. Interactions between the Hepatitis C Virus Nonstructural 2 Protein and Host Adaptor Proteins 1 and 4 Orchestrate Virus Release. mBio 2018, 9. [Google Scholar] [CrossRef]
- Gastaminza, P.; Kapadia, S.B.; Chisari, F.V. Differential biophysical properties of infectious intracellular and secreted hepatitis C virus particles. J. Virol. 2006, 80, 11074–11081. [Google Scholar] [CrossRef]
- Denolly, S.; Granier, C.; Fontaine, N.; Pozzetto, B.; Bourlet, T.; Guérin, M.; Cosset, F.-L. A serum protein factor mediates maturation and apoB-association of HCV particles in the extracellular milieu. J. Hepatol. 2019, 70, 626–638. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Acosta, E.G.; Stoeck, I.K.; Long, G.; Hiet, M.-S.; Mueller, B.; Fackler, O.T.; Kallis, S.; Bartenschlager, R. Apolipoprotein E likely contributes to a maturation step of infectious hepatitis C virus particles and interacts with viral envelope glycoproteins. J. Virol. 2014, 88, 12422–12437. [Google Scholar] [CrossRef]
- Mingorance, L.; Castro, V.; Ávila-Pérez, G.; Calvo, G.; Rodríguez, M.J.; Carrascosa, J.L.; Pérez-del-Pulgar, S.; Forns, X.; Gastaminza, P. Host phosphatidic acid phosphatase lipin1 is rate limiting for functional hepatitis C virus replicase complex formation. PLoS Pathog. 2018, 14, e1007284. [Google Scholar] [CrossRef]
- Takeuchi, K.; Reue, K. Biochemistry, physiology, and genetics of GPAT, AGPAT, and lipin enzymes in triglyceride synthesis. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E1195–E1209. [Google Scholar] [CrossRef]
- Huang, H.; Gao, Q.; Peng, X.; Choi, S.-Y.; Sarma, K.; Ren, H.; Morris, A.J.; Frohman, M.A. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev. Cell 2011, 20, 376–387. [Google Scholar] [CrossRef]
- Ha, K.D.; Clarke, B.A.; Brown, W.J. Regulation of the Golgi complex by phospholipid remodeling enzymes. Biochim. Et Biophys. Acta (Bba) - Mol. Cell. Biol. Lipids 2012, 1821, 1078–1088. [Google Scholar] [CrossRef][Green Version]
- Péterfy, M.; Phan, J.; Xu, P.; Reue, K. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 2001, 27, 121–124. [Google Scholar] [CrossRef]
- Donkor, J.; Sariahmetoglu, M.; Dewald, J.; Brindley, D.N.; Reue, K. Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J. Biol. Chem. 2007, 282, 3450–3457. [Google Scholar] [CrossRef]
- Brindley, D.N.; Pilquil, C.; Sariahmetoglu, M.; Reue, K. Phosphatidate degradation: Phosphatidate phosphatases (lipins) and lipid phosphate phosphatases. Biochim. Biophys. Acta 2009, 1791, 956–961. [Google Scholar] [CrossRef]
- Eaton, J.M.; Mullins, G.R.; Brindley, D.N.; Harris, T.E. Phosphorylation of Lipin 1 and Charge on the Phosphatidic Acid Head Group Control Its Phosphatidic Acid Phosphatase Activity and Membrane Association. J. Biol. Chem. 2013, 288, 9933–9945. [Google Scholar] [CrossRef]
- Zhang, P.; Reue, K. Lipin proteins and glycerolipid metabolism: Roles at the ER membrane and beyond. BBA- Biomembranes 2017, 1859, 1583–1595. [Google Scholar] [CrossRef]
- Peterson, T.R.; Sengupta, S.S.; Harris, T.E.; Carmack, A.E.; Kang, S.A.; Balderas, E.; Guertin, D.A.; Madden, K.L.; Carpenter, A.E.; Finck, B.N.; et al. mTOR Complex 1 Regulates Lipin 1 Localization to Control the SREBP Pathway. Cell 2011, 146, 408–420. [Google Scholar] [CrossRef]
