Lamins in Lung Cancer: Biomarkers and Key Factors for Disease Progression through miR-9 Regulation?
Abstract
1. Introduction
2. Lamins’ Expression in Normal and Cancer Lung Tissues
2.1. Physiological Lamins’ Expression
2.2. Lamins’ Expression Depending on Lung Cancer Histological Subtypes
2.3. Potential Link between the Loss of A-Type Lamins and Nuclear Deformability and Metastatic Potential Enhancement
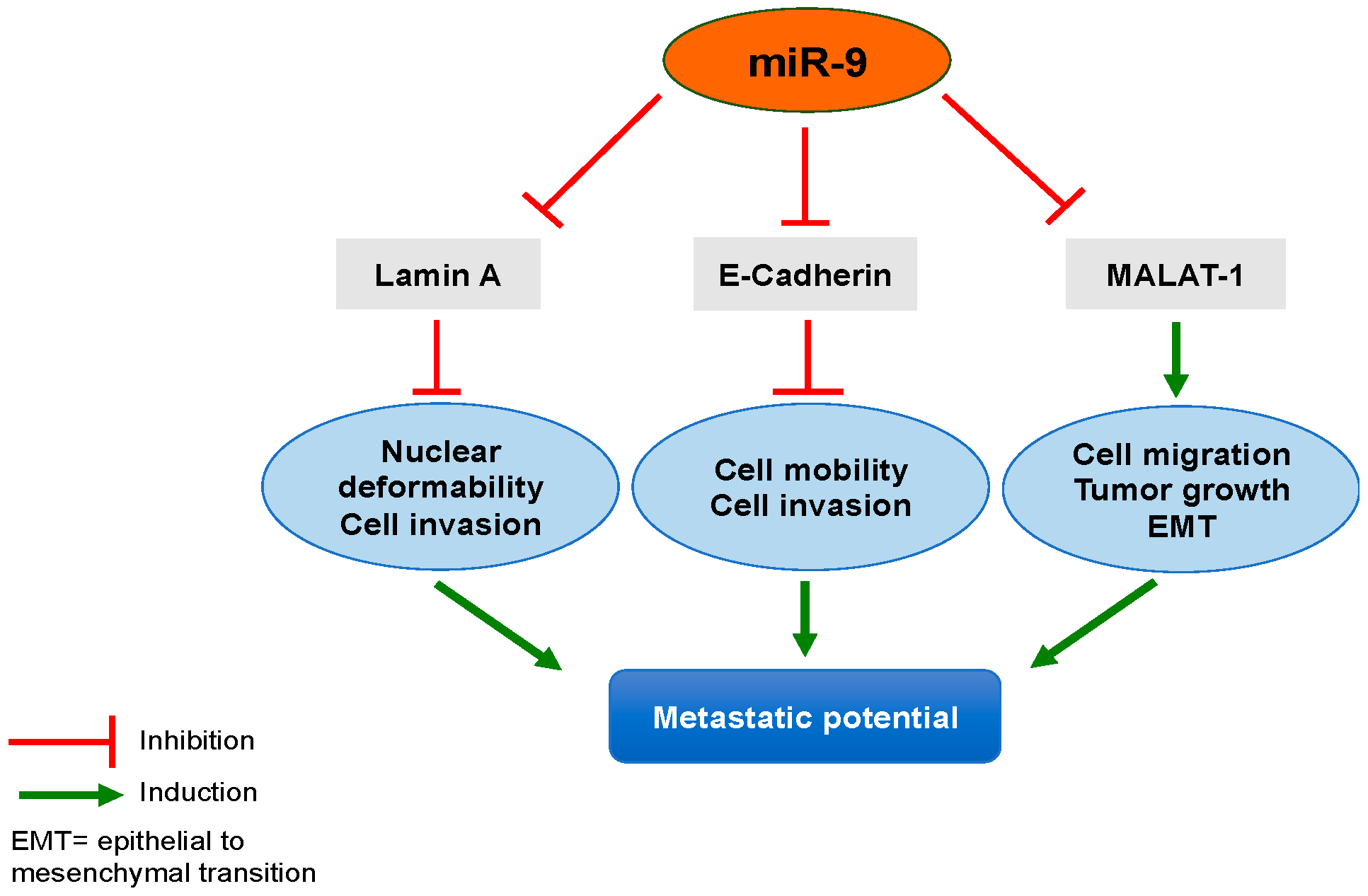
3. Potential Mechanisms of A-Type Lamins Regulation in Lung Cancer Implicating miR-9
3.1. MicroRNAs
3.2. MicroRNAs and Lung Cancer
3.3. miR-9 in Lung Cancer
3.4. miR-9 and A-Type Lamins
4. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lozano, R.; Naghavi, M.; Foreman, K.; Lim, S.; Shibuya, K.; Aboyans, V.; Abraham, J.; Adair, T.; Aggarwal, R.; Ahn, S.Y.; et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2095–2128. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Novello, S.; Barlesi, F.; Califano, R.; Cufer, T.; Ekman, S.; Levra, M.G.; Kerr, K.; Popat, S.; Reck, M.; Senan, S.; et al. Metastatic non-small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2016, 27, v1–v27. [Google Scholar] [CrossRef] [PubMed]
- Shea, M.; Costa, D.B.; Rangachari, D. Management of advanced non-small cell lung cancers with known mutations or rearrangements: Latest evidence and treatment approaches. Ther. Adv. Respir. Dis. 2016, 10, 113–129. [Google Scholar] [CrossRef] [PubMed]
- Barlesi, F.; Mazieres, J.; Merlio, J.-P.; Debieuvre, D.; Mosser, J.; Lena, H.; Ouafik, L.; Besse, B.; Rouquette, I.; Westeel, V.; et al. Routine molecular profiling of patients with advanced non-small-cell lung cancer: Results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT). Lancet 2016, 387, 1415–1426. [Google Scholar] [CrossRef]
- Capo-Chichi, C.D.; Cai, K.Q.; Smedberg, J.; Ganjei-Azar, P.; Godwin, A.K.; Xu, X.-X. Loss of A-type lamin expression compromises nuclear envelope integrity in breast cancer. Chin. J. Cancer 2011, 30, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Zink, D.; Fische, A.H.; Nickerson, J.A. Nuclear structure in cancer cells. Nat. Rev. Cancer 2004, 4, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Butin-Israeli, V.; Adam, S.A.; Goldman, A.E.; Goldman, R.D. Nuclear lamin functions and disease. Trends Genet. 2012, 28, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Lammerding, J.; Schulze, P.C.; Takahashi, T.; Kozlov, S.; Sullivan, T.; Kamm, R.D.; Stewart, C.L.; Lee, R.T. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J. Clin. Investig. 2004, 113, 370–378. [Google Scholar] [CrossRef] [PubMed]
- Lammerding, J.; Fong, L.G.; Ji, J.Y.; Reue, K.; Stewart, C.L.; Young, S.G.; Lee, R.T. Lamins A and C but Not Lamin B1 Regulate Nuclear Mechanics. J. Biol. Chem. 2006, 281, 25768–25780. [Google Scholar] [CrossRef] [PubMed]
- Redwood, A.B.; Perkins, S.M.; Vanderwaal, R.P.; Feng, Z.; Biehl, K.J.; Gonzalez-Suarez, I.; Morgado-Palacin, L.; Shi, W.; Sage, J.; Roti-Roti, J.L.; et al. A dual role for A-type lamins in DNA double-strand break repair. Cell Cycle 2011, 10, 2549–2560. [Google Scholar] [CrossRef] [PubMed]
- Butin-Israeli, V.; Adam, S.A.; Goldman, R.D. Regulation of Nucleotide Excision Repair by Nuclear Lamin B1. PLoS ONE 2013, 8, e69169. [Google Scholar] [CrossRef] [PubMed]
- Mahen, R.; Hattori, H.; Lee, M.; Sharma, P.; Jeyasekharan, A.D.; Venkitaraman, A.R. A-Type Lamins Maintain the Positional Stability of DNA Damage Repair Foci in Mammalian Nuclei. PLoS ONE 2013, 8, e61893. [Google Scholar] [CrossRef] [PubMed]
- Broers, J.L.V.; Ramaekers, F.C.S. The Role of the Nuclear Lamina in Cancer and Apoptosis. In Cancer Biology and the Nuclear Envelope; Schirmer, E.C., de las Heras, J.I., Eds.; Springer: New York, NY, USA, 2014; Volume 773, pp. 27–48. ISBN 9781489980311. [Google Scholar]
- Ungricht, R.; Kutay, U. Mechanisms and functions of nuclear envelope remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 229–245. [Google Scholar] [CrossRef] [PubMed]
- Machiels, B.M.; Zorenc, A.H.; Endert, J.M.; Kuijpers, H.J.; van Eys, G.J.; Ramaekers, F.C.; Broers, J.L. An alternative splicing product of the lamin A/C gene lacks exon 10. J. Biol. Chem. 1996, 271, 9249–9253. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Worman, H.J. Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C. J. Biol. Chem. 1993, 268, 16321–16326. [Google Scholar] [PubMed]
