Absence of the Tks4 Scaffold Protein Induces Epithelial-Mesenchymal Transition-Like Changes in Human Colon Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
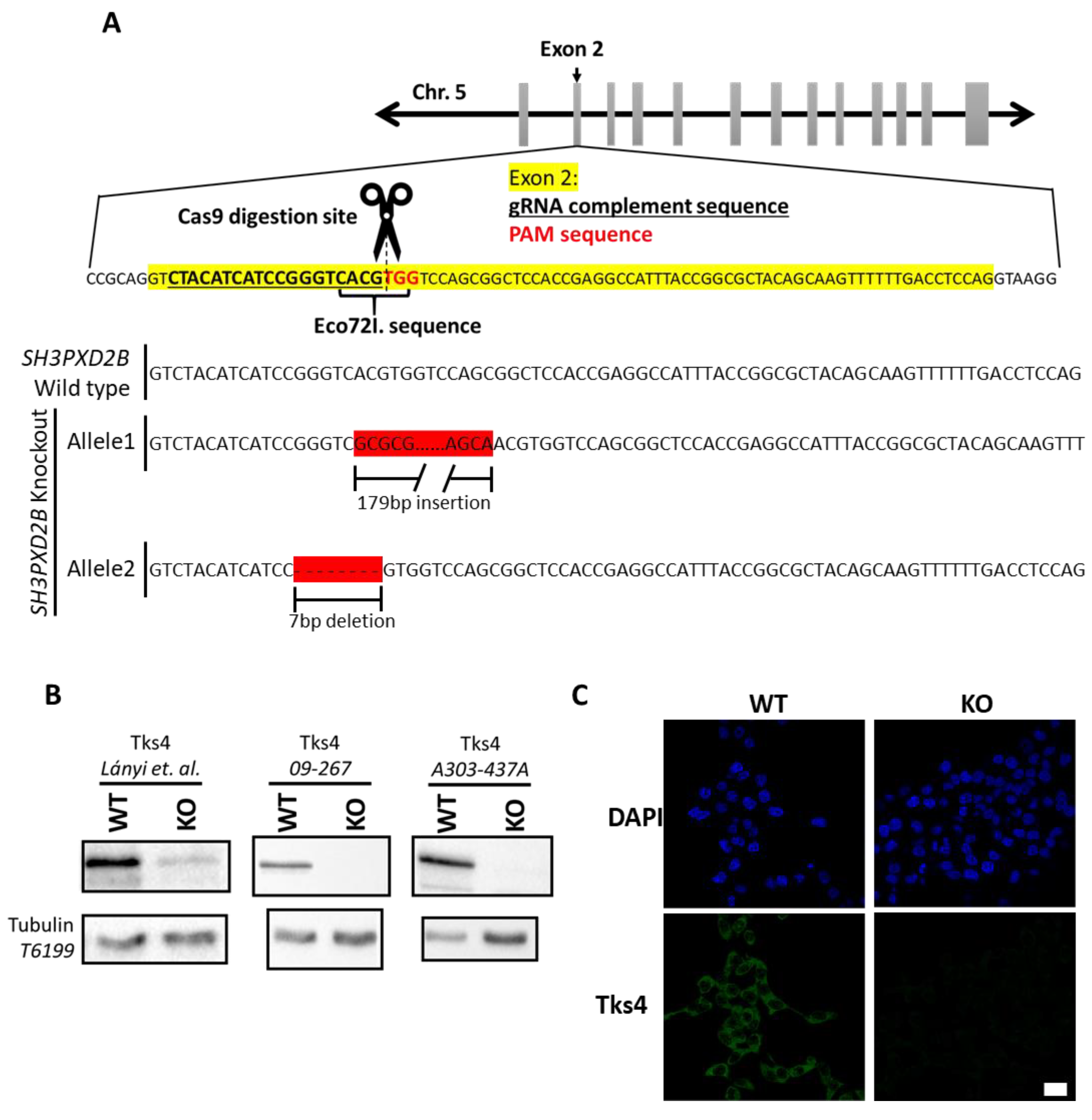
2.1. CRISPR/Cas9-Mediated Engineering of the HCT116 Cell Genome
2.2. Cell Adhesion Assays
2.3. Antibodies
2.4. Confocal Microscopy
2.5. Extraction of Total RNA and RT-PCR Analysis
2.6. Western Blot Analysis
2.7. Construction of the pCAGIG_hTks4 Rescue Plasmid and Transient Tks4 Expression
2.8. Automated Microscopy
2.9. Segmentation of Cell Covered Area
2.10. Optical Flow Analysis
2.11. Individual Cell Movement
2.12. Spheroid Formation
2.13. 3D Collagen Invasion Assay
2.14. Statistical Analyses
3. Results
3.1. Generation of Tks4 Knockout HCT116 Cells
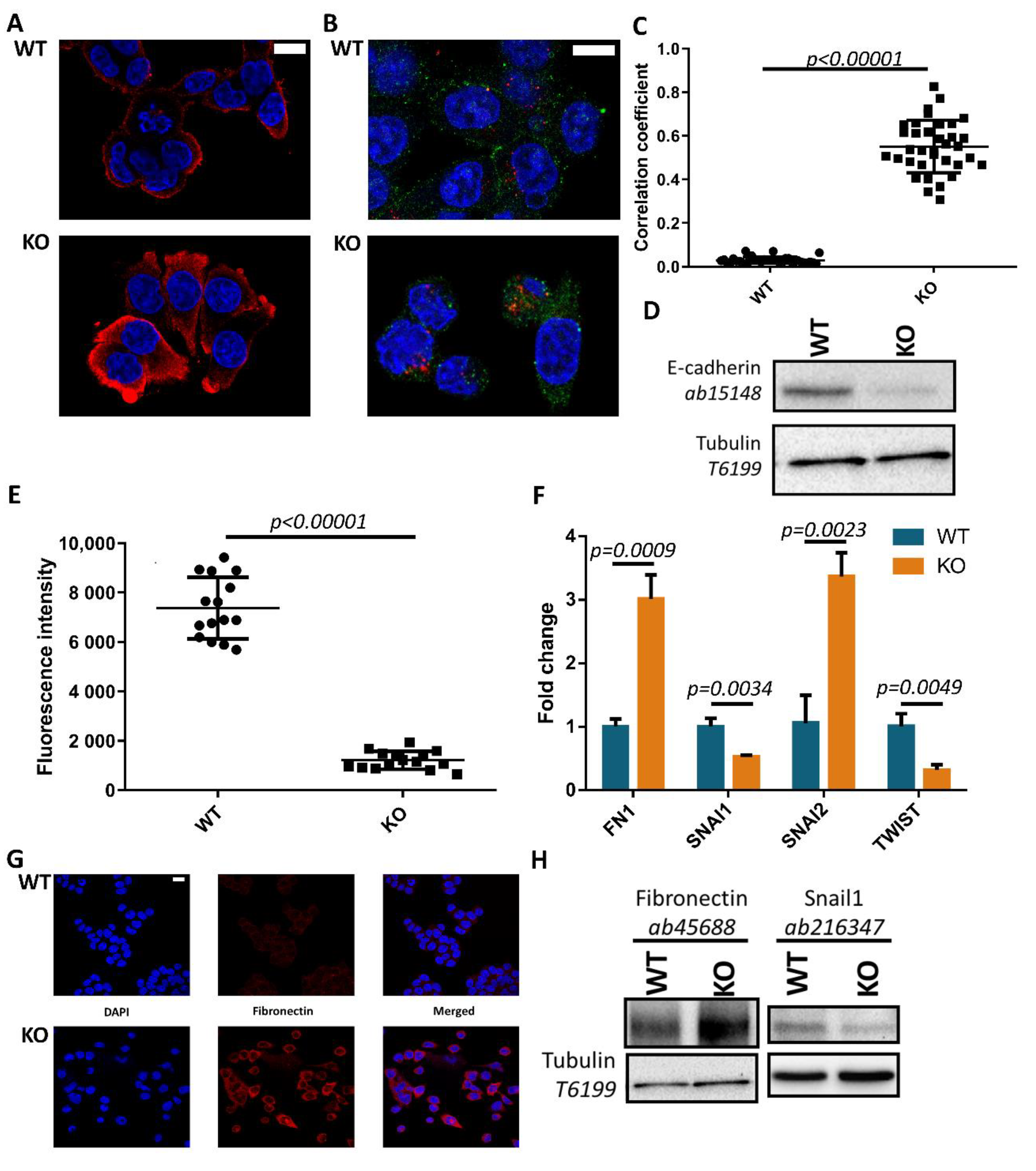
3.2. Tks4-KO Cells Show Morphological Changes and Increased Expression of Fibronectin and Snail2 Transcription Factor
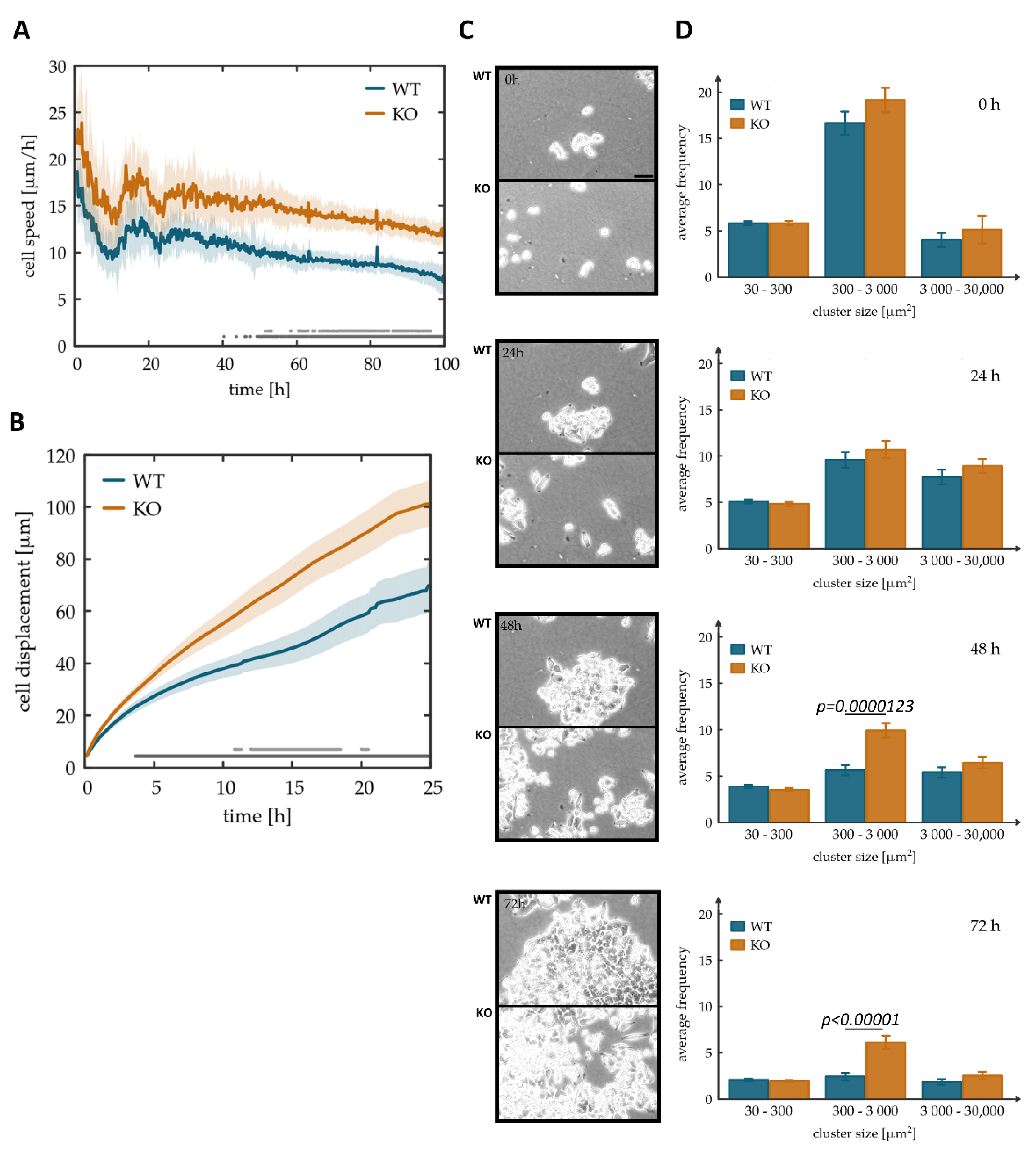
3.3. Tks4-KO Cells Showed Increased Motility and Detachment from Each Other in Monolayer Cultures
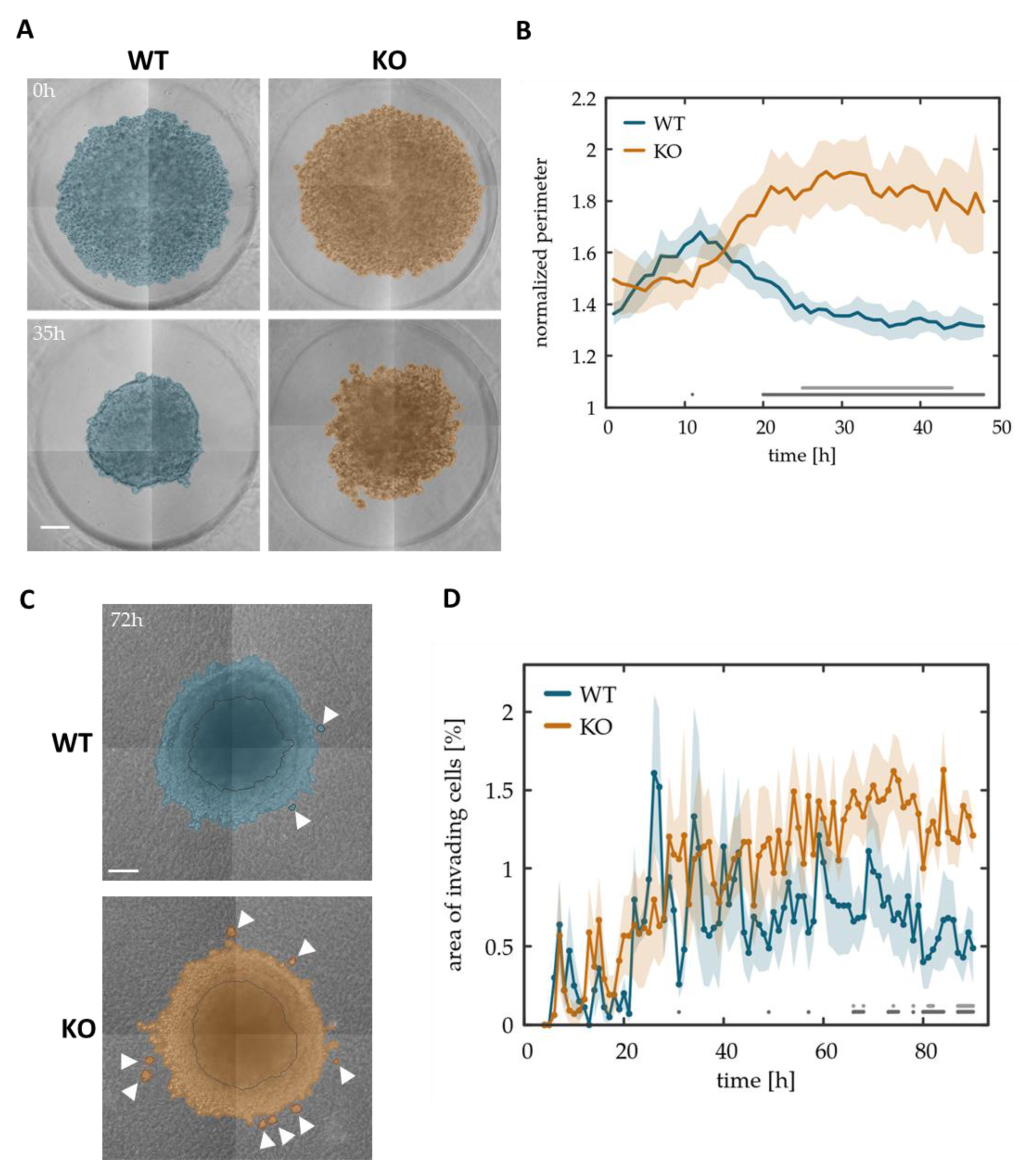
3.4. Tks4-KO Cells Show Decreased Spheroid Forming Potential and Display Increased Invasiveness in Collagen Matrix
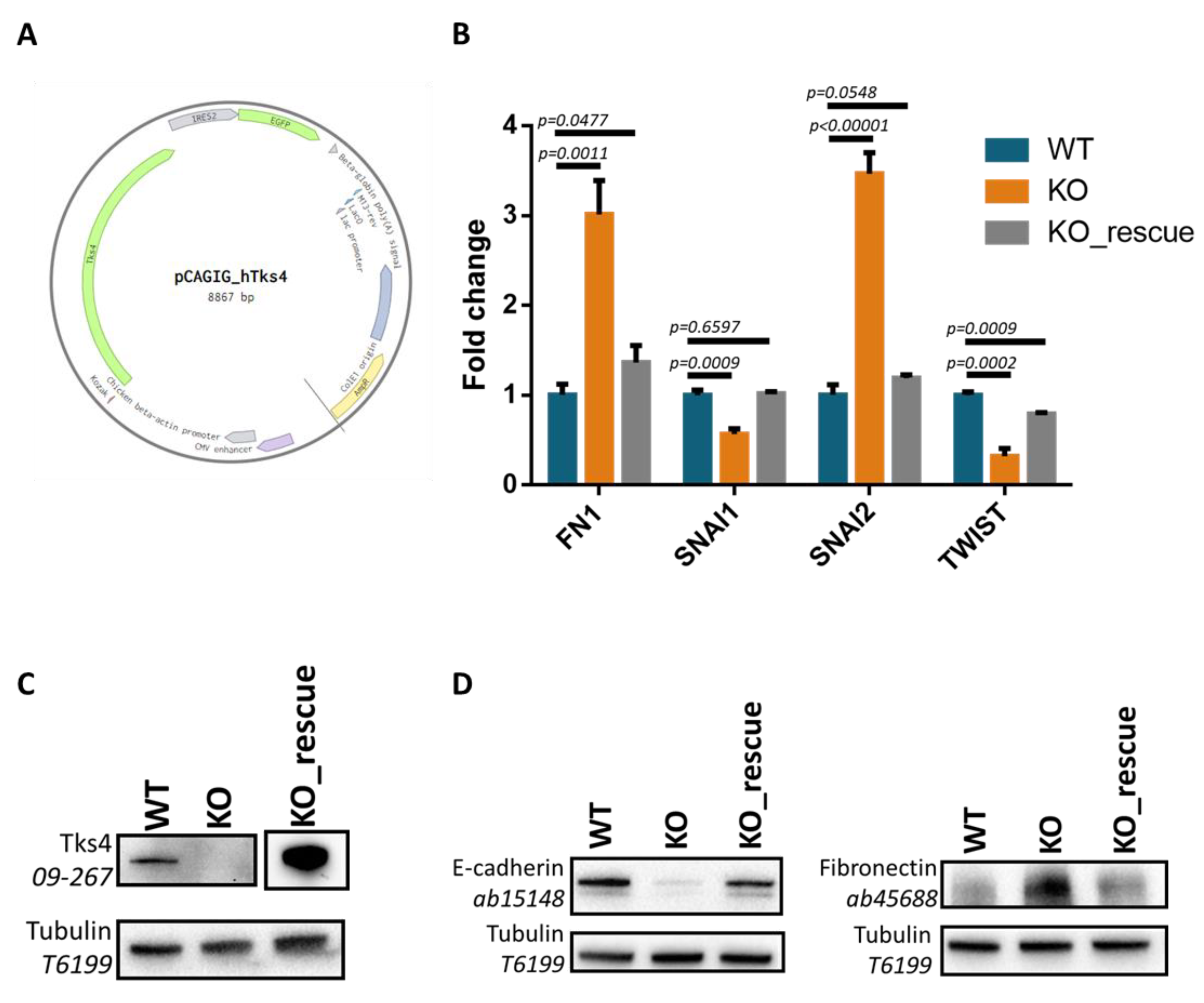
3.5. Reintroducing Tks4 Rescues Altered FN1 and EMT Transcription Factor Expression Levels
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Thiery, J.P.; Acloque, H.; Huang, R.Y.J.; Nieto, M.A. Epithelial-Mesenchymal Transitions in Development and Disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef] [PubMed]
- Acloque, H.; Adams, M.S.; Fishwick, K.; Bronner-Fraser, M.; Nieto, M.A. Epithelial-mesenchymal transitions: The importance of changing cell state in development and disease. J. Clin. Invest. 2009, 119, 1438–1449. [Google Scholar] [CrossRef] [PubMed]
- Burdsal, C.A.; Damsky, C.H.; Pedersen, R.A. The role of E-cadherin and integrins in mesoderm differentiation and migration at the mammalian primitive streak. Development 1993, 118, 829–844. [Google Scholar] [PubMed]
- Yeung, K.T.; Yang, J. Epithelial-mesenchymal transition in tumor metastasis. Mol. Oncol. 2017, 11, 28–39. [Google Scholar] [CrossRef]