- Eaton, J.M.; Takkellapati, S.; Lawrence, R.T.; McQueeney, K.E.; Boroda, S.; Mullins, G.R.; Sherwood, S.G.; Finck, B.N.; Villén, J.; Harris, T.E. Lipin 2 binds phosphatidic acid by the electrostatic hydrogen bond switch mechanism independent of phosphorylation. J. Biol. Chem. 2014, 289, 18055–18066. [Google Scholar] [CrossRef]
- Boroda, S.; Takkellapati, S.; Lawrence, R.T.; Entwisle, S.W.; Pearson, J.M.; Granade, M.E.; Mullins, G.R.; Eaton, J.M.; Villén, J.; Harris, T.E. The Phosphatidic Acid-Binding, Polybasic Domain is Responsible for the Differences in the Phosphoregulation of Lipins 1 and 3. J. Biol. Chem. 2017, 292, 20481–20493. [Google Scholar] [CrossRef]
- Zeharia, A.; Shaag, A.; Houtkooper, R.H.; Hindi, T.; de Lonlay, P.; Erez, G.; Hubert, L.; Saada, A.; de Keyzer, Y.; Eshel, G.; et al. Mutations in LPIN1 cause recurrent acute myoglobinuria in childhood. Am. J. Hum. Genet. 2008, 83, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, P.J.; Chen, S.; Tayeh, M.K.; Ochoa, L.; Leal, S.M.; Pelet, A.; Munnich, A.; Lyonnet, S.; Majeed, H.A.; El-Shanti, H. Homozygous mutations in LPIN2 are responsible for the syndrome of chronic recurrent multifocal osteomyelitis and congenital dyserythropoietic anaemia (Majeed syndrome). J. Med. Genet. 2005, 42, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, J.R.; Donkor, J.; Zhang, P.; Csaki, L.S.; Vergnes, L.; Lee, J.M.; Dewald, J.; Brindley, D.N.; Atti, E.; Tetradis, S.; et al. Mouse lipin-1 and lipin-2 cooperate to maintain glycerolipid homeostasis in liver and aging cerebellum. Proc. Natl. Acad. Sci. USA 2012, 109, E2486–E2495. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, H.; Taketa, K.; Miyano, K.; Yamane, T.; Sato, J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982, 42, 3858–3863. [Google Scholar]
- Zhong, J.; Gastaminza, P.; Cheng, G.; Kapadia, S.; Kato, T.; Burton, D.R.; Wieland, S.F.; Uprichard, S.L.; Wakita, T.; Chisari, F.V. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 2005, 102, 9294–9299. [Google Scholar] [CrossRef]
- DuBridge, R.B.; Tang, P.; Hsia, H.C.; Leong, P.M.; Miller, J.H.; Calos, M.P. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 1987, 7, 379–387. [Google Scholar] [CrossRef]
- Zhong, J.; Gastaminza, P.; Chung, J.; Stamataki, Z.; Isogawa, M.; Cheng, G.; McKeating, J.A.; Chisari, F.V. Persistent hepatitis C virus infection in vitro: Coevolution of virus and host. J. Virol 2006, 80, 11082–11093. [Google Scholar] [CrossRef]
- Gualano, R.C.; Pryor, M.J.; Cauchi, M.R.; Wright, P.J.; Davidson, A.D. Identification of a major determinant of mouse neurovirulence of dengue virus type 2 using stably cloned genomic-length cDNA. J. Gen. Virol. 1998, 79, 437–446. [Google Scholar] [CrossRef]
- Mutso, M.; Saul, S.; Rausalu, K.; Susova, O.; Žusinaite, E.; Mahalingam, S.; Merits, A. Reverse genetic system, genetically stable reporter viruses and packaged subgenomic replicon based on a Brazilian Zika virus isolate. J. Gen. Virol. 2017, 98, 2712–2724. [Google Scholar] [CrossRef]
- Décembre, E.; Assil, S.; Hillaire, M.L.B.; Dejnirattisai, W.; Mongkolsapaya, J.; Screaton, G.R.; Davidson, A.D.; Dreux, M. Sensing of immature particles produced by dengue virus infected cells induces an antiviral response by plasmacytoid dendritic cells. PLoS Pathog. 2014, 10, e1004434. [Google Scholar] [CrossRef]