- Dechat, T.; Pfleghaar, K.; Sengupta, K.; Shimi, T.; Shumaker, D.K.; Solimando, L.; Goldman, R.D. Nuclear lamins: Major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008, 22, 832–853. [Google Scholar] [CrossRef] [PubMed]
- Frangioni, J.V.; Neel, B.G. Use of a general purpose mammalian expression vector for studying intracellular protein targeting: Identification of critical residues in the nuclear lamin A/C nuclear localization signal. J. Cell Sci. 1993, 105 Pt 2, 481–488. [Google Scholar] [PubMed]
- Stuurman, N.; Heins, S.; Aebi, U. Nuclear Lamins: Their Structure, Assembly, and Interactions. J. Struct. Biol. 1998, 122, 42–66. [Google Scholar] [CrossRef] [PubMed]
- Cau, P.; Navarro, C.; Harhouri, K.; Roll, P.; Sigaudy, S.; Kaspi, E.; Perrin, S.; De Sandre-Giovannoli, A.; Lévy, N. WITHDRAWN: Nuclear matrix, nuclear envelope and premature aging syndromes in a translational research perspective. Semin. Cell Dev. Biol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Prokocimer, M.; Davidovich, M.; Nissim-Rafinia, M.; Wiesel-Motiuk, N.; Bar, D.Z.; Barkan, R.; Meshorer, E.; Gruenbaum, Y. Nuclear lamins: Key regulators of nuclear structure and activities. J. Cell. Mol. Med. 2009, 13, 1059–1085. [Google Scholar] [CrossRef] [PubMed]
- Broers, J.L.V.; Ramaekers, F.C.S.; Bonne, G.; Yaou, R.B.; Hutchison, C.J. Nuclear Lamins: Laminopathies and Their Role in Premature Ageing. Physiol. Rev. 2006, 86, 967–1008. [Google Scholar] [CrossRef] [PubMed]
- Gerace, L.; Burke, B. Functional Organization of the Nuclear Envelope. Annu. Rev. Cell Biol. 1988, 4, 335–374. [Google Scholar] [CrossRef] [PubMed]
- Krohne, G.; Benavente, R. The nuclear lamins. A multigene family of proteins in evolution and differentiation. Exp. Cell Res. 1986, 162, 1–10. [Google Scholar] [CrossRef]
- Nigg, E.A. Assembly and cell cycle dynamics of the nuclear lamina. Semin. Cell Biol. 1992, 3, 245–253. [Google Scholar] [CrossRef]
- Broers, J.L.; Machiels, B.M.; Kuijpers, H.J.; Smedts, F.; van den Kieboom, R.; Raymond, Y.; Ramaekers, F.C. A- and B-type lamins are differentially expressed in normal human tissues. Histochem. Cell Biol. 1997, 107, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Oguchi, M.; Sagara, J.; Matsumoto, K.; Saida, T.; Taniguchi, S. Expression of lamins depends on epidermal differentiation and transformation. Br. J. Dermatol. 2002, 147, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Irianto, J.; Pfeifer, C.R.; Ivanovska, I.L.; Swift, J.; Discher, D.E. Nuclear lamins in cancer. Cell. Mol. Bioeng. 2016, 9, 258–267. [Google Scholar] [CrossRef] [PubMed]
- Swift, J.; Ivanovska, I.L.; Buxboim, A.; Harada, T.; Dingal, P.C.D.P.; Pinter, J.; Pajerowski, J.D.; Spinler, K.R.; Shin, J.-W.; Tewari, M.; et al. Nuclear Lamin-A Scales with Tissue Stiffness and Enhances Matrix-Directed Differentiation. Science 2013, 341, 1240104. [Google Scholar] [CrossRef] [PubMed]
- Frankel, D.; Delecourt, V.; Harhouri, K.; De Sandre-Giovannoli, A.; Lévy, N.; Kaspi, E.; Roll, P. MicroRNAs in hereditary and sporadic premature aging syndromes and other laminopathies. Aging Cell 2018, e12766. [Google Scholar] [CrossRef] [PubMed]
- De Sandre-Giovannoli, A. Lamin A Truncation in Hutchinson-Gilford Progeria. Science 2003, 300, 2055. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, M.; Brown, W.T.; Gordon, L.B.; Glynn, M.W.; Singer, J.; Scott, L.; Erdos, M.R.; Robbins, C.M.; Moses, T.Y.; Berglund, P.; et al. Recurrent de novo point mutations in lamin A cause Hutchinson–Gilford progeria syndrome. Nature 2003, 423, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Kubben, N.; Misteli, T. Shared molecular and cellular mechanisms of premature ageing and ageing-associated diseases. Nat. Rev. Mol. Cell Biol. 2017, 18, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, P.; Scaffidi, P.; Markert, E.; Lee, J.-H.; Rane, S.; Misteli, T. Transformation Resistance in a Premature Aging Disorder Identifies a Tumor-Protective Function of BRD4. Cell Rep. 2014, 9, 248–260. [Google Scholar] [CrossRef] [PubMed]