- Yan, C.; Grimm, W.A.; Garner, W.L.; Qin, L.; Travis, T.; Tan, N.; Han, Y.-P. Epithelial to Mesenchymal Transition in Human Skin Wound Healing Is Induced by Tumor Necrosis Factor-α through Bone Morphogenic Protein-2. Am. J. Pathol. 2010, 176, 2247–2258. [Google Scholar] [CrossRef]
- Boyer, B.; Thiery, J.P. Epithelium-mesenchyme interconversion as example of epithelial plasticity. Apmis 1993, 101, 257–268. [Google Scholar] [CrossRef]
- Hay, E.D. An overview of epithelio-mesenchymal transformation. Acta Anat. 1995, 154, 8–20. [Google Scholar] [CrossRef]
- Cano, A.; Pérez-Moreno, M.A.; Rodrigo, I.; Locascio, A.; Blanco, M.J.; del Barrio, M.G.; Portillo, F.; Nieto, M.A. The transcription factor Snail controls epithelial–mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2000, 2, 76–83. [Google Scholar] [CrossRef]
- Batlle, E.; Sancho, E.; Francí, C.; Domínguez, D.; Monfar, M.; Baulida, J.; García de Herreros, A. The transcription factor Snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2000, 2, 84–89. [Google Scholar] [CrossRef]
- Mistry, D.S.; Chen, Y.; Wang, Y.; Zhang, K.; Sen, G.L. SNAI2 controls the undifferentiated state of human epidermal progenitor cells. Stem Cells 2014, 32, 3209–3218. [Google Scholar] [CrossRef]
- Bax, N.A.M.; Pijnappels, D.A.; van Oorschot, A.A.M.; Winter, E.M.; de Vries, A.A.F.; van Tuyn, J.; Braun, J.; Maas, S.; Schalij, M.J.; Atsma, D.E.; et al. Epithelial-to-mesenchymal transformation alters electrical conductivity of human epicardial cells. J. Cell. Mol. Med. 2011, 15, 2675–2683. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Cai, Y.; Liu, J.; Wang, Z.; Wu, Q.; Zhang, Z.; Yang, C.J.; Yuan, L.; Ouyang, G. Twist2 contributes to breast cancer progression by promoting an epithelial–mesenchymal transition and cancer stem-like cell self-renewal. Oncogene 2011, 30, 4707–4720. [Google Scholar] [CrossRef] [PubMed]
- Porta-de-la-Riva, M.; Stanisavljevic, J.; Curto, J.; Francí, C.; Díaz, V.M.; García de Herreros, A.; Baulida, J. TFCP2c/LSF/LBP-1c is required for Snail1-induced fibronectin gene expression. Biochem. J. 2011, 435, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Docherty, N.G. TGF-beta1-induced EMT can occur independently of its proapoptotic effects and is aided by EGF receptor activation. AJP Ren. Physiol. 2005, 290, F1202–F1212. [Google Scholar] [CrossRef] [PubMed]