- Assil, S.; Coléon, S.; Dong, C.; Décembre, E.; Sherry, L.; Allatif, O.; Webster, B.; Dreux, M. Plasmacytoid Dendritic Cells and Infected Cells Form an Interferogenic Synapse Required for Antiviral Responses. Cell Host Microbe 2019, 25, 730–745. [Google Scholar] [CrossRef] [PubMed]
- Dull, T.; Zufferey, R.; Kelly, M.; Mandel, R.J.; Nguyen, M.; Trono, D.; Naldini, L. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 1998, 72, 8463–8471. [Google Scholar] [PubMed]
- Friesland, M.; Mingorance, L.; Chung, J.; Chisari, F.V.; Gastaminza, P. Sigma-1 Receptor Regulates Early Steps of Viral RNA Replication at the Onset of Hepatitis C Virus Infection. J. Virol. 2013, 87, 6377–6390. [Google Scholar] [CrossRef] [PubMed]
- Levitz, S.M.; Diamond, R.D. A rapid colorimetric assay of fungal viability with the tetrazolium salt MTT. J. Infect. Dis. 1985, 152, 938–945. [Google Scholar] [CrossRef]
- Gastaminza, P.; Whitten-Bauer, C.; Chisari, F.V. Unbiased probing of the entire hepatitis C virus life cycle identifies clinical compounds that target multiple aspects of the infection. Proc. Natl. Acad. Sci. USA 2010, 107, 291–296. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Law, M.; Maruyama, T.; Lewis, J.; Giang, E.; Tarr, A.W.; Stamataki, Z.; Gastaminza, P.; Chisari, F.V.; Jones, I.M.; Fox, R.I.; et al. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat. Med. 2008, 14, 25–27. [Google Scholar] [CrossRef]
- Gastaminza, P.; Cheng, G.; Wieland, S.; Zhong, J.; Liao, W.; Chisari, F.V. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J. Virol. 2008, 82, 2120–2129. [Google Scholar] [CrossRef]
- Gentry, M.K.; Henchal, E.A.; McCown, J.M.; Brandt, W.E.; Dalrymple, J.M. Identification of distinct antigenic determinants on dengue-2 virus using monoclonal antibodies. Am. J. Trop. Med. Hyg. 1982, 31, 548–555. [Google Scholar] [CrossRef]
- Bartosch, B.; Dubuisson, J.; Cosset, F.-L. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 2003, 197, 633–642. [Google Scholar] [CrossRef]
- Hatziioannou, T.; Valsesia-Wittmann, S.; Russell, S.J.; Cosset, F.L. Incorporation of fowl plague virus hemagglutinin into murine leukemia virus particles and analysis of the infectivity of the pseudotyped retroviruses. J. Virol. 1998, 72, 5313–5317. [Google Scholar] [PubMed]
- Sandrin, V.; Boson, B.; Salmon, P.; Gay, W.; Nègre, D.; Le Grand, R.; Trono, D.; Cosset, F.-L. Lentiviral vectors pseudotyped with a modified RD114 envelope glycoprotein show increased stability in sera and augmented transduction of primary lymphocytes and CD34+ cells derived from human and nonhuman primates. Blood 2002, 100, 823–832. [Google Scholar] [CrossRef] [PubMed]
- Mingorance, L.; Friesland, M.; Coto-Llerena, M.; Pérez-del-Pulgar, S.; Boix, L.; López-Oliva, J.M.; Bruix, J.; Forns, X.; Gastaminza, P. Selective inhibition of hepatitis C virus infection by hydroxyzine and benztropine. Antimicrob. Agents Chemother. 2014, 58, 3451–3460. [Google Scholar] [CrossRef] [PubMed]
- Steinmann, E.; Brohm, C.; Kallis, S.; Bartenschlager, R.; Pietschmann, T. Efficient trans-encapsidation of hepatitis C virus RNAs into infectious virus-like particles. J. Virol. 2008, 82, 7034–7046. [Google Scholar] [CrossRef] [PubMed]
- Sofia, M.J.; Bao, D.; Chang, W.; Du, J.; Nagarathnam, D.; Rachakonda, S.; Reddy, P.G.; Ross, B.S.; Wang, P.; Zhang, H.-R.; et al. Discovery of a β-d-2’-deoxy-2‘-α-fluoro-2′-β-C-methyluridine nucleotide prodrug (PSI-7977) for the treatment of hepatitis C virus. J. Med. Chem. 2010, 53, 7202–7218. [Google Scholar] [CrossRef]