- Sakthivel, K.M.; Sehgal, P. A novel role of lamins from genetic disease to cancer biomarkers. Oncol. Rev. 2016, 10. [Google Scholar] [CrossRef] [PubMed]
- Belt, E.J.T.; Fijneman, R.J.A.; van den Berg, E.G.; Bril, H.; Delis-van Diemen, P.M.; Tijssen, M.; van Essen, H.F.; de Lange-de Klerk, E.S.M.; Beliën, J.A.M.; Stockmann, H.B.A.C.; et al. Loss of lamin A/C expression in stage II and III colon cancer is associated with disease recurrence. Eur. J. Cancer 2011, 47, 1837–1845. [Google Scholar] [CrossRef] [PubMed]
- Gong, G.; Chen, P.; Li, L.; Tan, H.; Zhou, J.; Zhou, Y.; Yang, X.; Wu, X. Loss of lamin A but not lamin C expression in epithelial ovarian cancer cells is associated with metastasis and poor prognosis. Pathol. Res. Pract. 2015, 211, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, A.; Hieda, M.; Yokoyama, Y.; Nishioka, Y.; Yoshidome, K.; Tsujimoto, M.; Matsuura, N. Global loss of a nuclear lamina component, lamin A/C, and LINC complex components SUN1, SUN2, and nesprin-2 in breast cancer. Cancer Med. 2015, 4, 1547–1557. [Google Scholar] [CrossRef] [PubMed]
- Saarinen, I.; Mirtti, T.; Seikkula, H.; Boström, P.J.; Taimen, P. Differential Predictive Roles of A- and B-Type Nuclear Lamins in Prostate Cancer Progression. PLoS ONE 2015, 10, e0140671. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Wu, L.; Weng, D.; Xu, D.; Geng, J.; Zhao, F. Reduced expression of lamin A/C correlates with poor histological differentiation and prognosis in primary gastric carcinoma. J. Exp. Clin. Cancer Res. 2009, 28, 8. [Google Scholar] [CrossRef] [PubMed]
- Cicchillitti, L.; Corrado, G.; Carosi, M.; Dabrowska, M. E.; Loria, R.; Falcioni, R.; Cutillo, G.; Piaggio, G.; Vizza, E. Prognostic role of NF-YA splicing isoforms and Lamin A status in low grade endometrial cancer. Oncotarget 2017, 8, 7935–7945. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Schäfer, G.; Bu, H.; Zhang, Y.; Zhang, Y.; Klocker, H. Lamin A/C protein is overexpressed in tissue-invading prostate cancer and promotes prostate cancer cell growth, migration and invasion through the PI3K/AKT/PTEN pathway. Carcinogenesis 2012, 33, 751–759. [Google Scholar] [CrossRef] [PubMed]
- Willis, N.D.; Cox, T.R.; Rahman-Casañs, S.F.; Smits, K.; Przyborski, S.A.; van den Brandt, P.; van Engeland, M.; Weijenberg, M.; Wilson, R.G.; de Bruïne, A.; et al. Lamin A/C Is a Risk Biomarker in Colorectal Cancer. PLoS ONE 2008, 3, e2988. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, R.; Cho, K.R.; Thomas, D.G.; Gossner, G.; Liu, J.R.; Giordano, T.J.; Shedden, K.A.; Misek, D.E.; Lubman, D.M. Differential Protein Mapping of Ovarian Serous Adenocarcinomas: Identification of Potential Markers for Distinct Tumor Stage. J. Proteome Res. 2009, 8, 1452–1463. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, S.H.; Mabry, M.; Jasti, R.; Shaper, J.H. Differential expression of nuclear envelope lamins A and C in human lung cancer cell lines. Cancer Res. 1991, 51, 581–586. [Google Scholar] [PubMed]
- Broers, J.L.; Raymond, Y.; Rot, M.K.; Kuijpers, H.; Wagenaar, S.S.; Ramaekers, F.C. Nuclear A-type lamins are differentially expressed in human lung cancer subtypes. Am. J. Pathol. 1993, 143, 211–220. [Google Scholar] [PubMed]
- Moss, S.F.; Krivosheyev, V.; de Souza, A.; Chin, K.; Gaetz, H.P.; Chaudhary, N.; Worman, H.J.; Holt, P.R. Decreased and aberrant nuclear lamin expression in gastrointestinal tract neoplasms. Gut 1999, 45, 723–729. [Google Scholar] [CrossRef] [PubMed]
- Machiels, B.M.; Broers, J.L.; Raymond, Y.; de Ley, L.; Kuijpers, H.J.; Caberg, N.E.; Ramaekers, F.C. Abnormal A-type lamin organization in a human lung carcinoma cell line. Eur. J. Cell Biol. 1995, 67, 328–335. [Google Scholar] [PubMed]
- Kaspi, E.; Frankel, D.; Guinde, J.; Perrin, S.; Laroumagne, S.; Robaglia-Schlupp, A.; Ostacolo, K.; Harhouri, K.; Tazi-Mezalek, R.; Micallef, J.; et al. Low lamin A expression in lung adenocarcinoma cells from pleural effusions is a pejorative factor associated with high number of metastatic sites and poor Performance status. PLoS ONE 2017, 12, e0183136. [Google Scholar] [CrossRef] [PubMed]
- Liao, G.; Wang, M.; Ou, Y.; Zhao, Y. IGF-1-induced epithelial–mesenchymal transition in MCF-7 cells is mediated by MUC1. Cell. Signal. 2014, 26, 2131–2137. [Google Scholar] [CrossRef] [PubMed]