- Zavadil, J.; Cermak, L.; Soto-Nieves, N.; Böttinger, E.P. Integration of TGF-β/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition. EMBO J. 2004, 23, 1155–1165. [Google Scholar] [CrossRef]
- Leong, K.G.; Niessen, K.; Kulic, I.; Raouf, A.; Eaves, C.; Pollet, I.; Karsan, A. Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin. J. Exp. Med. 2007, 204, 2935–2948. [Google Scholar] [CrossRef]
- Yook, J.I.; Li, X.-Y.; Ota, I.; Hu, C.; Kim, H.S.; Kim, N.H.; Cha, S.Y.; Ryu, J.K.; Choi, Y.J.; Kim, J.; et al. A Wnt–Axin2–GSK3β cascade regulates Snail1 activity in breast cancer cells. Nat. Cell Biol. 2006, 8, 1398–1406. [Google Scholar] [CrossRef]
- Fodde, R.; Brabletz, T. Wnt/β-catenin signaling in cancer stemness and malignant behavior. Curr. Opin. Cell Biol. 2007, 19, 150–158. [Google Scholar] [CrossRef]
- Sahlgren, C.; Gustafsson, M.V.; Jin, S.; Poellinger, L.; Lendahl, U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc. Natl. Acad. Sci. USA 2008, 105, 6392–6397. [Google Scholar] [CrossRef]
- Kim, J.; Kong, J.; Chang, H.; Kim, H.; Kim, A. EGF induces epithelial-mesenchymal transition through phospho-Smad2/3-Snail signaling pathway in breast cancer cells. Oncotarget 2016, 7, 85021–85032. [Google Scholar] [CrossRef]
- Zeke, A.; Lukács, M.; Lim, W.A.; Reményi, A. Scaffolds: Interaction platforms for cellular signalling circuits. Trends Cell Biol. 2009, 19, 364–374. [Google Scholar] [CrossRef] [PubMed]
- Buday, L.; Tompa, P. Functional classification of scaffold proteins and related molecules. FEBS J. 2010, 277, 4348–4355. [Google Scholar] [CrossRef] [PubMed]
- Buschman, M.D.; Bromann, P.A.; Cejudo-Martin, P.; Wen, F.; Pass, I.; Courtneidge, S.A. The Novel Adaptor Protein Tks4 (SH3PXD2B) Is Required for Functional Podosome Formation. Mol. Biol. Cell 2009, 20, 1302–1311. [Google Scholar] [CrossRef] [PubMed]
- Bögel, G.; Gujdár, A.; Geiszt, M.; Lányi, Á.; Fekete, A.; Sipeki, S.; Downward, J.; Buday, L. Frank-ter Haar syndrome protein Tks4 regulates epidermal growth factor-dependent cell migration. J. Biol. Chem. 2012, 287, 31321–31329. [Google Scholar] [CrossRef]
- Dülk, M.; Kudlik, G.; Fekete, A.; Ernszt, D.; Kvell, K.; Pongrácz, J.E.; Merő, B.L.; Szeder, B.; Radnai, L.; Geiszt, M.; et al. The scaffold protein Tks4 is required for the differentiation of mesenchymal stromal cells (MSCs) into adipogenic and osteogenic lineages. Sci. Rep. 2016, 6, 34280. [Google Scholar] [CrossRef]