- Capul, A.A.; De La Torre, J.C. A cell-based luciferase assay amenable to high-throughput screening of inhibitors of arenavirus budding. Virology 2008, 382, 107–114. [Google Scholar] [CrossRef]
- Sciaky, N.; Presley, J.; Smith, C.; Zaal, K.J.; Cole, N.; Moreira, J.E.; Terasaki, M.; Siggia, E.; Lippincott-Schwartz, J. Golgi tubule traffic and the effects of brefeldin A visualized in living cells. J. Cell. Biol. 1997, 139, 1137–1155. [Google Scholar] [CrossRef]
- Lavillette, D.; Tarr, A.W.; Voisset, C.; Donot, P.; Bartosch, B.; Bain, C.; Patel, A.H.; Dubuisson, J.; Ball, J.K.; Cosset, F.-L. Characterization of host-range and cell entry properties of the major genotypes and subtypes of hepatitis C virus. Hepatology 2005, 41, 265–274. [Google Scholar] [CrossRef]
- Bankaitis, V.A.; Garcia-Mata, R.; Mousley, C.J. Golgi membrane dynamics and lipid metabolism. Curr. Biol. 2012, 22, R414–R424. [Google Scholar] [CrossRef]
- Carrasco, S.; Merida, I. Diacylglycerol-dependent binding recruits PKCtheta and RasGRP1 C1 domains to specific subcellular localizations in living T lymphocytes. Mol. Biol. Cell 2004, 15, 2932–2942. [Google Scholar] [CrossRef]
- Riva, L.; Dubuisson, J. Similarities and Differences Between HCV Pseudoparticle (HCVpp) and Cell Culture HCV (HCVcc) in the Study of HCV. Methods Mol. Biol. 2019, 1911, 33–45. [Google Scholar] [PubMed]
- Benedicto, I.; Gondar, V.; Molina-Jiménez, F.; García-Buey, L.; López-Cabrera, M.; Gastaminza, P.; Majano, P.L. Clathrin mediates infectious hepatitis C virus particle egress. J. Virol. 2015, 89, 4180–4190. [Google Scholar] [CrossRef] [PubMed]
- Takacs, C.N.; Andréo, U.; Dao Thi, V.L.; Wu, X.; Gleason, C.E.; Itano, M.S.; Spitz-Becker, G.S.; Belote, R.L.; Hedin, B.R.; Scull, M.A.; et al. Differential Regulation of Lipoprotein and Hepatitis C Virus Secretion by Rab1b. Cell. Rep. 2017, 21, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.D.; Johnsen, I.B.; Stiberg, K.A.; Sherstova, T.; Wakita, T.; Richard, G.M.; Kandasamy, R.K.; Meurs, E.F.; Anthonsen, M.W. Hepatitis C virus triggers Golgi fragmentation and autophagy through the immunity-related GTPase, M. Proc. Natl. Acad. Sci. USA 2017, 114, E3462–E3471. [Google Scholar] [CrossRef] [PubMed]
- Hsu, N.-Y.; Ilnytska, O.; Belov, G.; Santiana, M.; Chen, Y.-H.; Takvorian, P.M.; Pau, C.; van der Schaar, H.; Kaushik-Basu, N.; Balla, T.; et al. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 2010, 141, 799–811. [Google Scholar] [CrossRef]
- Neufeldt, C.J.; Cortese, M.; Acosta, E.G.; Bartenschlager, R. Rewiring cellular networks by members of the Flaviviridae family. Nat. Rev. Microbiol. 2018, 16, 125–142. [Google Scholar] [CrossRef]
- Reiss, S.; Rebhan, I.; Backes, P.; Romero-Brey, I.; Erfle, H.; Matula, P.; Kaderali, L.; Poenisch, M.; Blankenburg, H.; Hiet, M.-S.; et al. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe 2011, 9, 32–45. [Google Scholar] [CrossRef]
- Bayer, K.; Banning, C.; Bruss, V.; Wiltzer-Bach, L.; Schindler, M. Hepatitis C Virus Is Released via a Noncanonical Secretory Route. J. Virol. 2016, 90, 10558–10573. [Google Scholar] [CrossRef]
- Corless, L.; Crump, C.M.; Griffin, S.D.C.; Harris, M. Vps4 and the ESCRT-III complex are required for the release of infectious hepatitis C virus particles. J. Gen. Virol. 2010, 91, 362–372. [Google Scholar] [CrossRef]