- Nath, S.; Mukherjee, P. MUC1: A multifaceted oncoprotein with a key role in cancer progression. Trends Mol. Med. 2014, 20, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Hutchison, C.J.; Worman, H.J. A-type lamins: Guardians of the soma? Nat. Cell Biol. 2004, 6, 1062–1067. [Google Scholar] [CrossRef] [PubMed]
- Guilak, F.; Tedrow, J.R.; Burgkart, R. Viscoelastic Properties of the Cell Nucleus. Biochem. Biophys. Res. Commun. 2000, 269, 781–786. [Google Scholar] [CrossRef] [PubMed]
- Davidson, P.M.; Sliz, J.; Isermann, P.; Denais, C.; Lammerding, J. Design of a microfluidic device to quantify dynamic intra-nuclear deformation during cell migration through confining environments. Integr. Biol. 2015, 7, 1534–1546. [Google Scholar] [CrossRef] [PubMed]
- Yabuki, M.; Miyake, T.; Doi, Y.; Fujiwara, T.; Hamazaki, K.; Yoshioka, T.; Horton, A.A.; Utsumi, K. Role of nuclear lamins in nuclear segmentation of human neutrophils. Physiol. Chem. Phys. Med. NMR 1999, 31, 77–84. [Google Scholar] [PubMed]
- Pajerowski, J.D.; Dahl, K.N.; Zhong, F.L.; Sammak, P.J.; Discher, D.E. Physical plasticity of the nucleus in stem cell differentiation. Proc. Natl. Acad. Sci. USA 2007, 104, 15619–15624. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.-W.; Chen, Y.; Kriegstein, A.R.; Vallee, R.B. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J. Cell Biol. 2005, 170, 935–945. [Google Scholar] [CrossRef] [PubMed]
- Denais, C.; Lammerding, J. Nuclear Mechanics in Cancer. In Cancer Biology and the Nuclear Envelope; Schirmer, E.C., de las Heras, J.I., Eds.; Springer: New York, NY, USA, 2014; Volume 773, pp. 435–470. ISBN 9781489980311. [Google Scholar]
- Lu, Q.-Y.; Yang, Y.; Jin, Y.S.; Zhang, Z.-F.; Heber, D.; Li, F.P.; Dubinett, S.M.; Sondej, M.A.; Loo, J.A.; Rao, J.Y. Effects of green tea extract on lung cancer A549 cells: Proteomic identification of proteins associated with cell migration. Proteomics 2009, 9, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; Kim, V.N. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Hutvagner, G. A Cellular Function for the RNA-Interference Enzyme Dicer in the Maturation of the let-7 Small Temporal RNA. Science 2001, 293, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Krek, A.; Grün, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; da Piedade, I.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Place, R.F.; Li, L.-C.; Pookot, D.; Noonan, E.J.; Dahiya, R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl. Acad. Sci. USA 2008, 105, 1608–1613. [Google Scholar] [CrossRef] [PubMed]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—MicroRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.-K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Investig. New Drugs 2017, 35, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Slaby, O.; Laga, R.; Sedlacek, O. Therapeutic targeting of non-coding RNAs in cancer. Biochem. J. 2017, 474, 4219–4251. [Google Scholar] [CrossRef] [PubMed]
- Wilk, G.; Braun, R. Integrative analysis reveals disrupted pathways regulated by microRNAs in cancer. Nucleic Acids Res. 2018, 46, 1089–1101. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Wen, S.; Zhang, Y.; Shi, X.; Zhu, Y.; Xu, Y.; Lv, H.; Wang, G. Identification of dysregulated long non-coding RNAs/microRNAs/mRNAs in TNM I stage lung adenocarcinoma. Oncotarget 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.-L.; Hung, J.-Y.; Lee, Y.-L.; Chen, F.-W.; Chang, K.-F.; Chang, W.-A.; Tsai, Y.-M.; Chong, I.-W.; Kuo, P.-L. Identification of novel gene expression signature in lung adenocarcinoma by using next-generation sequencing data and bioinformatics analysis. Oncotarget 2017, 8, 104831. [Google Scholar] [CrossRef] [PubMed]
- Uddin, A.; Chakraborty, S. Role of miRNAs in lung cancer. J. Cell. Physiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, S.; Okamoto, A.; Nikaido, T.; Saito, M.; Takao, M.; Yanaihara, N.; Takakura, S.; Ochiai, K.; Tanaka, T. Mesenchymal to epithelial transition in the human ovarian surface epithelium focusing on inclusion cysts. Oncol. Rep. 2009, 21, 1209–1214. [Google Scholar] [CrossRef] [PubMed]