- Vas, V.; Kovács, T.; Körmendi, S.; Bródy, A.; Kudlik, G.; Szeder, B.; Mező, D.; Kállai, D.; Koprivanacz, K.; Merő, B.L.; et al. Significance of the Tks4 scaffold protein in bone tissue homeostasis. Sci. Rep. 2019, 9, 5781. [Google Scholar] [CrossRef]
- Iizuka, S.; Abdullah, C.; Buschman, M.D.; Diaz, B.; Courtneidge, S.A. The role of Tks adaptor proteins in invadopodia formation, growth and metastasis of melanoma. Oncotarget 2016, 7. [Google Scholar] [CrossRef]
- Murphy, D.A.; Courtneidge, S.A. The “ins” and “outs” of podosomes and invadopodia: Characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 2011, 12, 413–426. [Google Scholar] [CrossRef]
- Vas, V.; Háhner, T.; Kudlik, G.; Ernszt, D.; Kvell, K.; Kuti, D.; Kovács, K.J.; Tóvári, J.; Trexler, M.; Merő, B.L.; et al. Analysis of Tks4 Knockout Mice Suggests a Role for Tks4 in Adipose Tissue Homeostasis in the Context of Beigeing. Cells 2019, 8, 831. [Google Scholar] [CrossRef]
- Iqbal, Z.; Cejudo-Martin, P.; de Brouwer, A.; van der Zwaag, B.; Ruiz-Lozano, P.; Scimia, M.C.; Lindsey, J.D.; Weinreb, R.; Albrecht, B.; Megarbane, A.; et al. Disruption of the Podosome Adaptor Protein TKS4 (SH3PXD2B) Causes the Skeletal Dysplasia, Eye, and Cardiac Abnormalities of Frank-Ter Haar Syndrome. Am. J. Hum. Genet. 2010, 86, 254–261. [Google Scholar] [CrossRef]
- Dundar, M.; Saatci, C.; Tasdemir, S.; Akcakus, M.; Caglayan, A.O.; Ozkul, Y. Frank-ter Haar syndrome with unusual clinical features. Eur. J. Med. Genet. 2009. [Google Scholar] [CrossRef] [PubMed]
- Saeed, M.; Shair, Q.A.; Saleem, S.M. Frank-Ter Haar Syndrome. J. Coll. Physicians Surg. Pakistan 2011, 21, 252–253. [Google Scholar]
- Femitha, P.; Joy, R.; Gane, B.D.; Adhisivam, B.; Bhat, B.V. Frank -Ter Haar Syndrome in a Newborn. Indian J. Pediatr. 2012, 79, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Köse, T.E.; İşler, C.; Şenel, Ş.N.; Şitilci, T.; Özcan, İ.; Aksakallı, N. Frank–ter Haar syndrome—additional findings? Dentomaxillofacial Radiol. 2016, 45, 20150119. [Google Scholar] [CrossRef]