- Choi, S.-Y.; Huang, P.; Jenkins, G.M.; Chan, D.C.; Schiller, J.; Frohman, M.A. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat. Cell. Biol. 2006, 8, 1255–1262. [Google Scholar] [CrossRef]
- Roth, M.G. Molecular mechanisms of PLD function in membrane traffic. Traffic 2008, 9, 1233–1239. [Google Scholar] [CrossRef] [PubMed]
- Almena, M.; Mérida, I. Shaping up the membrane: Diacylglycerol coordinates spatial orientation of signaling. Trends Biochem. Sci. 2011, 36, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Nelson, R.K.; Frohman, M.A. Physiological and pathophysiological roles for phospholipase D. J. Lipid Res. 2015, 56, 2229–2237. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Naslavsky, N.; Caplan, S. Diacylglycerol kinases in membrane trafficking. Cell. Logist. 2015, 5, e1078431. [Google Scholar] [CrossRef]
- Kadamur, G.; Ross, E.M. Mammalian phospholipase C. Annu. Rev. Physiol. 2013, 75, 127–154. [Google Scholar] [CrossRef]
- Carrasco, S.; Mérida, I. Diacylglycerol, when simplicity becomes complex. Trends Biochem. Sci. 2007, 32, 27–36. [Google Scholar] [CrossRef]
- Starr, M.L.; Fratti, R.A. The Participation of Regulatory Lipids in Vacuole Homotypic Fusion. Trends Biochem. Sci. 2019, 44, 546–554. [Google Scholar] [CrossRef]
- Adachi, Y.; Itoh, K.; Yamada, T.; Cerveny, K.L.; Suzuki, T.L.; Macdonald, P.; Frohman, M.A.; Ramachandran, R.; Iijima, M.; Sesaki, H. Coincident Phosphatidic Acid Interaction Restrains Drp1 in Mitochondrial Division. Mol. Cell 2016, 63, 1034–1043. [Google Scholar] [CrossRef]
- Fernández-Ulibarri, I.; Vilella, M.; Lázaro-Diéguez, F.; Sarri, E.; Martínez, S.E.; Jiménez, N.; Claro, E.; Mérida, I.; Burger, K.N.J.; Egea, G. Diacylglycerol is required for the formation of COPI vesicles in the Golgi-to-ER transport pathway. Mol. Biol. Cell 2007, 18, 3250–3263. [Google Scholar] [CrossRef]
- Asp, L.; Kartberg, F.; Fernandez-Rodriguez, J.; Smedh, M.; Elsner, M.; Laporte, F.; Bárcena, M.; Jansen, K.A.; Valentijn, J.A.; Koster, A.J.; et al. Early stages of Golgi vesicle and tubule formation require diacylglycerol. Mol. Biol. Cell 2009, 20, 780–790. [Google Scholar] [CrossRef]
- Liu, G.-H.; Gerace, L. Sumoylation regulates nuclear localization of lipin-1alpha in neuronal cells. PLoS ONE 2009, 4, e7031. [Google Scholar] [CrossRef][Green Version]







© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castro, V.; Calvo, G.; Ávila-Pérez, G.; Dreux, M.; Gastaminza, P. Differential Roles of Lipin1 and Lipin2 in the Hepatitis C Virus Replication Cycle. Cells 2019, 8, 1456. https://doi.org/10.3390/cells8111456
Castro V, Calvo G, Ávila-Pérez G, Dreux M, Gastaminza P. Differential Roles of Lipin1 and Lipin2 in the Hepatitis C Virus Replication Cycle. Cells. 2019; 8(11):1456. https://doi.org/10.3390/cells8111456
Chicago/Turabian StyleCastro, Victoria, Gema Calvo, Ginés Ávila-Pérez, Marlène Dreux, and Pablo Gastaminza. 2019. "Differential Roles of Lipin1 and Lipin2 in the Hepatitis C Virus Replication Cycle" Cells 8, no. 11: 1456. https://doi.org/10.3390/cells8111456
APA StyleCastro, V., Calvo, G., Ávila-Pérez, G., Dreux, M., & Gastaminza, P. (2019). Differential Roles of Lipin1 and Lipin2 in the Hepatitis C Virus Replication Cycle. Cells, 8(11), 1456. https://doi.org/10.3390/cells8111456