- Zhai, X.; Zhu, H.; Wang, W.; Zhang, S.; Zhang, Y.; Mao, G. Abnormal expression of EMT-related proteins, S100A4, vimentin and E-cadherin, is correlated with clinicopathological features and prognosis in HCC. Med. Oncol. 2014, 31, 970. [Google Scholar] [CrossRef] [PubMed]
- Gout, S.; Brambilla, E.; Boudria, A.; Drissi, R.; Lantuejoul, S.; Gazzeri, S.; Eymin, B. Abnormal Expression of the Pre-mRNA Splicing Regulators SRSF1, SRSF2, SRPK1 and SRPK2 in Non Small Cell Lung Carcinoma. PLoS ONE 2012, 7, e46539. [Google Scholar] [CrossRef] [PubMed]
- Mongroo, P.S.; Rustgi, A.K. The role of the miR-200 family in epithelial-mesenchymal transition. Cancer Biol. Ther. 2010, 10, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Lang, Y.; Xu, S.; Ma, J.; Wu, J.; Jin, S.; Cao, S.; Yu, Y. MicroRNA-429 induces tumorigenesis of human non-small cell lung cancer cells and targets multiple tumor suppressor genes. Biochem. Biophys. Res. Commun. 2014, 450, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chen, K.; Zhou, Y.; Hu, Z.; Chen, S.; Huang, Y. Application of serum microRNA-9-5p, 21-5p, and 223-3p combined with tumor markers in the diagnosis of non-small-cell lung cancer in Yunnan in southwestern China. OncoTargets Ther. 2018, 11, 587–597. [Google Scholar] [CrossRef] [PubMed]
- Nadal, E.; Truini, A.; Nakata, A.; Lin, J.; Reddy, R.M.; Chang, A.C.; Ramnath, N.; Gotoh, N.; Beer, D.G.; Chen, G. A Novel Serum 4-microRNA Signature for Lung Cancer Detection. Sci. Rep. 2015, 5, 12464. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; He, J.; Chen, D.; Zhang, B.; Xu, L.; Ma, H.; Liu, X.; Zhang, Y.; Le, H. Expression of miR-29c, miR-93, and miR-429 as Potential Biomarkers for Detection of Early Stage Non-Small Lung Cancer. PLoS ONE 2014, 9, e87780. [Google Scholar] [CrossRef] [PubMed]
- Lujambio, A.; Calin, G.A.; Villanueva, A.; Ropero, S.; Sanchez-Cespedes, M.; Blanco, D.; Montuenga, L.M.; Rossi, S.; Nicoloso, M.S.; Faller, W.J.; et al. A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl. Acad. Sci. USA 2008, 105, 13556–13561. [Google Scholar] [CrossRef] [PubMed]
- Senyuk, V.; Zhang, Y.; Liu, Y.; Ming, M.; Premanand, K.; Zhou, L.; Chen, P.; Chen, J.; Rowley, J.D.; Nucifora, G.; et al. Critical role of miR-9 in myelopoiesis and EVI1-induced leukemogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 5594–5599. [Google Scholar] [CrossRef] [PubMed]
- Sromek, M.; Glogowski, M.; Chechlinska, M.; Kulinczak, M.; Szafron, L.; Zakrzewska, K.; Owczarek, J.; Wisniewski, P.; Wlodarczyk, R.; Talarek, L.; et al. Changes in plasma miR-9, miR-16, miR-205 and miR-486 levels after non-small cell lung cancer resection. Cell. Oncol. 2017, 40, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Volinia, S.; Calin, G.A.; Liu, C.-G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [PubMed]
- Crawford, M.; Batte, K.; Yu, L.; Wu, X.; Nuovo, G.J.; Marsh, C.B.; Otterson, G.A.; Nana-Sinkam, S.P. MicroRNA 133B targets pro-survival molecules MCL-1 and BCL2L2 in lung cancer. Biochem. Biophys. Res. Commun. 2009, 388, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Võsa, U.; Vooder, T.; Kolde, R.; Fischer, K.; Välk, K.; Tõnisson, N.; Roosipuu, R.; Vilo, J.; Metspalu, A.; Annilo, T. Identification of miR-374a as a prognostic marker for survival in patients with early-stage nonsmall cell lung cancer. Genes Chromosom. Cancer 2011, 50, 812–822. [Google Scholar] [CrossRef] [PubMed]
- Heller, G.; Weinzierl, M.; Noll, C.; Babinsky, V.; Ziegler, B.; Altenberger, C.; Minichsdorfer, C.; Lang, G.; Dome, B.; End-Pfutzenreuter, A.; et al. Genome-Wide miRNA Expression Profiling Identifies miR-9-3 and miR-193a as Targets for DNA Methylation in Non-Small Cell Lung Cancers. Clin. Cancer Res. 2012, 18, 1619–1629. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Chen, X.-F.; Shu, Y.-Q. Prediction of non-small cell lung cancer metastasis-associated microRNAs using bioinformatics. Am. J. Cancer Res. 2015, 5, 32–51. [Google Scholar] [PubMed]