- Zrhidri, A.; Jaouad, I.C.; Lyahyai, J.; Raymond, L.; Egéa, G.; Taoudi, M.; El Mouatassim, S.; Sefiani, A. Identification of two novel SH3PXD2B gene mutations in Frank-Ter Haar syndrome by exome sequencing: Case report and review of the literature. Gene 2017, 628, 190–193. [Google Scholar] [CrossRef]
- Seals, D.F.; Azucena, E.F.; Pass, I.; Tesfay, L.; Gordon, R.; Woodrow, M.; Resau, J.H.; Courtneidge, S.A. The adaptor protein Tks5/Fish is required for podosome formation and function, and for the protease-driven invasion of cancer cells. Cancer Cell 2005, 7, 155–165. [Google Scholar] [CrossRef]
- Stylli, S.S.; Stacey, T.T.; Kaye, A.H.; Lock, P. Prognostic significance of Tks5 expression in gliomas. J. Clin. Neurosci. 2012, 19, 436–442. [Google Scholar] [CrossRef]
- Li, C.M.-C.; Chen, G.; Dayton, T.L.; Kim-Kiselak, C.; Hoersch, S.; Whittaker, C.A.; Bronson, R.T.; Beer, D.G.; Winslow, M.M.; Jacks, T. Differential Tks5 isoform expression contributes to metastatic invasion of lung adenocarcinoma. Genes Dev. 2013, 27, 1557–1567. [Google Scholar] [CrossRef]
- Burger, K.L.; Learman, B.S.; Boucherle, A.K.; Sirintrapun, S.J.; Isom, S.; Díaz, B.; Courtneidge, S.A.; Seals, D.F. Src-dependent Tks5 phosphorylation regulates invadopodia-associated invasion in prostate cancer cells. Prostate 2014, 74, 134–148. [Google Scholar] [CrossRef]
- Blouw, B.; Patel, M.; Iizuka, S.; Abdullah, C.; You, W.K.; Huang, X.; Li, J.-L.; Diaz, B.; Stallcup, W.B.; Courtneidge, S.A. The Invadopodia Scaffold Protein Tks5 Is Required for the Growth of Human Breast Cancer Cells In Vitro and In Vivo. PLoS ONE 2015, 10, e0121003. [Google Scholar] [CrossRef]
- Lányi, Á.; Baráth, M.; Péterfi, Z.; Bögel, G.; Orient, A.; Simon, T.; Petrovszki, E.; Kis-Tóth, K.; Sirokmány, G.; Rajnavölgyi, É.; et al. The homolog of the five SH3-domain protein (HOFI/SH3PXD2B) regulates lamellipodia formation and cell spreading. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Kenong, W.; Gauthier, D.; Levine, M.D. Live cell image segmentation. IEEE Trans. Biomed. Eng. 1995, 42, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Neufeld, Z.; von Witt, W.; Lakatos, D.; Wang, J.; Hegedus, B.; Czirok, A. The role of Allee effect in modelling post resection recurrence of glioblastoma. PLOS Comput. Biol. 2017, 13, e1005818. [Google Scholar] [CrossRef] [PubMed]
- Zamir, E.A.; Czirok, A.; Cui, C.; Little, C.D.; Rongish, B.J. Mesodermal cell displacements during avian gastrulation are due to both individual cell-autonomous and convective tissue movements. Proc. Natl. Acad. Sci. USA 2006, 103, 19806–19811. [Google Scholar] [CrossRef]
- Czirok, A.; Isai, D.G.; Kosa, E.; Rajasingh, S.; Kinsey, W.; Neufeld, Z.; Rajasingh, J. Optical-flow based non-invasive analysis of cardiomyocyte contractility. Sci. Rep. 2017, 7, 10404. [Google Scholar] [CrossRef]
- Gönci, B.; Németh, V.; Balogh, E.; Szabó, B.; Dénes, Á.; Környei, Z.; Vicsek, T. Viral Epidemics in a Cell Culture: Novel High Resolution Data and Their Interpretation by a Percolation Theory Based Model. PLoS ONE 2010, 5, e15571. [Google Scholar] [CrossRef]
- Dunn, G.A.; Brown, A.F. A unified approach to analysing cell motility. J. Cell Sci. Suppl. 1987, 8, 81–102. [Google Scholar] [CrossRef]
- Maheshwari, G.; Lauffenburger, D.A. Deconstructing (and reconstructing) cell migration. Microsc. Res. Tech. 1998, 43, 358–368. [Google Scholar] [CrossRef]
- Gulyas, M.; Csiszer, M.; Mehes, E.; Czirok, A. Software tools for cell culture-related 3D printed structures. PLoS ONE 2018, 13, e0203203. [Google Scholar] [CrossRef]
- Palacios, F.; Tushir, J.S.; Fujita, Y.; D’Souza-Schorey, C. Lysosomal Targeting of E-Cadherin: A Unique Mechanism for the Down-Regulation of Cell-Cell Adhesion during Epithelial to Mesenchymal Transitions. Mol. Cell. Biol. 2005, 25, 389–402. [Google Scholar] [CrossRef]
- Diepenbruck, M.; Christofori, G. Epithelial–mesenchymal transition (EMT) and metastasis: Yes, no, maybe? Curr. Opin. Cell Biol. 2016, 43, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Li, W.; Wang, H. Roles of miRNA in the Initiation and Development of Colorectal Carcinoma. Curr. Pharm. Des. 2013, 19, 1253–1261. [Google Scholar] [PubMed]
- Maziveyi, M.; Dong, S.; Baranwal, S.; Alahari, S.K. Nischarin regulates focal adhesion and Invadopodia formation in breast cancer cells. Mol. Cancer 2018, 17, 21. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.H.; Liu, Y.; Gao, S.Y. Correlation between IL-7 genomic protein methylation level and acute myeloid leukemia. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 1196–1202. [Google Scholar]