- Kang, H.-W.; Crawford, M.; Fabbri, M.; Nuovo, G.; Garofalo, M.; Nana-Sinkam, S.P.; Friedman, A. A Mathematical Model for MicroRNA in Lung Cancer. PLoS ONE 2013, 8, e53663. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Ai, J.; Long, H.; Liu, W.; Wang, X.; Zuo, Y.; Li, Y.; Wu, Q.; Deng, Y. Integrative microRNA and gene profiling data analysis reveals novel biomarkers and mechanisms for lung cancer. Oncotarget 2016, 7, 8441. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhu, L.; Ma, Z.; Sun, G.; Luo, X.; Li, M.; Zhai, S.; Li, P.; Wang, X. Oncogenic miR-9 is a target of erlotinib in NSCLCs. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Liu, X.; Han, L.; Shen, H.; Liu, L.; Shu, Y. Up-regulation of miR-9 expression as a poor prognostic biomarker in patients with non-small cell lung cancer. Clin. Transl. Oncol. 2014, 16, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wu, Q.; Zhang, Y.; Zhang, H.-N.; Wang, Y.-B.; Wang, W. TGF-β1-induced epithelial–mesenchymal transition in lung cancer cells involves upregulation of miR-9 and downregulation of its target, E-cadherin. Cell. Mol. Biol. Lett. 2017, 22, 22. [Google Scholar] [CrossRef] [PubMed]
- Polley, E.; Kunkel, M.; Evans, D.; Silvers, T.; Delosh, R.; Laudeman, J.; Ogle, C.; Reinhart, R.; Selby, M.; Connelly, J.; et al. Small Cell Lung Cancer Screen of Oncology Drugs, Investigational Agents, and Gene and microRNA Expression. J. Natl. Cancer Inst. 2016, 108, djw122. [Google Scholar] [CrossRef] [PubMed]
- Muraoka, T.; Soh, J.; Toyooka, S.; Maki, Y.; Shien, K.; Furukawa, M.; Ueno, T.; Tanaka, N.; Yamamoto, H.; Asano, H.; et al. Impact of aberrant methylation of microRNA-9 family members on non-small cell lung cancers. Mol. Clin. Oncol. 2013, 1, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, B.; Han, L.; Li, X.; Tao, H.; Zhang, S.; Hu, Y. Demethylation of miR-9-3 and miR-193a Genes Suppresses Proliferation and Promotes Apoptosis in Non-Small Cell Lung Cancer Cell Lines. Cell. Physiol. Biochem. 2013, 32, 1707–1719. [Google Scholar] [CrossRef] [PubMed]
- Gwak, J.M.; Kim, H.J.; Kim, E.J.; Chung, Y.R.; Yun, S.; Seo, A.N.; Lee, H.J.; Park, S.Y. MicroRNA-9 is associated with epithelial-mesenchymal transition, breast cancer stem cell phenotype, and tumor progression in breast cancer. Breast Cancer Res. Treat. 2014, 147, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Young, J.; Prabhala, H.; Pan, E.; Mestdagh, P.; Muth, D.; Teruya-Feldstein, J.; Reinhardt, F.; Onder, T.T.; Valastyan, S.; et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 2010. [Google Scholar] [CrossRef] [PubMed]
- Seashols-Williams, S.J.; Budd, W.; Clark, G.C.; Wu, Q.; Daniel, R.; Dragoescu, E.; Zehner, Z.E. miR-9 Acts as an OncomiR in Prostate Cancer through Multiple Pathways That Drive Tumour Progression and Metastasis. PLoS ONE 2016, 11, e0159601. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Li, J.; Zhu, Y.; Dai, Y.; Zeng, T.; Liu, L.; Li, J.; Wang, H.; Qin, Y.; Zeng, M.; et al. MicroRNA-9 promotes tumor metastasis via repressing E-cadherin in esophageal squamous cell carcinoma. Oncotarget 2014, 5, 11669. [Google Scholar] [CrossRef] [PubMed]
- Yanaihara, N.; Noguchi, Y.; Saito, M.; Takenaka, M.; Takakura, S.; Yamada, K.; Okamoto, A. MicroRNA Gene Expression Signature Driven by miR-9 Overexpression in Ovarian Clear Cell Carcinoma. PLoS ONE 2016, 11, e0162584. [Google Scholar] [CrossRef] [PubMed]
- Ding, G.; Peng, Z.; Shang, J.; Kang, Y.; Ning, H.; Mao, C. LincRNA-p21 inhibits invasion and metastasis of hepatocellular carcinoma through miR-9/E-cadherin cascade signaling pathway molecular mechanism. OncoTargets Ther. 2017, 10, 3241–3247. [Google Scholar] [CrossRef] [PubMed]
- Yap, A.S. The morphogenetic role of cadherin cell adhesion molecules in human cancer: A thematic review. Cancer Investig. 1998, 16, 252–261. [Google Scholar] [CrossRef]
- Cowin, P.; Rowlands, T.M.; Hatsell, S.J. Cadherins and catenins in breast cancer. Curr. Opin. Cell Biol. 2005, 17, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Junghans, D.; Haas, I.G.; Kemler, R. Mammalian cadherins and protocadherins: About cell death, synapses and processing. Curr. Opin. Cell Biol. 2005, 17, 446–452. [Google Scholar] [CrossRef] [PubMed]
- Frixen, U.H.; Behrens, J.; Sachs, M.; Eberle, G.; Voss, B.; Warda, A.; Löchner, D.; Birchmeier, W. E-cadherin-mediated cell-cell adhesion prevents invasiveness of human carcinoma cells. J. Cell Biol. 1991, 113, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Vleminckx, K.; Vakaet, L.; Mareel, M.; Fiers, W.; van Roy, F. Genetic manipulation of E-cadherin expression by epithelial tumor cells reveals an invasion suppressor role. Cell 1991, 66, 107–119. [Google Scholar] [CrossRef]
- Perl, A.-K.; Wilgenbus, P.; Dahl, U.; Semb, H.; Christofori, G. A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature 1998, 392, 190–193. [Google Scholar] [CrossRef] [PubMed]
- Derksen, P.W.B.; Liu, X.; Saridin, F.; van der Gulden, H.; Zevenhoven, J.; Evers, B.; van Beijnum, J.R.; Griffioen, A.W.; Vink, J.; Krimpenfort, P.; et al. Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell 2006, 10, 437–449. [Google Scholar] [CrossRef] [PubMed]
- Onder, T.T.; Gupta, P.B.; Mani, S.A.; Yang, J.; Lander, E.S.; Weinberg, R.A. Loss of E-Cadherin Promotes Metastasis via Multiple Downstream Transcriptional Pathways. Cancer Res. 2008, 68, 3645–3654. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Miao, Y.; Zhang, X.; Liu, D.; Jiang, G.; Lin, X.; Han, Q.; Luan, L.; Xu, Z.; Wang, E. Btbd7 contributes to reduced E-cadherin expression and predicts poor prognosis in non-small cell lung cancer. BMC Cancer 2014, 14, 704. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Ma, W.; Li, Y.; Jiang, Y.; Ma, G.; Zhang, X.; Meng, L.; Du, J. Prognostic value of Twist, Snail and E-cadherin expression in pathological N0 non-small-cell lung cancer: A retrospective cohort study. Eur. J. Cardio-Thorac. Surg. 2018. [Google Scholar] [CrossRef] [PubMed]
- Sinkevicius, K.W.; Bellaria, K.J.; Barrios, J.; Pessina, P.; Gupta, M.; Brainson, C.F.; Bronson, R.T.; Kim, C.F. E-cadherin Loss Accelerates Tumor Progression and Metastasis in a Mouse Model of Lung Adenocarcinoma. Am. J. Respir. Cell Mol. Biol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Leucci, E.; Patella, F.; Waage, J.; Holmstrøm, K.; Lindow, M.; Porse, B.; Kauppinen, S.; Lund, A.H. microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci. Rep. 2013, 3, 2535. [Google Scholar] [CrossRef] [PubMed]
- Gutschner, T.; Hammerle, M.; Eissmann, M.; Hsu, J.; Kim, Y.; Hung, G.; Revenko, A.; Arun, G.; Stentrup, M.; Gross, M.; et al. The Noncoding RNA MALAT1 Is a Critical Regulator of the Metastasis Phenotype of Lung Cancer Cells. Cancer Res. 2013, 73, 1180–1189. [Google Scholar] [CrossRef] [PubMed]
- Gutschner, T.; Hämmerle, M.; Diederichs, S. MALAT1—A paradigm for long noncoding RNA function in cancer. J. Mol. Med. 2013, 91, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, L.H.; Spieker, T.; Koschmieder, S.; Humberg, J.; Jungen, D.; Bulk, E.; Hascher, A.; Wittmer, D.; Marra, A.; Hillejan, L.; et al. The Long Noncoding MALAT-1 RNA Indicates a Poor Prognosis in Non-small Cell Lung Cancer and Induces Migration and Tumor Growth. J. Thorac. Oncol. 2011, 6, 1984–1992. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Chen, L.; Wang, Y.; Jiang, X.; Xia, H.; Zhuang, Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J. Neuro-Oncol. 2015, 121, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Weber, D.; Johnen, G.; Casjens, S.; Bryk, O.; Pesch, B.; Jöckel, K.-H.; Kollmeier, J.; Brüning, T. Evaluation of long noncoding RNA MALAT1 as a candidate blood-based biomarker for the diagnosis of non-small cell lung cancer. BMC Res. Notes 2013, 6, 518. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.-J.; Coffinier, C.; Choe, Y.; Beigneux, A.P.; Davies, B.S.J.; Yang, S.H.; Barnes, R.H.; Hong, J.; Sun, T.; Pleasure, S.J.; et al. Regulation of prelamin A but not lamin C by miR-9, a brain-specific microRNA. Proc. Natl. Acad. Sci. USA 2012, 109, E423–E431. [Google Scholar] [CrossRef] [PubMed]
- Nissan, X.; Blondel, S.; Navarro, C.; Maury, Y.; Denis, C.; Girard, M.; Martinat, C.; De Sandre-Giovannoli, A.; Levy, N.; Peschanski, M. Unique Preservation of Neural Cells in Hutchinson-Gilford Progeria Syndrome Is Due to the Expression of the Neural-Specific miR-9 MicroRNA. Cell Rep. 2012, 2, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The Nuclear-Retained Noncoding RNA MALAT1 Regulates Alternative Splicing by Modulating SR Splicing Factor Phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef] [PubMed]