- Caires-dos-Santos, L.; da Silva, S.V.; Smuczek, B.; de Siqueira, A.S.; Cruz, K.S.P.; Barbuto, J.A.M.; Augusto, T.M.; Freitas, V.M.; Carvalho, H.F.; Jaeger, R.G. Laminin-derived peptide C16 regulates Tks expression and reactive oxygen species generation in human prostate cancer cells. J. Cell. Physiol. 2019, 1–12. [Google Scholar] [CrossRef]
- Gianni, D.; Taulet, N.; Zhang, H.; Dermardirossian, C.; Kister, J.; Martinez, L.; Roush, W.R.; Brown, S.J.; Bokoch, G.M.; Rosen, H. A Novel and Specific NADPH Oxidase-1 (Nox1) Small-Molecule Inhibitor Blocks the Formation of Funcional Invadopodia in Human Colon Cancer Cells. ACS Chem. Biol. 2010, 5, 981–993. [Google Scholar] [CrossRef]
- Schoumacher, M.; Goldman, R.D.; Louvard, D.; Vignjevic, D.M. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J. Cell Biol. 2010, 189, 541–556. [Google Scholar] [CrossRef]
- Welman, A.; Cawthome, C.; Ponce-Perez, L.; Barraclough, J.; Danson, S.; Murray, S.; Cummings, J.; Allen, T.D.; Dive, C. Increases in c-Src Expression Level and Activity Do Not Promote the Growth of Human Colorectal Carcinoma Cells In Vitro and In Vivo. Neoplasia 2006, 8, 905. [Google Scholar] [CrossRef]
- Pino, M.S.; Kikuchi, H.; Zeng, M.; Herraiz, M.; Sperduti, I.; Berger, D.; Park, D.; Iafrate, A.J.; Zukerberg, L.R.; Chung, D.C. Epithelial to Mesenchymal Transition Is Impaired in Colon Cancer Cells with Microsatellite Instability. Gastroenterology 2010, 138, 1406–1417. [Google Scholar] [CrossRef]
- Dülk, M.; Szeder, B.; Glatz, G.; Merö, B.L.; Koprivanacz, K.; Kudlik, G.; Vas, V.; Sipeki, S.; Cserkaszky, A.; Radnai, L.; et al. EGF Regulates the Interaction of Tks4 with Src through Its SH2 and SH3 Domains. Biochemistry 2018, 57, 4186–4196. [Google Scholar] [CrossRef]
- Wendum, D.; Clapéron, A.; Mergey, M.; Vignjevic, D.; Housset, C.; Paradis, V.; Fouassier, L.; Frazao, A.; Guedj, N.; Merabtene, F.; et al. EGF/EGFR axis contributes to the progression of cholangiocarcinoma through the induction of an epithelial-mesenchymal transition. J. Hepatol. 2014, 61, 325–332. [Google Scholar]