- Scaffidi, P. Lamin A-Dependent Nuclear Defects in Human Aging. Science 2006, 312, 1059–1063. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Roychowdhury-Saha, M.; Black, C.; Watt, A.T.; Marcusson, E.G.; Freier, S.M.; Edgington, T.S. Control of RNA processing by a large non-coding RNA over-expressed in carcinomas. FEBS Lett. 2011, 585, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Krainer, A.R. Emerging Functions of SRSF1, Splicing Factor and Oncoprotein, in RNA Metabolism and Cancer. Mol. Cancer Res. 2014, 12, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Eliav, M.; Golan-Gerstl, R.; Siegfried, Z.; Andersen, C.L.; Thorsen, K.; Ørntoft, T.F.; Mu, D.; Karni, R. The splicing factor SRSF6 is amplified and is an oncoprotein in lung and colon cancers: SRSF6 is amplified and is an oncoprotein in lung and colon cancers. J. Pathol. 2013, 229, 630–639. [Google Scholar] [CrossRef] [PubMed]
- Fischer, K.R.; Durrans, A.; Lee, S.; Sheng, J.; Li, F.; Wong, S.T.C.; Choi, H.; El Rayes, T.; Ryu, S.; Troeger, J.; et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 2015, 527, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Carstens, J.L.; Kim, J.; Scheible, M.; Kaye, J.; Sugimoto, H.; Wu, C.-C.; LeBleu, V.S.; Kalluri, R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 2015, 527, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, G.; Zhang, H.; Zhang, F.; Zhou, B.; Ning, F.; Wang, H.-S.; Cai, S.-H.; Du, J. Acquisition of epithelial–mesenchymal transition phenotype and cancer stem cell-like properties in cisplatin-resistant lung cancer cells through AKT/β-catenin/Snail signaling pathway. Eur. J. Pharmacol. 2014, 723, 156–166. [Google Scholar] [CrossRef] [PubMed]
| Cancer/Tumor Type | A-Type Lamins Expression | Prognostic Value | References |
|---|---|---|---|
| Gastric carcinoma | Decrease | Decreased overall survival | Wu et al., J. Exp. Clin. Cancer Res. 2009 |
| Cytoplasmic localization | / | Moss et al., Gut 1999 | |
| Breast carcinoma | Decrease | Decreased overall survival | Capo-Chichi et al., Chin. J. Cancer 2011 |
| Ovarian carcinoma | Increase | Higher stage tumours | Wang et al., J. Proteome Res. 2009 |
| Isolated decrease of lamin A | Decreased overall survival Increased number of metastatic sites | Gong et al., Pathol. Res. Pract. 2015 | |
| Endometrial carcinoma | Isolated decrease of lamin A | Tumor agressiveness | Cicchillitti et al., Oncotarget 2017 |
| Prostate adenocarcinoma | Increase | / | Kong et al., Carcinogenesis 2012 |
| Decrease | Increased risk for lymph node metastasis | Saarinen et al., PLoS ONE 2015 | |
| Colon carcinoma | Increase | Decreased overall survival | Willis et al., PLoS ONE 2008 |
| Decrease | Increase of disease recurrence | Belt et al., EJC 2011 | |
| Small Cell Lung carcinoma | Decrease | / | Broers et al., Adv. Exp. Med. Biol. 2014 Broers et al., Am. J. Pathol. 1993 Kaufmann et al., Cancer Res. 1991 |
| Lung adenocarcinoma | Cytoplasmic localization | / | Broers et al., Adv. Exp. Med. Biol. 2014 |
| Isolated decrease of lamin A | Increased number of metastatic sites Poor Performans status | Kaspi et al., PLoS ONE 2017 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guinde, J.; Frankel, D.; Perrin, S.; Delecourt, V.; Lévy, N.; Barlesi, F.; Astoul, P.; Roll, P.; Kaspi, E. Lamins in Lung Cancer: Biomarkers and Key Factors for Disease Progression through miR-9 Regulation? Cells 2018, 7, 78. https://doi.org/10.3390/cells7070078
Guinde J, Frankel D, Perrin S, Delecourt V, Lévy N, Barlesi F, Astoul P, Roll P, Kaspi E. Lamins in Lung Cancer: Biomarkers and Key Factors for Disease Progression through miR-9 Regulation? Cells. 2018; 7(7):78. https://doi.org/10.3390/cells7070078
Chicago/Turabian StyleGuinde, Julien, Diane Frankel, Sophie Perrin, Valérie Delecourt, Nicolas Lévy, Fabrice Barlesi, Philippe Astoul, Patrice Roll, and Elise Kaspi. 2018. "Lamins in Lung Cancer: Biomarkers and Key Factors for Disease Progression through miR-9 Regulation?" Cells 7, no. 7: 78. https://doi.org/10.3390/cells7070078
APA StyleGuinde, J., Frankel, D., Perrin, S., Delecourt, V., Lévy, N., Barlesi, F., Astoul, P., Roll, P., & Kaspi, E. (2018). Lamins in Lung Cancer: Biomarkers and Key Factors for Disease Progression through miR-9 Regulation? Cells, 7(7), 78. https://doi.org/10.3390/cells7070078