- Cheng, J.-C.; Auersperg, N.; Leung, P.C.K. EGF-Induced EMT and Invasiveness in Serous Borderline Ovarian Tumor Cells: A Possible Step in the Transition to Low-Grade Serous Carcinoma Cells? PLoS ONE 2012, 7, e34071. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L.; Li, D.-D.; Yang, C.-L.; Peng, R.-Q.; Guo, Y.-Q.; Zhang, X.-S.; Zhu, X.-F. Reactive oxygen species mediate oxaliplatin-induced epithelial-mesenchymal transition and invasive potential in colon cancer. Tumor Biol. 2016, 37, 8413–8423. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.-H.; Kim, S.-M.; Lee, C.-E. Mechanism of suppressors of cytokine signaling 1 inhibition of epithelial-mesenchymal transition signaling through ROS regulation in colon cancer cells: Suppression of Src leading to thioredoxin up-regulation. Oncotarget 2016, 7. [Google Scholar] [CrossRef]
- Leong, H.S.; Robertson, A.E.; Stoletov, K.; Leith, S.J.; Chin, C.A.; Chien, A.E.; Hague, M.N.; Ablack, A.; Carmine-Simmen, K.; McPherson, V.A.; et al. Invadopodia Are Required for Cancer Cell Extravasation and Are a Therapeutic Target for Metastasis. Cell Rep. 2014, 8, 1558–1570. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szeder, B.; Tárnoki-Zách, J.; Lakatos, D.; Vas, V.; Kudlik, G.; Merő, B.; Koprivanacz, K.; Bányai, L.; Hámori, L.; Róna, G.; et al. Absence of the Tks4 Scaffold Protein Induces Epithelial-Mesenchymal Transition-Like Changes in Human Colon Cancer Cells. Cells 2019, 8, 1343. https://doi.org/10.3390/cells8111343
Szeder B, Tárnoki-Zách J, Lakatos D, Vas V, Kudlik G, Merő B, Koprivanacz K, Bányai L, Hámori L, Róna G, et al. Absence of the Tks4 Scaffold Protein Induces Epithelial-Mesenchymal Transition-Like Changes in Human Colon Cancer Cells. Cells. 2019; 8(11):1343. https://doi.org/10.3390/cells8111343
Chicago/Turabian StyleSzeder, Bálint, Júlia Tárnoki-Zách, Dóra Lakatos, Virág Vas, Gyöngyi Kudlik, Balázs Merő, Kitti Koprivanacz, László Bányai, Lilla Hámori, Gergely Róna, and et al. 2019. "Absence of the Tks4 Scaffold Protein Induces Epithelial-Mesenchymal Transition-Like Changes in Human Colon Cancer Cells" Cells 8, no. 11: 1343. https://doi.org/10.3390/cells8111343
APA StyleSzeder, B., Tárnoki-Zách, J., Lakatos, D., Vas, V., Kudlik, G., Merő, B., Koprivanacz, K., Bányai, L., Hámori, L., Róna, G., Czirók, A., Füredi, A., & Buday, L. (2019). Absence of the Tks4 Scaffold Protein Induces Epithelial-Mesenchymal Transition-Like Changes in Human Colon Cancer Cells. Cells, 8(11), 1343. https://doi.org/10.3390/cells8111343






